Abstract
Background
Infiltrative growth pattern at the tumor margin has been associated with shorter patient survival. However, little is known on prognostic significance of tumor growth pattern, independent of tumoral molecular alterations and other histological features.
Methods
Utilizing a database of 1139 colon and rectal cancer patients in two prospective cohort studies, histological features including tumor growth pattern, tumor differentiation, lymphocytic reaction, mucinous component, and signet ring cell component were recorded by a single pathologist. Cox proportional hazards model was used to compute mortality hazard ratio (HR), adjusting for clinical, pathological and tumor molecular features, including microsatellite instability (MSI), the CpG island methylator phenotype, LINE-1 methylation, and KRAS, BRAF, and PIK3CA mutations.
Results
Among 1139 colorectal cancers, we observed expansile growth pattern in 372 tumors (33%), intermediate growth pattern in 610 tumors (54%), and infiltrative growth pattern in 157 tumors (14%). Compared to patients with expansile growth pattern, those with infiltrative growth pattern experienced shorter cancer-specific survival [log-rank p<0.0001; multivariate HR=1.74; 95% confidence interval (CI), 1.22-2.47] and overall survival [log-rank p<0.0001; multivariate HR=1.78; 95% CI, 1.33-2.39]. The prognostic association of infiltrative growth pattern was confined to stage I-III patients (Pinteraction with stage=0.0001).
Conclusions
Infiltrative growth pattern was associated with worse prognosis among stage I-III colorectal cancer patients, independent of other clinical, pathologic, and molecular characteristics.
INTRODUCTION
Tumor-host interactions play a critical role in tumor growth and progression. An infiltrative growth pattern at the tumor margin observed by pathologic examination is characterized by widespread dissemination of tumor cells into normal tissue structures with loss of a clear boundary between tumor and host tissue.1 Studies have shown that an infiltrative growth pattern is associated with shorter patient survival in colorectal cancer.2-10 However, the true nature of this association remains uncertain. An infiltrating growth pattern is inversely correlated with the presence of immune and inflammatory responses at the invasive tumor front.1, 4, 5 In fact, greater lymphocytic reaction to colorectal cancer has been associated with longer patient survival.11-15 Alternatively, infiltrative growth pattern may reflect specific tumor molecular alterations associated with aggressive tumor behavior. Indeed, studies16-20 have shown that infiltrative growth pattern is inversely associated with microsatellite instability (MSI), which has been associated with better patient survival.21-23 Because infiltrative growth pattern, lymphocytic reaction, and MSI have all been associated with prognosis, all of these variables can confound each other in survival analysis. To assess a prognostic role of tumor growth pattern independent of lymphocytic reaction and tumor molecular features, it is necessary to examine the lymphocytic reaction and tumor molecular features.
We therefore examined the prognostic role of tumor growth pattern, utilizing a database of colorectal cancer patients (N=1139) in two U.S. nationwide prospective cohort studies. Because patient characteristics and histological features as well as major tumor molecular features such as MSI, the CpG island methylator phenotype (CIMP), LINE-1 methylation, and KRAS, BRAF and PIK3CA mutations have been accumulated in our database, we were able to evaluate the effect of tumor growth pattern, independent of these potential confounders.
MATERIALS AND METHODS
Study population
We utilized the database of two U.S. nationwide prospective cohort studies, the Nurses’ Health Study (N=121,701 women followed since 1976) and the Health Professionals Follow-up Study (N=51,529 men followed since 1986).24, 25 Every two years, participants were sent follow-up questionnaires to identify newly diagnosed cancer in themselves and their first-degree relatives. When colorectal cancer was identified, study physicians reviewed medical records as well as recorded tumor location and pathological tumor-node-metastasis stage. We collected paraffin-embedded tissue blocks from hospitals where patients underwent tumor resections.25 We excluded cases preoperatively treated with radiation and/or chemotherapy. Based on the availability of tissue specimens for pathologic analyses, we included a total of 1139 stage I to IV colorectal cancer cases diagnosed up to 2006 (Table 1). Patients were observed until death or January 1, 2011, whichever came first. Deaths were ascertained by the National Death Index. The cause of death was assigned by physicians unaware of pathologic, molecular or outcome data. Written informed consent was obtained from all study subjects. This study was approved by the Human Subjects Committees at Brigham and Women’s Hospital and the Harvard School of Public Health.
