Abstract
Kidney fibrosis is a common process that leads to the progression of kidney diseases. We used an integrated computational/experimental systems biology approach to identify upstream protein kinases that regulate gene expression changes in kidneys of HIV-1 transgenic mice (Tg26), which have both tubulo-interstitial fibrosis and glomerulosclerosis. We identified the homeo-domain interacting protein kinase 2 (HIPK2) as a key regulator of kidney fibrosis. HIPK2 was upregulated in kidneys of Tg26 and patients with various kidney diseases. HIV infection increased the protein level of HIPK2 by promoting oxidative stress, which inhibited SIAH1-mediated proteasomal degradation of HIPK2. HIPK2 induced apoptosis and expression of epithelial-mesenchymal trans-differentiation markers in kidney epithelial cells by activating p53, TGF-β/Smad3, and Wnt/Notch pathways. Knockout of HIPK2 improved renal function and attenuated proteinuria and kidney fibrosis in Tg26 as well as in other animal models of kidney fibrosis. We conclude that HIPK2 is a potential target for anti-fibrosis therapy.
Keywords: HIPK2, tubular epithelial cells, HIV, fibrosis, systems biology
Introduction
Kidney fibrosis is a final common pathogenic process for many forms of chronic renal diseases including HIV-associated nephropathy (HIVAN), which is a leading cause of renal disease among HIV-infected African-Americans1. Collapsing focal segmental glomerulosclerosis (FSGS), tubulointerstitial inflammation and fibrosis, tubular dilatation with microcyst formation, and renal tubular epithelial cell (RTEC) apoptosis are key pathologic features of HIVAN2. HIV directly infects RTEC, but the cellular response to the infection remains poorly defined3. A HIV transgenic mouse model (Tg26), which recapitulates pathologic changes observed in human disease, have been used to study HIVAN pathogenesis4,5. While prior studies have identified individual protein regulators, protein-protein interactions, and cell signaling pathways altered in HIVAN, systematic evaluation of these changes has not been attempted.
Transcriptomic measurements are reflective of quantitative changes in mRNA levels but do not provide direct understanding of upstream regulatory mechanisms that are responsible for the observed changes in gene expression. Identifying potential regulatory mechanisms responsible for changes in gene expression under disease conditions could help to reveal disease mechanisms and enable discovery of drug targets. Here, we present an integrative experimental and computational approach for identifying and ranking protein kinases that are likely responsible for observed changes in mRNA expression and transcription factor (TF) activation. We computationally predicted and then experimentally validated that HIPK2 is critically involved in kidney fibrosis.
Results
Identification of HIPK2 as a potential upstream kinase mediating the pathogenesis of HIVAN
We confirmed the renal histopathology (Supplementary Table a) and determined differentially expressed genes in the kidney cortex of Tg26 compared to matched wild type (WT) littermates: 434 genes were significantly up-regulated and 72 genes were down-regulated (T-test with FDR p<0.01). Upstream TFs likely to be responsible for the observed changes in gene expression were computationally deduced by two methods. First, TRANSFAC matrices6 were used to scan the promoter region of genes to identify TFs with over-represented binding sites among the 434 up-regulated genes. Nine TFs were identified (Supplementary Excel file). We identified only 1 TF when the same approach was applied to the 72 down-regulated genes. Thus, subsequent analyses utilized only up-regulated genes. We used a second computational approach (ChIP Enrichment Analysis, ChEA7) to identify upstream TFs and found 62 TFs (Supplementary Excel file). We experimentally profiled protein/DNA interactions that were differentially regulated between Tg26 and WT kidneys using a protein/DNA binding array (Fig. 1 and Supplementary Fig. 1a). Twelve up-regulated and 14 down-regulated binding interactions for 35 TFs were identified (Supplementary Table b and Supplementary Excel file). Some consensus binding motifs on the protein/DNA array were linked to several TFs. The activity of selected TFs was confirmed by electrophoretic mobility shift assay (Supplementary Fig. 1b). We generated a selectively combined TF list by including the top five TFs from the TRANSFAC analysis, the top five TFs from ChEA, as well as TFs that were predicted by either TRANSFAC or ChEA and also identified by the protein/DNA interaction array (Fig. 1 and Supplementary Excel file). These 18 TFs are potentially responsible for the pattern of gene expression in the Tg26 kidney.
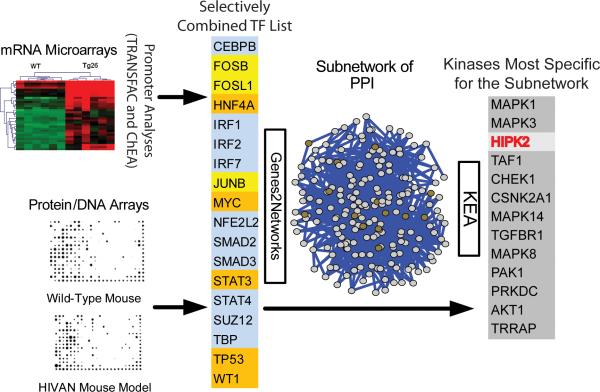
Figure 1. Identification of key signaling pathways activated in HIVAN.
Two methods of computational promoter analysis (TRANSFAC and ChEA) and protein/DNA interaction array were used to identify eighteen transcription factors (TFs) differentially activated in kidneys of Tg26 mice. Top 5 TFs that are highly ranked in either of the promoter analyses are in blue (n=10); TFs that are in common between TRANSFAC and the protein/DNA interaction array are in yellow (n=3); TFs that are in common between ChEA and the protein/DNA interaction array are in orange (n=5). We generated a subnetwork of proteins that interact with this selectively combined list of 18 TFs using Genes2Networks. Proteins within the subnetwork (n=205) were used as inputs for Kinase Enrichment Analysis (KEA) to identify protein kinases that are likely responsible for the phosphorylation of proteins within the subnetwork. HIPK2 is the third most highly ranked kinase (red).
We linked these TFs to upstream regulatory mechanisms by constructing a protein-protein interaction subnetwork based on known interactions (Genes2Networks8). This subnetwork has 205 nodes and is enriched in co-regulators and other TFs that physically interact with the 18 TFs (Fig 1 and Supplementary Excel file). We further linked protein kinases that are likely to phosphorylate proteins within the subnetwork by Kinase Enrichment Analysis (KEA)9. Consistent with our previous studies10, MAPK3 and MAPK1 (the canonical MAP-kinases ERK1 and ERK2) are highly ranked, whereas the less known kinase HIPK2 is third on the list (Fig 1). HIPK2 has 14 substrates within the subnetwork, which is almost half of all the known substrates for HIPK2 (n=34, p-value of 2.2E-09. Supplementary Excel file).
HIPK2 is a conserved serine/threonine nuclear kinase that regulates gene expression by phosphorylating TFs and accessory components of the transcriptional machinery11. It is activated in response to morphogenic signals that regulate cell differentiation and apoptosis in neurons12. In response to DNA-damaging agents, HIPK2 promotes apoptosis by phosphorylating and activating p5313. HIPK2 also mediates the activation of Wnt14, Notch15, and TGFβ-induced signaling16,17. Since p53 and TGFβ are known mediators of apoptosis for RTEC18-20 and TGFβ, Notch and Wnt/β-catenin pathways promote renal fibrosis21-24 we hypothesized that HIPK2, a previously unrecognized kinase in the context of kidney disease, contributes to kidney epithelial cell injury and renal fibrosis in HIVAN.
HIPK2 expression is increased in kidneys of Tg26 mice and in HIV-infected kidney cells
To test our hypothesis, we examined the expression of HIPK2 in Tg26 kidneys. Immunostaining of HIPK2 revealed that it is increased mostly in the tubular compartment and only mildly in glomeruli of Tg26 (Fig. 2a). This was confirmed by Western blot analysis of protein lysates from the renal cortex, which contains mostly tubules, and from isolated glomeruli (Fig. 2b). The expression of HIPK2 mRNA in the renal cortex and in isolated glomeruli was not statistically different between Tg26 and WT (Fig. 2c), suggesting that HIPK2 expression was regulated at the post-transcriptional level. A corresponding increase in HIPK2 kinase activity was observed in Tg26 kidneys (Fig. 2d). Expression of HIPK1 and HIPK3 was not different between Tg26 and WT kidneys (Data not shown).
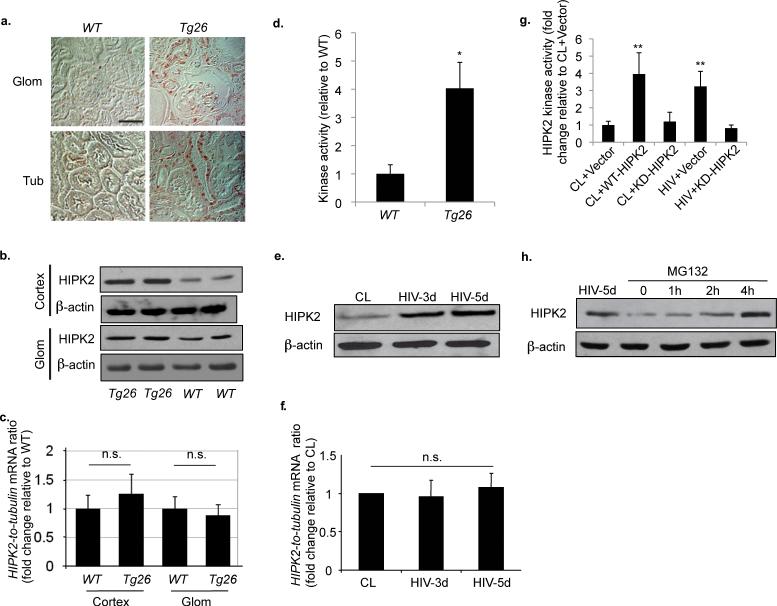
Figure 2. HIV induces HIPK2 expression in kidney cells.
(a) Immunostaining of HIPK2 in kidneys from Tg26 mice and wild type (WT) littermates. n=5 for WT and Tg26. (b) Western blots of HIPK2 and β-actin in protein lysates from the kidney cortex (Cortex) and isolated glomeruli (Glom) of two Tg26 and two WT animals. (c) HIPK2 expression in Cortex and Glom samples from Tg26 and WT as analyzed by real-time PCR, n=5. (d) HIPK2 kinase activity in kidneys of Tg26 and WT mice, n=5, *p<0.001. Western blots (e) and real-time PCR (f) for HIPK2 in human renal tubular epithelial cells (hRTEC) infected with a control virus (CL) or a pseudotyped HIV-1 virus (HIV) for 3 and 5 days (HIV-3d and HIV-5d), n=4. (g) HIPK2 kinase activity in hRTEC infected with either CL or HIV and transfected with an empty expression vector (vector), a wild-type HIPK2 vector (WT-HIPK2) or a kinase dead HIPK2 vector (KD-HIPK2). n=4, p**<0.01 compared to CL+Vector. (h) Western of HIPK2 for hRTEC infected with HIV for 5 days (HIV-5d) or treated with MG132 for 0, 1, 2, and 4 hours. Representative blots of three independent experiments are shown for all western blots. Error bars represent s.d. Scale bar, 50μm. n.s. = not significant.
To examine whether HIV infection of kidney cells was responsible for the induction of HIPK2 activity, we infected primary human renal tubular epithelial cells (hRTEC) with HIV. HIPK2 protein expression was increased, but mRNA level was unchanged, in HIV-infected cells (Fig. 2e,f). HIPK2 kinase activity was increased by HIV infection and suppressed by a kinase-dead mutant of HIPK2 (KD-HIPK2) (Fig. 2g). To examine the post-translational regulation of HIPK2 by proteasomal degradation, we treated cells with MG132, which is a specific 26S proteosome inhibitor, and found that HIPK2 was significantly increased (Fig. 2h), which suggests that HIV infection increased HIPK2 by reducing HIPK2 proteasomal degradation.
SIAH1 is an upstream regulator of HIPK2
Next, we determined how HIV up-regulates HIPK2 protein levels. DNA damage enhances HIPK2 protein level by inhibiting the ubiquitin ligase seven in absentia homologue-1 ( SIAH1), which facilitates HIPK2 polyubiquitination and proteasomal degradation25. HIV infection promotes oxidative stress26 and DNA damage27 in kidney cells. Since SIAH1 expression was reduced in our gene expression microarray data from the Tg26 renal cortex, we sought to examine whether HIV-induced up-regulation of HIPK2 was due to a reduction in SIAH1.
We first confirmed that SIAH1 expression was diminished in the Tg26 renal cortex by real-time PCR (Fig. 3a) and western blot (Fig. 3b). An inverse relationship between SIAH1 and HIPK2 expression was observed by western blot (Fig. 3b) and by immunostaining in adjacent kidney sections (Fig. 3c). The weak background staining in Fig. 3c is likely due to short development of the stain. Next, we determined whether HIV infection reduces SIAH1. We found that infection of hRTEC by HIV suppressed SIAH1 protein and mRNA expression (Fig. 3d,e). To confirm the role of SIAH1 in the regulation of HIPK2, we either over-expressed or silenced SIAH1 in hRTEC by transfecting a SIAH1 expression construct or a specific siRNA, respectively. Transfection efficiency is shown in Supplementary Fig 1c-e. HIPK2 was increased by SIAH1 knock-down and reduced by SIAH1 over-expression (Fig. 3f), indicating that SIAH1 acts upstream of HIPK2. To determine the mechanisms by which HIV infection suppressed SIAH1 we treated hRTEC with H2O2 and adriamycin and found that both treatments reduced SIAH1 and increased HIPK2 (Fig. 3g,h). Pretreatment with an oxidant scavenger, N-acetylcysteine, abrogated HIV-induced HIPK2 up-regulation and restored SIAH1 in hRTEC (Fig. 3i). Treatment with either H2O2 or adriamycin did not affect the mRNA level of HIPK2 in hRTEC (Data not shown).
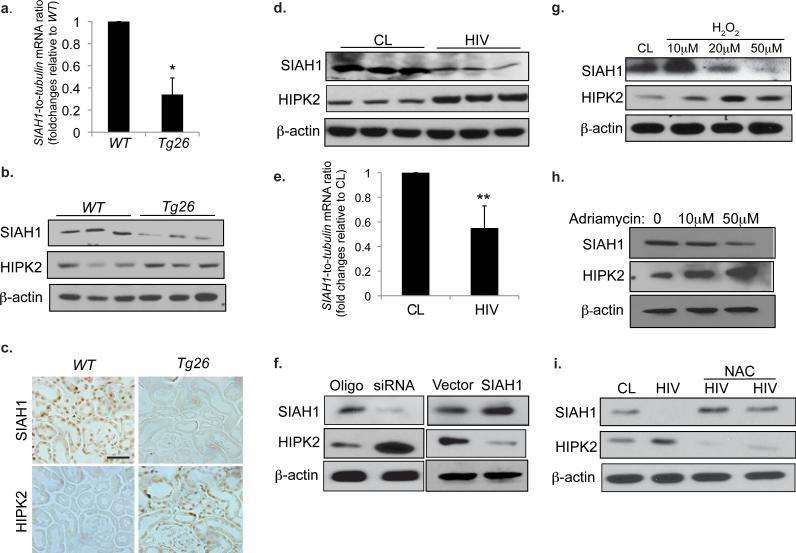
Figure 3. SIAH1 is an upstream regulator of HIPK2 expression.
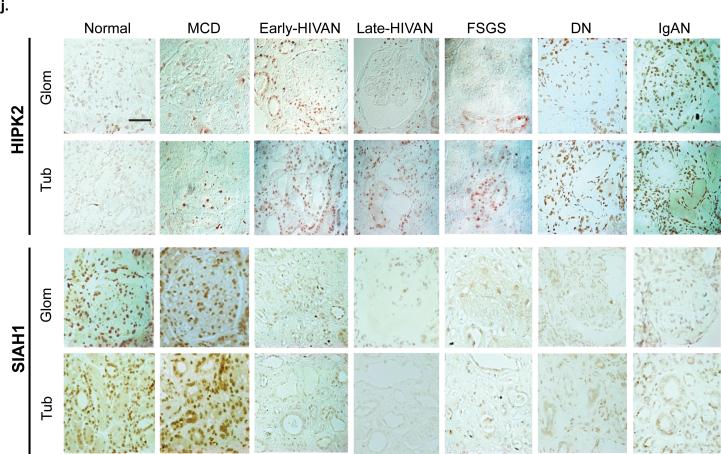
(a) Expression of SIAH1 in the renal cortex of WT and Tg26 mice as determined by real-time PCR, n=5. (b) Western blots of SIAH1, HIPK2 and β-actin in the kidney cortex of WT and Tg26 mice, n=3. (c) Immunostaining of SIAH1 and HIPK2 in adjacent kidney sections of Tg26 and WT mice. (d) Western blots of SIAH1, HIPK2, and β-actin in human renal tubular epithelial cells (hRTECs) infected with a control virus (CL) or a pseudotyped HIV-1 virus (HIV). (e) Expression of SIAH1 in hRTEC infected with CL or HIV as quantified by real-time PCR, n=4 (f) Western blots of SIAH-1, HIPK2, and β-actin in hRTEC transfected with a specific siRNA to knockdown SIAH1 (siRNA), a negative control siRNA oligo (Oligo), an empty expression vector (Vector), or a SIAH1 expression vector (SIAH1). Western blots of hRTECs cultured in the presence of H2O2 (0, 10, 20, 50 μM) for 24 hr (g), treated with adriamycin (0, 10, and 50μM) for 24 hr (h), or infected with CL or HIV and then cultured in the presence and absence of N-acetylcystein (NAC; 10mM) for 3 days (i). (j) Immunostaining of HIPK2 and SIAH1 in patients with minimal change disease (MCD), HIVAN (early and late stages), idiopathic focal segmental glomerulosclerosis (FSGS), diabetic nephropathy (DN), IgA nephropathy (IgAN), and normal nephrectomy samples (Normal). Representative fields of glomeruli (Glom) and tubules (Tub) are shown. Semi-quantitative scoring of staining results is summarized in Supplementary Table c. All error bars represent s.d. *p<0.01 compared to WT. **p<0.05 compared to CL. Scale bar, 50μm
Expression of SIAH1 and HIPK2 in human kidneys
We corroborated the in vitro findings by examining the renal expression of SIAH1 and HIPK2 in different kidney diseases. HIPK2 nuclear staining was markedly increased in kidneys of patients with HIVAN, FSGS, diabetic nephropathy (DN), and severe IgA nephropathy (IgAN) compared to patients with minimal change disease (MCD) and normal kidneys (n=5) (Fig 3j: upper panels). However, no significant change in HIPK2 staining was noted in MCD and Normal. Consistent with these findings, tubule-interstitial injury/fibrosis and GS were present in HIVAN, FSGS, DN, and IgAN, but not in MCD. In both early and late HIVAN kidneys, HIPK2 staining localized mainly to tubules with a less pronounced increase in glomerular staining. Glomerular HIPK2 expression was significantly higher in DN and IgAN compared to HIVAN. Focal increase in HIPK2 expression was observed in patients with early FSGS. SIAH1 staining was reduced in diseased kidneys where HIPK2 expression was increased (Fig 3j: lower panels). This inverse relationship between HIPK2 and SIAH1 staining was obvious in tubular area, but not in glomerular area of FSGS and HIVAN kidneys. Further reduction of HIPK2 staining in late HIVAN glomeruli did not correlate with SIAH1 expression, suggesting that HIPK2 may be regulated here by mechanisms independent of SIAH1. Semi-quantitative scoring of immunostaining data is summarized in Supplementary Table c.
Role of HIPK2 in HIV-induced apoptosis and expression of EMT markers in hRTEC
Since RTEC apoptosis is a hallmark of tubulo-interstitial injury in HIVAN and HIPK2 is known to mediate apoptosis, we sought to determine the role of HIPK2 in HIV-induced apoptosis of RTEC. Over-expression of KD-HIPK2 in hRTEC significantly attenuated HIV-induced apoptosis (Fig. 4a). In addition, KD-HIPK2 over-expression or siRNA-mediated knockdown of HIPK2 abrogated HIV-induced caspase 3 activity (Fig. 4b).
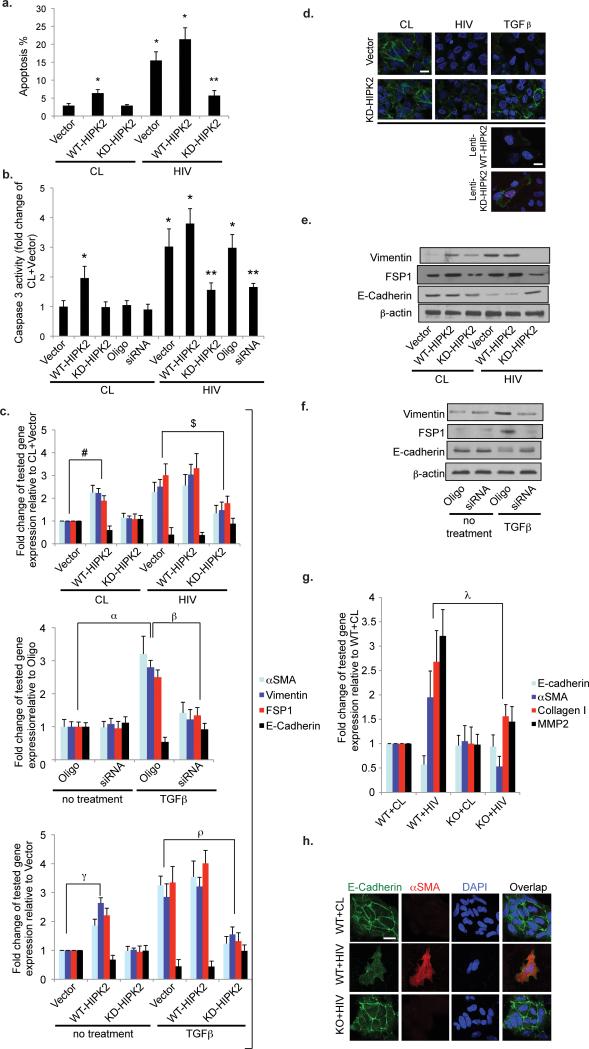
Figure 4. HIPK2 mediates HIV-induced apoptosis and expression of EMT markers in human renal tubular epithelial cells (hRTEC).
(a) Apoptosis was measured by flowcytometry after labeling with Annexin V and propidium iodide in hRTEC infected with either a control virus (CL) or a HIV-1 pseudotyped virus (HIV) and then transfected with an empty expression vector (Vector), a wild-type HIPK2 vector (WT-HIPK2), or a kinase-dead HIPK2 vector (KD-HIPK2). (b) Caspase 3 activity of hRTEC infected with CL or HIV and then transfected with Vector, WT-HIPK2, KD-HIPK2, a negative control siRNA (Oligo), or a specific siRNA to knockdown HIPK2 (siRNA). n=4 for all groups. (c, upper panel) Expression of epithelial mesenchymal transition (EMT) markers in hRTEC infected with HIV or CL virus and then transfected with Vector, WT-HIPK2, or KD-HIPK2. Fold change in expression relative to the CL+Vector group is shown. n=4 for all groups. (c. middle panel) Gene expression of hRTEC transfected with Oligo or siRNA and then treated with or without TGFβ (5 ng/ml). n=4 for all groups. (c lower panel) Gene expression in hRTEC transfected with WT-HIPK2, KD-HIPK2 or Vector and then incubated with or without TGFβ (5 ng/ml). n=4 for all groups. Immunofluorescent staining of E-cadherin in hRTEC transfected with Vector or KD-HIPK2 and then infected with either CL or HIV or treated with TGFβ (d, two upper panels). Immunostaining of E-Cadherin in hRTECs transduced with lentivectors to overexpress a fusion protein of mCherry with WT-HIPK2 (Lenti-WT-HIPK2) or with KD-HIPK2 (Lenti-KD-HIPK2) and treated with TGFβ (d, lower panel). E-Cadherin is green, Nuclear staining with DAPI is blue, mCherry fusion proteins are magenta. Representative results are shown. Scale bar, 10μm. Western blots of hRTEC cultured in the same condition as Fig 4c, upper panel (e) and as Fig 4c, middle panel (f). Representative blots of three independent experiments are shown. (g) Expression of targeted genes in primary renal tubular cells isolated from HIPK2 knockout mice (KO) and their WT (WT) littermates were infected with CL or HIV for 3 days and then quantified by real-time PCR. n=4 for all groups. (h) Immunofluorescent staining of E-Cadherin, alphaSMA, and DAPI in primary RTEC isolated from WT mice and infected with CL or HIV or from KO mice infected with HIV. αSMA = alpha smooth muscle actin. Scale bar 10μm. FSP1 = fibroblast-specific protein 1. TGFβ = transforming growth factor-β. DAPI = 4',6-diamidino-2-phenylindole. * p<0.01 compared to CL+Vector. ** p<0.01 compared to HIV+Vector. For each of the tested genes, # p<0.01 CL+Vector vs. CL+WT-HIPK2; $ p<0.05 HIV+Vector vs. HIV+KD-HIPK2; α p<0.01 Oligo vs. Oligo+TGFβ; β p<0.05 Oligo+TGFβ vs. siRNA+TGFβ; γ p<0.01 Vector vs. WT-HIPK2; ρ p<0.05 TGFβ+Vector vs. TGFβ+ KD-HIPK2; λ p<0.01 WT+HIV vs. KO+HIV.
Since HIPK2 interacts with the TGFβ pathway, a well-known mediator of epithelial-mesenchymal trans-differentiation (EMT) for RTEC in vitro and kidney fibrosis in vivo, we examined the effects of HIPK2 on the expression of EMT markers in hRTEC. We found that WT-HIPK2 over-expression or TGFβ treatment induced the expression of EMT markers compared to control cells, while over-expression of KD-HIPK2 diminished the expression of EMT markers in HIV-infected cells (Fig 4c). EMT is characterized by gain of αSMA, FSP-1, and vimentin and loss of E-cadherin. In addition, suppression of HIPK2 activity by either KD-HIPK2 over-expression or siRNA-mediated knockdown prevented TGFβ–induced expression of EMT markers (Fig. 4c). Over-expression of KD-HIPK2 restored the expression of E-cadherin in hRTEC treated with TGFβ or infected with HIV (Fig 4d, upper panels). hRTECs have reduced E-cadherin when expressing a mCherry-WT-HIPK2 fusion gene and protected from TGFβ-induced loss of E-cadherin when expressing a mCherry-KD-HIPK2-fusion gene (Fig 4d. lower panels). The effects of HIPK2 on the expression of EMT markers was further confirmed by western blot (Fig 4e,f). We also isolated primary RTEC from HIPK2 knockout (KO) mice and WT littermates. HIV infection induced EMT markers in WT, but not in KO RTECs (Fig 4g,h).
Role of HIPK2 mediated downstream signaling pathways in hRTEC
Signaling through p53 and TGFβ are known mediator of RTEC apoptosis18-20. We found that signaling components of p53 and TGF-β pathways were among 14 HIPK2 substrates enriched in our subnetwork of protein-protein interactions. Therefore, we sought to determine whether HIPK2 mediated HIV-induced activation of these pathways in hRTEC. For these studies, we used an immortalized proximal tubular epithelial cell line, HK2 cells, because of the limited availability of primary hRTEC and the large number of cells required for these experiments. First, we confirmed that HIPK2 mediated HIV-induced expression of EMT markers in HK2 cells (Supplementary Fig. 2a). We found that KD-HIPK2 inhibited TGFβ-induced Smad3 phosphorylation (Fig. 5a) suggesting that HIPK2 is required for TGFβ-induced Smad3 phosphorylation. The interaction of HIPK2 and Smad3 was confirmed by co-immunoprecipitation (Supplementary Fig. 2b). Smad3 phosphorylation was enhanced by HIV infection, which was abrogated by KD-HIPK2 over-expression (Fig. 5b). HIV infection also induced p53 phosphorylation in hRTECs, which was blocked by over-expression of KD-HIPK2 (Fig 5c). In addition, we found that HIV infection induced the expression of known target genes of Wnt-β-catenin and Notch pathways, which were blocked by over-expression of KD-HIPK2 (Fig. 5d,e). HIPK2 is known to up-regulate Wnt-mediated transcription by phosphorylating TCF3, a transcriptional repressor, but inhibit Wnt-mediated transcription by phosphorylating LEF1, a transcriptional activator28. Consistent with this, we found that WT-HIPK2 overexpression increased TCF3 phosphorylation, but not LEF1 in hRTEC (Supplementary Fig. 2c). These results demonstrate that HIPK2 mediates HIV-induced activation of p53, TGFβ, and Wnt/Notch pathways in RTECs.
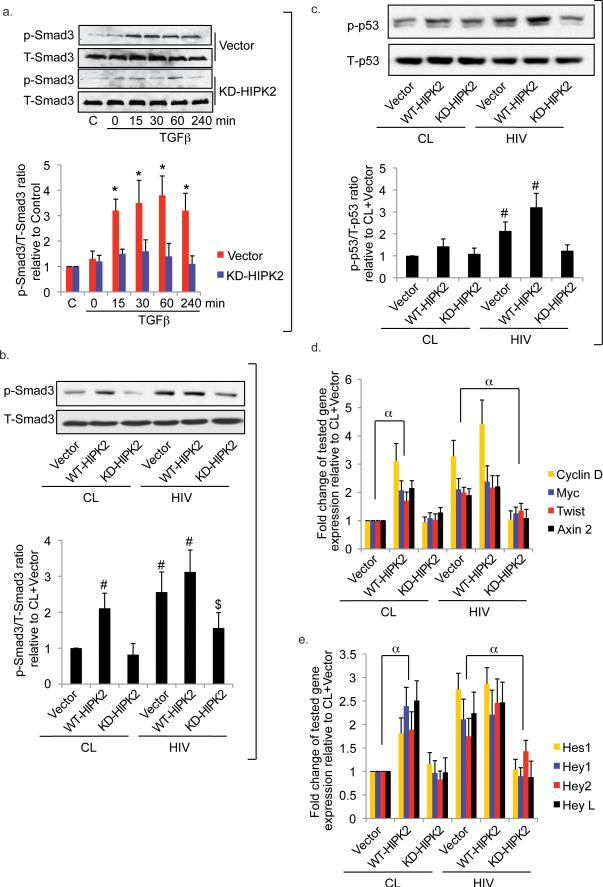
Figure 5. HIPK2 mediates downstream signaling pathways in RTEC.
Western blots (a. upper panel) of phospho- and total Smad3 (p-Smad3 and T-Smad3) using nuclear lysate of HK2 cells transfected with either an empty expression vector (Vector) or a kinase dead HIPK2 (KD-HIPK2) expression vector and then stimulated with TGFβ (5 ng/ml) for 0, 15, 30, 60, or 240 min. (a. lower panel) Ratios of densitometric measurements of p-Smad3 to T-Smad3 band intensity. Western blots (b. upper panel) and densitometric ratios (b. lower panel) of p-Smad3 to T-Smad3 in HK2 infected with either a control (CL) virus or a HIV-1 pseudotyped virus (HIV) prior to transfection with an empty expression vector (Vector), a wild-type HIPK2 vector (WT), or a kinase-dead HIPK2 vector (KD). Western blots (c. upper panel) and densitometric ratios (c. lower panel) of phospho-p53 (p-p53) to total p53 (T-p53) in primary hRTEC. Expression of genes involved in Wnt-β-catenin (d) and Notch (e) signaling pathways from primary hRTECs. * p<0.05 compared to the corresponding KD-HIPK2 group. # p<0.05 compared to the CL+Vector group. $ p<0.05 compared to the HIV+Vector group. α p<0.05 for all tested genes between the indicated groups. Representative blots shown. C = Cells without treatment.
To identify additional downstream HIPK2-mediated signaling pathways we profiled protein/DNA interactions and pattern of gene expression in human embryonic kidney (HEK) cells with HIPK2 over-expression. HEK cell line was chosen for its high efficiency of transfection. Fourteen TFs were up-regulated and 15 TFs were down-regulated by HIPK2 over-expression in protein/DNA interaction arrays (Supplementary Table d). Of the up-regulated interactions, NFkB and Stat3 are known to be activated in Tg26 kidney. p53, and a downstream nuclear mediator of the TGFβ pathway—Smad3, were also identified as up-regulated TFs. Furthermore, Snail29 and Ets130—two TFs known to mediate kidney fibrosis—were also up-regulated. Promoter analysis of differentially expressed genes in microarray identified an enrichment of NFkB and Stat3 target genes (Supplementary Table e). To confirm that NFkB is a key TF downstream of HIPK2 we examined NFkB activation by luciferase reporter assay. In hRTEC, KD-HIPK2 overexpression suppressed TNF-α-induced NFkB activation (TNF-α vs. no treatment: 10.32±1.22, TNF-α+KD-HIPK2 vs. no treatment+KD-HIPK2: 2.41±0.65, n=4, p<0.01 for TNF-α-treated cells vs. TNF-α+KD-HIPK2). We also confirmed that KD-HIPK2 over-expression inhibited—while WT-HIPK2 over-expression induced—the expression of NFkB-targeted genes in HIV-infected hRTEC (Supplementary Fig. 2d). Pathway analysis of genes up-regulated by HIPK2 using Wiki-pathways31 showed that TGFβ receptor and Wnt signaling pathways were both highly ranked (Supplementary Table 2e). These unbiased analyses confirmed that TGFβ/Smad3, Wnt, p53, and NFKB are key pathways downstream of HIPK2.
Knockout of HIPK2 attenuates kidney fibrosis in Tg26
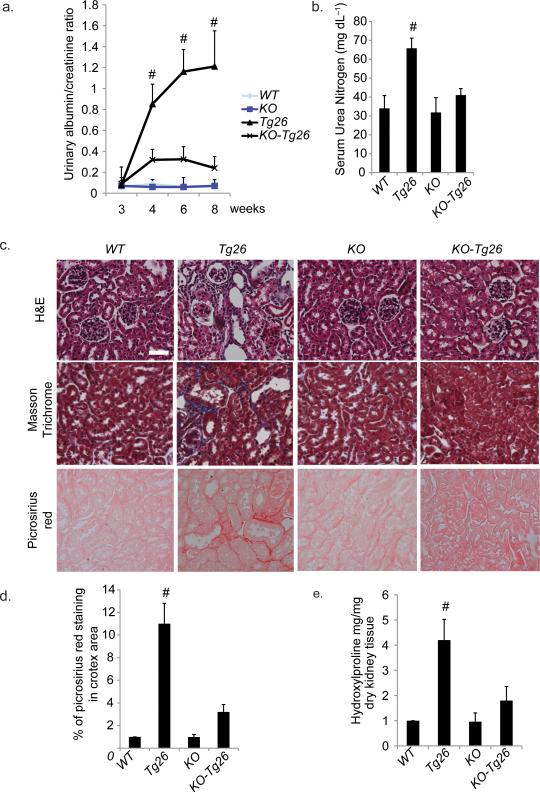
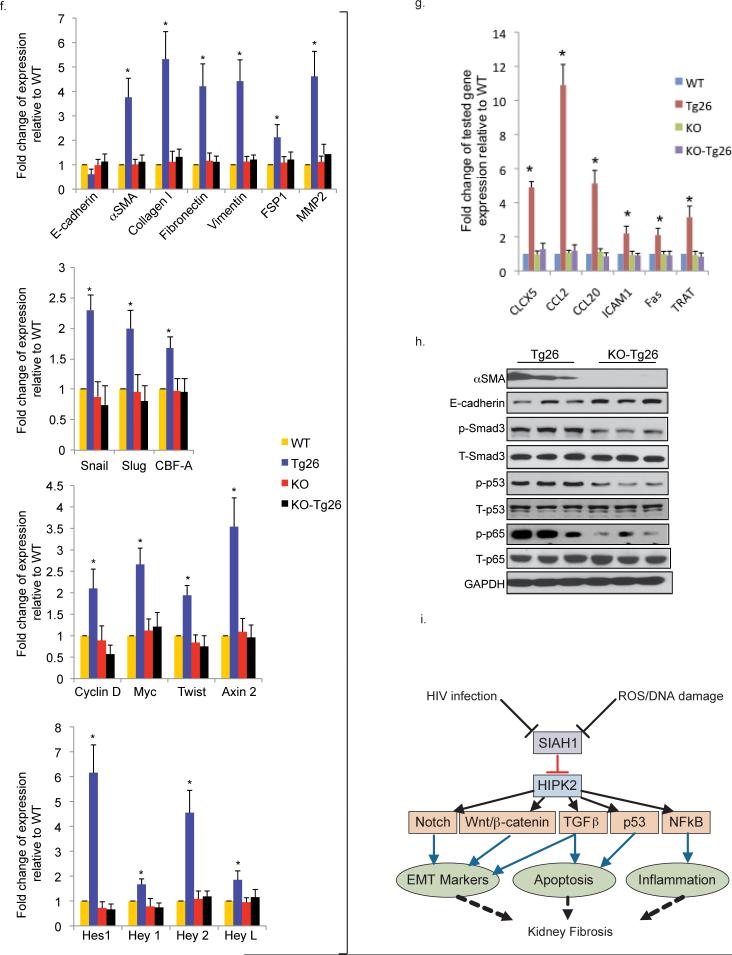
So far, we identified HIPK2 as a key regulator of RTEC apoptosis and expression of EMT markers in vitro. However, controversy exists in whether EMT of RTEC in-vitro correlates with renal fibrosis in-vivo32. To verify the in vivo role of HIPK2, we generated double transgenic animals that were HIPK2 knockout (KO) and expressed the HIV1 transgene (KO-Tg26). KO animals have no obvious renal phenotype33,34. Tg26 mice typically develop proteinuria from 4 weeks of age. KO-Tg26 had reduced proteinuria compared to Tg26 mice (Fig 6a). KO-Tg26 mice had a lower level of serum urea nitrogen, which is a marker of renal function, compared to Tg26 mice (Fig 6b). Tubulointerstitial injury and fibrosis were also significantly attenuated in KO-Tg26 compared to Tg26 (Fig. 6c,d and Supplementary Table f). Reduction in renal fibrosis was confirmed by measurement of collagen content within the renal cortex (Fig 6e). Podocyte hyperplasia and glomerulosclerosis were also improved in KO-Tg26 compared to Tg26 (Supplementary Table f). The expression of EMT markers was significantly increased in kidney cortex of Tg26 compared to WT, but not in KO-Tg26 (Fig. 6f top two panels, 6h, and Supplementary Fig. 2e top two panels). To determine whether signaling pathways downstream of HIPK2 were reduced in KO-Tg26 mice, we examined the expression of Wnt/Notch and NFkB target genes by real-time PCR. Expression of Wnt/Notch (Fig. 6f bottom two panels) and NFkB (Fig. 6g) targeted genes was higher in the kidney cortex of Tg26 compared to KO-Tg26 mice. Phosphorylation of p53, Smad3, and NFkB was also increased in the kidney of Tg26 mice, compared to KO-Tg26 mice (Fig. 6h). We also confirmed the increase in nuclear staining of β-catenin, which is a marker of EMT35, and phospho-Smad3 in kidneys of Tg26 compared to KO-Tg26 mice (Supplementary Fig. 2e bottom two panels). Together, these findings showed that knockout of HIPK2 abolished HIV-induced activation of p53, TGF-β/Smad3, and Wnt/Notch pathways in the kidney cortex of Tg26 mice. We also confirmed that expression of the HIV nef gene was not reduced in KO-Tg26 mice (data not shown) to ensure that reduction of HIPK2 did not interfere with HIV viral gene expression as a confounding experimental factor.
Figure 6. Knockout of HIPK2 prevents kidney injury in Tg26.
(a) Urine albumin to creatinine ratio of Tg26 and wildtype (WT) mice with and without HIPK2 knockout (KO) at 3, 4, 6 and 8 weeks of age. (b) Serum urea nitrogen levels of mice at sacrifice. (c) H&E, Masson Trichrome, and Picrosirius red staining of kidney sections. Representative pictures are shown. Scale bar 50μm. (d) Quantification of percentage area with picrosirius red staining by morphometric analysis. (e) Quantification of hydroxyproline content in kidney tissue. Expression of markers of epithelial-mesenchymal transition (f. upper two panels) and target genes of the Wnt-β-catenin (f. third panel), Notch (f. bottle panel) and NFkB (g) pathways in the renal cortex. (h) Western blots for α-smooth muscle actin (αSMA), E-cadherin, phospho- and total-Smad3, p53, and p65, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Representative blots from 3 mice in each group are shown. (i) A schematic that summarizes the flow of signals from initial insults (i.e. HIV infection or oxidative stress) to the suppression of SIAH1, expression of HIPK2, and activation of multiple signaling pathways involved in kidney fibrosis. # p<0.05 compared to KO-Tg26. n=6 for all groups. * p<0.05 compared to other groups (WT, KO, and KO-Tg26).
Role of HIPK2 in mediating HIV-induced podocyte dedifferentiation/EMT
Since HIPK2 is increased in glomeruli of Tg26 and KO-Tg26 mice had less proteinuria, podocyte hyperplasia, and glomerulosclerosis, we sought to determine the role of HIPK2 in HIV-induced podocyte de-differentiation and EMT. Glomerular expression of EMT markers was increased and of podocyte differentiation markers was suppressed in Tg26 compared to WT, but not in KO-Tg26 (Supplementary Fig. 3a-c). Glomerular expression of Wnt/Notch/NFkB-targeted genes was enhanced in Tg26 compared to WT, but not in KO-Tg26 (Supplementary Fig. 3d,e). HIPK2 expression remained unchanged (Supplementary Fig. 3d). Smad3 phosphorylation is increased in Tg26 compared KO-Tg26 glomeruli (Supplementary Fig. 3f). We also confirmed that KD-HIPK2 over-expression suppressed desmin and Snail1, restored nephrin and ZO-1, inhibited HIV-induced expression of Wnt/Notch and NFkB-targeted genes, and attenuated HIV-induced Smad3 phosphorylation in podocytes (Supplementary Fig. 3g-j). These data validated the role of HIPK2 in HIV-induced activation of pro-fibrotic pathways in podocytes.
Role of ROS in regulation of SIAH1/HIPK2 expression and kidney fibrosis
To investigate the cellular effects of NAC treatment we treated HIV-infected hRTEC with NAC and found that NAC treatment attenuated HIV-induced increase in caspase 3 activity and EMT marker expression (Supplementary Fig 3a,b). Treatment of Tg26 mice with NAC partially attenuated proteinuria, serum urea nitrogen elevation, and kidney fibrosis in Tg26 mice (Supplementary Fig. 4c-f). By western blot, we confirmed that NAC increased SIAH1, suppressed HIPK2, and increased the ratio of reduced to oxidized glutathione in renal cortices of Tg26 compared to vehicle-treated Tg26 renal cortices (Supplementary Fig 4g,h). These findings confirmed that ROS mediate HIV-induced HIPK2 expression and kidney fibrosis in Tg26 mice.
Role of HIPK2 in tubulointerstitial fibrosis (TIF) induced by unilateral ureteral obstruction (UUO)
Since our in vitro studies suggest that HIPK2 mediates TGFβ-induced expression of EMT markers in RTEC, we hypothesized that HIPK2 might have a more general role in renal fibrosis. We tested this hypothesis in a well-established model of renal fibrosis—unilateral ureteral obstruction (UUO). We found that HIPK2 was significantly increased in the tubulo-interstitium of WT kidney with UUO compared to the sham-operated WT kidney (Supplementary Fig. 5a top panel). UUO caused significant fibrosis in WT kidney compared to KO kidney as confirmed by both Masson Trichrome and picrosirius red staining (Supplementary Fig. 5a bottom panels). SIAH1 was reduced and HIPK2 was increased in WT-UUO kidneys (Supplementary Fig. 5b). Renal fibrosis was confirmed by quantification of morphometric measurement of picrosirius red staining and collagen content in the renal cortex (Supplementary Fig. 5c). Renal expression of EMT markers (Supplementary Fig. 5d,e) and nuclear β-catenin and p-Smad3 staining (Supplementary Fig. 5f top and bottom panel, respectively) were increased in WT-UUO compared to WT-Sham, but not in KO-UUO kidneys. Smad3 phosphorylation was further confirmed by western blot analysis (Supplementary Fig. 5g). We also examined the expression of Wnt/Notch target genes, which were upregulated in WT-UUO compared to WT-Sham, but not in KO-UUO (Supplementary Fig 5h). These data indicated that HIPK2 knockout in vivo prevents TIF and the expression of EMT markers in the UUO model.
Role of HIPK2 in folic acid-induced kidney fibrosis model
We also examined the role of HIPK2 in a third model of renal fibrosis—folic acid (FA) induced renal fibrosis36-38. We found that KO mice are protected from FA induced kidney fibrosis as demonstrated by renal histology and measurement of kidney hydroxylproline content (Supplementary Fig. 6a,b). Serum urea nitrogen level and renal expression of EMT markers were similarly reduced in KO+FA compared to WT+FA animals (Supplementary Fig. 6c, d).
Discussion
In this study, we integrated experimental and computational approaches to identify upstream protein kinases responsible for observed gene regulation changes and connected it to a disease state. With this approach, we identified MAPK1 (ERK2), MAPK3 (ERK1), and HIPK2 as the top three protein kinases mediating gene expression in the Tg26 kidney. We previously showed that ERK mediates HIV-induced podocyte proliferation and RTEC apoptosis10,39. Here, we identified HIPK2 as a critical regulator of EMT in vitro and kidney fibrosis in vivo. Our data suggest that both ERK and HIPK2 are critical for HIV-induced kidney disease. Further studies are required to determine the crosstalk of ERK and HIPK2 pathways.
We confirmed that HIPK2 expression is increased not only in kidneys of Tg26 mice and patients with HIVAN, but also in kidneys of patients with FSGS, DN and IgAN. This supports a more general role for HIPK2 in human kidney diseases, which was corroborated by the fact that HIPK2 mediates renal fibrosis in three established animal models of renal fibrosis (Tg26, UUO and FA).
We found that HIPK2 mediates HIV-induced apoptosis and expression of EMT markers in cultured hRTEC. Apoptosis and EMT of RTEC leading to renal fibrosis have been described in many kidney diseases including HIVAN40,41,42,43. Recent studies suggest that complete EMT of RTEC may not occur in vivo. However, RTEC can gain EMT markers (epithelial plasticity in fibrogenesis) after injury44,45. Our data suggest that HIPK2 mediates expression of EMT markers in RTEC both in vitro and in vivo.
Our data indicate that HIPK2 is increased in RTEC as well as glomeruli of diseased kidney. It is possible that fibroblasts and/or pericytes also express HIPK2. We found that HIPK2 also mediates HIV-induced dedifferentiation of podocytes and the expression of EMT markers. Consistent with this, knockout of HIPK2 significantly reduced proteinuria, podocyte hyperplasia, and glomerulosclerosis in Tg26 mice. Future studies are needed to determine whether HIPK2 is required for the activation of fibroblasts and/or pericytes or if HIPK2 mediates RTEC-fibroblast crosstalk.
In this study we focused on the intracellular mechanisms of HIPK2 regulation in diseased kidneys. We found that HIV decreases SIAH1 expression through DNA damage and oxidative stress, leading to the accumulation of HIPK2. In addition, we confirmed that inhibition of ROS by NAC restores Siah1 expression, reduces HIPK2 protein level, and attenuates kidney injury/fibrosis in Tg26 mice. Although renal protective effects of NAC in human kidney disease remain controversial46, our data indicate that anti-ROS therapy could be a potential approach to treat patients with kidney fibrosis. Our findings are consistent with prior studies that poly-ubiquitination promotes HIPK2 degradation47 and DNA-damage and oxidative stress activate HIPK248.
We confirmed that HIPK2 mediates TGFβ- and HIV-induced pro-apoptotic and pro-fibrotic pathways in RTEC including p53, Smad3, and Notch/Wnt. Previous study showed that HIPK2 can either repress or promote Wnt-β-catenin-mediated transcriptional activation 34,14. Consistent with this, recent studies suggest that HIPK2 could affect Wnt pathway in a context-dependent manner28. HIPK2 up-regulates Wnt-mediated transcription by phosphorylating TCF3, a transcriptional repressor, but inhibites Wnt-mediated transcription by phosphorylating LEF1, a transcriptional activator28. Consistent with this, we found that overexpression of WT-HIPK2 increases TCF3 phosphorylation, but not LEF1 in RTEC. NFkB-mediated inflammation is known to cause tissue injury/fibrosis. Our studies also suggest that HIPK is a key regulator of the NFkB pathway, which is a novel finding that has never been explored previously. These findings, summarized in Fig 6j, demonstrate that signals from initial insults, such as DNA damage or oxidative stress, increase HIPK2 expression to activate multiple signaling pathways that are involved in kidney fibrosis. HIPK2 is a logical therapeutic drug target for kidney fibrosis and protein kinases are “drug-able” targets.
In conclusion, we presented here a systems approach to identifying upstream protein kinases based on genome-wide mRNA expression microarrays and DNA/protein arrays. We identified and confirmed that HIPK2, a protein kinase previously unrecognized in kidney disease, plays a critical role in renal fibrosis. We elucidated the regulation of HIPK2 in kidney disease and downstream signaling pathways that mediate HIPK2-induced apoptosis and the expression of EMT markers in kidney cells. We believe that HIPK2 could be a novel therapeutic target for kidney disease.
Methods
Methods and associated references are available in the online version of the paper.
Supplementary Material
Acknowledgement
The authors acknowledge and thank G.L. Gusella for providing the lentiviral construct and E. Huang for providing the HIPK2 knockout mice. We also thank R. Iyengar and P. Klotman for critical suggestions on experimental design and for other valuable contributions. JCH is supported by NIH 1R01DK078897, and VA Merit Award; JCH and AM are supported by NIH 1R01DK088541, NIH P01-DK-56492, and 1RC4DK090860; AM is supported by RC2OD006536 and P50GM071558; PYC is supported by NIH 5K08DK082760. CN are supported by Chinese 973 fund 2012CB517601.
Online Methods
Cells
HEK293 and HK2 cells were obtained from ATCC1. Primary human renal epithelial cells were obtained from PromoCell. Conditionally immortalized murine podocytes were cultured as described2. Primary renal tubular epithelial cells were isolated from mouse kidneys as described in Supplementary Methods.
Gene Delivery
For HIV infection, pNL4–3: Δ G/P-EGFP, a gag/pol-deleted HIV-1 construct that contains EGFP in the gag open reading frame, and pHR'-IRES-EGFP, a control EGFP construct, were used to generate VSV-G–pseudotyped virus3. Lonza Nucleofector Technology (Basic Neucleofector kit for Primary Mammalian Epithelial Cells, Program T20) was used for transfection of renal epithelial cells. For in vitro experiments, cells were infected with HIV pseudotyped virus or control virus for 2 days and then transfected with expression constructs (i.e. WT-HIPK2, KD-HIPK2, or siRNAs) for 3 more days. For experiments where cells were treated with TGFβ, cells were transfected with expression constructs or siRNAs 3 days prior to TGFβ treatment for 48h.
Constructs and siRNAs
siRNA for SIAH1 (Qiagen, FlexiTube siRNA SI03037839) and HIPK2 were purchased from Thermo Scientific (Dharmacon ON-TARGET siRNA). Negative Control siRNAs were used as negative experimental controls. WT and KD HIPK2 constructs were obtained from Dr. RH Goodman, Oregon Health and Science University, Portland4. SIAH1 expression vector was purchased from Open Biosystems. Lentivectors encoding for fusion proteins of mCherry with HIPK2 (WT and KD) were generated as described in Supplementary Methods.
Apoptosis analysis
Annexin V-FITC Apoptosis Detection Kit (BD Bioscience), FACSCalibur Flow cytometer (BD Biosciences) and CellQuest software (BD Biosciences) were used. Caspase 3 activity was measured using a Human Active Caspase-3 Immunoassay kit from R&D Systems, Inc.
Real-time PCR
Quantitative real-time PCR was performed using Qiagen QuantiTect One Step RTPCR SYBR green kit (Qiagen). Data were analyzed by the 2 -ΔΔCT method and presented as fold change relative to a control sample after normalization against the expression of housekeeping genes.
Western blot
Western blot was performed as described in the Supplementary Methods. Following antibodies were used: antibody to mouse HIPK2 (Santa-Cruz), to human HIPK2 (Abcam), to SIAH1 (Novus), to phospho- or total Smad3, p53, p65, LEF, vimentin, Snail, E-Cadherin, ZO-1 and TCF (Cell Signaling Technology), to FSP-1 (Milipore), to nephrin (a gift from Dr. Larry Holzman), to synaptopodin (a gift from Dr. Peter Mundel), and to β-actin and GAPDH (Sigma).
Immunostaining of cells and tissue
Immunostaining of cells and tissue are described in Supplementary methods. Archival human kidney biopsies were collected at Columbia University under a protocol approved by its Institutional Review Board.
HIPK2 kinase activity assay
HIPK2 activity was measured in nuclear protein extracts following a protocol provided by Millipore (see Supplementary Method).
NFkB reporter assay
NFkB reporter assay was performed as described in Supplementary Methods.
Animal studies
The IACUC committee of Mount Sinai School of Medicine approved all animal studies. HIPK2–/– (KO) mice were obtained from Dr. Eric Huang5. KO mice were backcrossed to FVB/N for 9 generations and then crossed to Tg26 mice to generate HIPK2+/–;Tg26 mice. Subsequent crossings of HIPK2+/–;Tg26 to HIPK2+/–;Tg26 generated HIPK2–/–;Tg26 (KO-Tg26), HIPK2+/–;Tg26, HIPK+/+;Tg26 (Tg26) mice, and corresponding non-Tg26 littermates. UUO and folic acid induced acute kidney injury models were created as described in Supplementary Methods. Tg26 mice and their age and sex-matched control littermates were fed n-acetylcysteine at 300mg/kg body weight or vehicle as described6 by daily gavage.
Kidney histology
Histologic analyses on PAS-stained slides were scored for the glomerulosclerosis index, podocyte hypertrophy and hyperplasia, as well as tubulointerstitial changes as described7. Masson Trichrome and Picrosirius Red staining were performed to assess fibrosis8. Renal cortical collagen content was determined by morphometric measurement of areas stained with picrosirius red9.
Affymatrix/Panomics protein/DNA arrays
Protein/DNA arrays (Combo Array, Affymatrix) were performed using nuclear proteins of kidneys from Tg26 and WT mice (n=5) following manufacturer's protocol. Data was analyzed as described in Supplementary Methods.
Microarray studies
Affymatrix gene expression microarrays were performed at the Mount Sinai Institution Microarray Core Facility and analyzed as described in Supplementary Methods.
Quantitative determination of tissue hydroxyproline
Hydroxyproline content in the kidney hydrolysates was quantified as described10 and expressed as mg mg–1 dry kidney weight.
Measurement of GSH/GSSG ratio
GSH to GSSG ratio was measured as described in Supplementary Methods.
Isolation of glomeruli from mice for western blot and real-time PCR
Mouse glomeruli were isolated as described11,12. The purity of the glomerular isolate was verified by microscopy and western blotting for synaptopodin, nephrin, and WT-1.
Statistical analysis
Data were expressed as mean ± standard deviation (X±SD). Unpaired T-test was used to analyze data between 2 groups after determination of data distribution. The Bonferroni correction was used when more than two groups were present. Statistical significance was considered when p<0.05.
Computational network analysis
Promoter analysis was conducted by utilizing the TRANSFAC and ChEA. Protein interaction subnetworks were created using Gene2Networks. Kinase enrichment analysis was conducted using KEA. For details see Supplementary Methods.
Footnotes
Author contributions:
J.C.H., A.M., P.Y.C., Y.J., N.C. and K.R. designed the research project; Y.J., K.R., Y.F., Y.Z., M.R., H.X., and P.Y.C. performed the experiments; P.Y.C and Y.D. prepared lentiviral constructs for HIPK2; V.D. analyzed pathologic findings; A.M., E.Y.C., and A.M. performed bioinformatics and systems biology analyses; J.C.H., P.Y.C, and A.M. wrote the manuscript and all author contributed to the preparation of the manuscript.
Competing interest statement:
The authors declare that they have no competing financial interests.
Accession codes:
The microarray data was deposited to GEO (GSE35226) [NCBI tracking system #16249557].
Reference
- 1.Wyatt CM, Klotman PE. HIV-associated nephropathy in the era of antiretroviral therapy. Am J Med. 2007;120:488–492. doi: 10.1016/j.amjmed.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 2.Leventhal JS, Ross MJ. Pathogenesis of HIV-associated nephropathy. Semin Nephrol. 2008;28:523–534. doi: 10.1016/j.semnephrol.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Bruggeman LA, et al. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol. 2000;11:2079–2087. doi: 10.1681/ASN.V11112079. [DOI] [PubMed] [Google Scholar]
- 4.Kopp JB, et al. Progressive glomerulosclerosis and enhanced renal accumulation of basement membrane components in mice transgenic for human immunodeficiency virus type 1 genes. Proc Natl Acad Sci U S A. 1992;89:1577–1581. doi: 10.1073/pnas.89.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickie P, et al. HIV-associated nephropathy in transgenic mice expressing HIV-1 genes. Virology. 1991;185:109–119. doi: 10.1016/0042-6822(91)90759-5. [DOI] [PubMed] [Google Scholar]
- 6.Matys V, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger SI, Posner JM, Ma'ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He JC, et al. Nef stimulates proliferation of glomerular podocytes through activation of Src-dependent Stat3 and MAPK1,2 pathways. J Clin Invest. 2004;114:643–651. doi: 10.1172/JCI21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calzado MA, Renner F, Roscic A, Schmitz ML. HIPK2: a versatile switchboard regulating the transcription machinery and cell death. Cell Cycle. 2007;6:139–143. doi: 10.4161/cc.6.2.3788. [DOI] [PubMed] [Google Scholar]
- 12.Isono K, et al. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol Cell Biol. 2006;26:2758–2771. doi: 10.1128/MCB.26.7.2758-2771.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Orazi G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 14.Lee W, Swarup S, Chen J, Ishitani T, Verheyen EM. Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of beta-catenin/Arm and stimulation of target gene expression. Development. 2009;136:241–251. doi: 10.1242/dev.025460. [DOI] [PubMed] [Google Scholar]
- 15.Lee W, Andrews BC, Faust M, Walldorf U, Verheyen EM. Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev Biol. 2009;325:263–272. doi: 10.1016/j.ydbio.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, et al. Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat Neurosci. 2007;10:77–86. doi: 10.1038/nn1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann TG, Stollberg N, Schmitz ML, Will H. HIPK2 regulates transforming growth factor-beta-induced c-Jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res. 2003;63:8271–8277. [PubMed] [Google Scholar]
- 18.Seth R, Yang C, Kaushal V, Shah SV, Kaushal GP. p53-dependent caspase-2 activation in mitochondrial release of apoptosis-inducing factor and its role in renal tubular epithelial cell injury. J Biol Chem. 2005;280:31230–31239. doi: 10.1074/jbc.M503305200. [DOI] [PubMed] [Google Scholar]
- 19.Yoo J, et al. Transforming growth factor-beta-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. J Biol Chem. 2003;278:43001–43007. doi: 10.1074/jbc.M307869200. [DOI] [PubMed] [Google Scholar]
- 20.Inazaki K, et al. Smad3 deficiency attenuates renal fibrosis, inflammation,and apoptosis after unilateral ureteral obstruction. Kidney Int. 2004;66:597–604. doi: 10.1111/j.1523-1755.2004.00779.x. [DOI] [PubMed] [Google Scholar]
- 21.Zeisberg M, et al. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 22.Vincent T, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 25.Winter M, et al. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol. 2008;10:812–824. doi: 10.1038/ncb1743. [DOI] [PubMed] [Google Scholar]
- 26.Husain M, et al. Inhibition of p66ShcA longevity gene rescues podocytes from HIV-1-induced oxidative stress and apoptosis. J Biol Chem. 2009;284:16648–16658. doi: 10.1074/jbc.M109.008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenstiel PE, et al. HIV-1 Vpr activates the DNA damage response in renal tubule epithelial cells. AIDS. 2009;23:2054–2056. doi: 10.1097/QAD.0b013e32833088a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hikasa H, Sokol SY. Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J Biol Chem. 286:12093–12100. doi: 10.1074/jbc.M110.185280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boutet A, et al. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J. 2006;25:5603–5613. doi: 10.1038/sj.emboj.7601421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizui M, et al. Transcription factor Ets-1 is essential for mesangial matrix remodeling. Kidney Int. 2006;70:298–305. doi: 10.1038/sj.ki.5001541. [DOI] [PubMed] [Google Scholar]
- 31.Pico AR, et al. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriz W, Kaissling B, Le Hir M. Epithelial-mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? J Clin Invest. 2011;121:468–474. doi: 10.1172/JCI44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 34.Wei G, et al. HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc Natl Acad Sci U S A. 2007;104:13040–13045. doi: 10.1073/pnas.0703213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertig A. [Epithelial-mesenchymal transition of the renal graft]. Nephrol Ther. 2008;4(Suppl 1):S25–S28. doi: 10.1016/S1769-7255(08)73648-4. [DOI] [PubMed] [Google Scholar]
- 36.Long DA, et al. Angiopoietin-1 therapy enhances fibrosis and inflammation following folic acid-induced acute renal injury. Kidney Int. 2008;74:300–309. doi: 10.1038/ki.2008.179. [DOI] [PubMed] [Google Scholar]
- 37.Doi K, et al. Attenuation of folic acid-induced renal inflammatory injury in platelet-activating factor receptor-deficient mice. Am J Pathol. 2006;168:1413–1424. doi: 10.2353/ajpath.2006.050634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bielesz B, et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 120:4040–4054. doi: 10.1172/JCI43025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyder A, et al. HIV-1 viral protein r induces ERK and caspase-8-dependent apoptosis in renal tubular epithelial cells. AIDS. 24:1107–1119. doi: 10.1097/QAD.0b013e328337b0ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav A, et al. HIVAN phenotype: consequence of epithelial mesenchymal transdifferentiation. Am J Physiol Renal Physiol. 2009 doi: 10.1152/ajprenal.00415.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross MJ, et al. Role of ubiquitin-like protein FAT10 in epithelial apoptosis in renal disease. J Am Soc Nephrol. 2006;17:996–1004. doi: 10.1681/ASN.2005070692. [DOI] [PubMed] [Google Scholar]
- 42.Docherty NG, O'Sullivan OE, Healy DA, Fitzpatrick JM, Watson RW. Evidence that inhibition of tubular cell apoptosis protects against renal damage and development of fibrosis following ureteric obstruction. Am J Physiol Renal Physiol. 2006;290:F4–13. doi: 10.1152/ajprenal.00045.2005. [DOI] [PubMed] [Google Scholar]
- 43.Zeisberg M, Kalluri R. The role of epithelial-to-mesenchymal transition in renal fibrosis. J Mol Med. 2004;82:175–181. doi: 10.1007/s00109-003-0517-9. [DOI] [PubMed] [Google Scholar]
- 44.Quaggin SE, Kapus A. Scar wars: mapping the fate of epithelial-mesenchymal-myofibroblast transition. Kidney Int. 2011 doi: 10.1038/ki.2011.77. [DOI] [PubMed] [Google Scholar]
- 45.Hertig A, Flier SN, Kalluri R. Contribution of epithelial plasticity to renal transplantation-associated fibrosis. Transplant Proc. 2010;42:S7–12. doi: 10.1016/j.transproceed.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mainra R, Gallo K, Moist L. Effect of N-acetylcysteine on renal function in patients with chronic kidney disease. Nephrology (Carlton) 2007;12:510–513. doi: 10.1111/j.1440-1797.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 47.Di Stefano V, Blandino G, Sacchi A, Soddu S, D'Orazi G. HIPK2 neutralizes MDM2 inhibition rescuing p53 transcriptional activity and apoptotic function. Oncogene. 2004;23:5185–5192. doi: 10.1038/sj.onc.1207656. [DOI] [PubMed] [Google Scholar]
- 48.Rinaldo C, Prodosmo A, Siepi F, Soddu S. HIPK2: a multitalented partner for transcription factors in DNA damage response and development. Biochem Cell Biol. 2007;85:411–418. doi: 10.1139/O07-071. [DOI] [PubMed] [Google Scholar]
- 1.Rosenstiel PE, et al. HIV-1 Vpr inhibits cytokinesis in human proximal tubule cells. Kidney Int. 2008;74:1049–1058. doi: 10.1038/ki.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He JC, et al. Nef stimulates proliferation of glomerular podocytes through activation of Src-dependent Stat3 and MAPK1,2 pathways. J Clin Invest. 2004;114:643–651. doi: 10.1172/JCI21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross MJ, et al. Role of ubiquitin-like protein FAT10 in epithelial apoptosis in renal disease. J Am Soc Nephrol. 2006;17:996–1004. doi: 10.1681/ASN.2005070692. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 5.Winter M, et al. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol. 2008;10:812–824. doi: 10.1038/ncb1743. [DOI] [PubMed] [Google Scholar]
- 6.Ivanovski O, et al. The antioxidant N-acetylcysteine prevents accelerated atherosclerosis in uremic apolipoprotein E knockout mice. Kidney Int. 2005;67:2288–2294. doi: 10.1111/j.1523-1755.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 7.D'Agati V. Pathologic classification of focal segmental glomerulosclerosis. Semin Nephrol. 2003;23:117–134. doi: 10.1053/snep.2003.50012. [DOI] [PubMed] [Google Scholar]
- 8.He W, et al. Wnt/beta-catenin signaling promotes renal interstitial fibrosis. J Am Soc Nephrol. 2009;20:765–776. doi: 10.1681/ASN.2008060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junqueira LC, Bignolas G, Brentani RR. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem J. 1979;11:447–455. doi: 10.1007/BF01002772. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Dai C, Liu Y. Hepatocyte growth factor gene therapy and angiotensin II blockade synergistically attenuate renal interstitial fibrosis in mice. J Am Soc Nephrol. 2002;13:2464–2477. doi: 10.1097/01.asn.0000031827.16102.c1. [DOI] [PubMed] [Google Scholar]
- 11.Takemoto M, et al. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161:799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratnam KK, et al. Role of the retinoic acid receptor-alpha in HIV-associated nephropathy. Kidney Int. 79:624–634. doi: 10.1038/ki.2010.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.