Table 1.
Clinical, pathologic or molecular characteristics according to tumor growth pattern in colorectal cancer
| Tumor growth pattern |
|||||
|---|---|---|---|---|---|
| Clinical, pathologic or molecular feature |
Total N | expansile | intermediate | infiltrative | p value |
| All cases | 1139 | 372 (33%) | 610 (54%) | 157 (14%) | |
| Sex | 0.64 | ||||
| Male | 498 (44%) | 170 (46%) | 260 (43%) | 68 (43%) | |
| Female | 641 (56%) | 202 (54%) | 350 (57%) | 89 (57%) | |
| Mean age ± SD | 68.6 ± 8.9 | 69.0 ± 9.0 | 68.3 ± 8.7 | 68.7 ± 9.5 | 0.43 |
| Family history of colorectal cancer in any first degree relative |
0.74 | ||||
| (−) | 930 (82%) | 307 (83%) | 493 (81%) | 130 (83%) | |
| (+) | 209 (18%) | 65 (17%) | 117 (19%) | 27 (17%) | |
| Year of diagnosis | 0.29 | ||||
| Prior to 1998 | 562 (49%) | 173 (47%) | 314 (51%) | 75 (48%) | |
| 1998-2006 | 577 (51%) | 199 (53%) | 296 (49%) | 82 (52%) | |
| Body mass index (kg/m2) | 0.18 | ||||
| <30 | 924 (81%) | 291 (78%) | 505 (83%) | 128 (82%) | |
| ≤30 | 213 (19%) | 81 (22%) | 104 (17%) | 28 (18%) | |
| Tumor location | 0.18 | ||||
| Proximal colon (cecum to transverse) | 569 (50%) | 174 (47%) | 311 (51%) | 84 (54%) | |
| Distal colon (splenic flexure to sigmoid) |
331 (29%) | 109 (29%) | 185 (30%) | 37 (24%) | |
| Rectum | 233 (21%) | 87 (24%) | 112 (18%) | 34 (22%) | |
| Disease stage | <0.0001 | ||||
| I | 257 (23%) | 144 (39%) | 110 (18%) | 3 (1.9%) | |
| II | 348 (31%) | 111 (30%) | 208 (34%) | 29 (18%) | |
| III | 311 (27%) | 65 (17%) | 190 (31%) | 56 (36%) | |
| IV | 142 (12%) | 23 (6.2%) | 62 (10%) | 57 (36%) | |
| Unknown | 81 (7.1%) | 29 (7.8%) | 40 (6.6%) | 12 (7.6%) | |
| Tumor differentiation | 0.058 | ||||
| Well to moderate | 1025 (90%) | 339 (91%) | 553 (91%) | 133 (85%) | |
| Poor | 114 (10%) | 33 (8.9%) | 57 (9.3%) | 24 (15%) | |
| Mucinous component | 0.017 | ||||
| 0% | 674 (59%) | 202 (54%) | 380 (62%) | 92 (59%) | |
| 1-50% | 337 (30%) | 115 (31%) | 168 (28%) | 54 (34%) | |
| >50% | 128 (11%) | 55 (15%) | 62 (10%) | 11 (7.0%) | |
| Signet ring cell component | 0.0067 | ||||
| 0% | 1008 (89%) | 336 (90%) | 540 (89%) | 132 (84%) | |
| 1-50% | 112 (9.8%) | 32 (8.6%) | 66 (10%) | 17 (11%) | |
| >50% | 19 (1.7%) | 4 (1.1%) | 7 (1.2%) | 8 (5.1%) | |
| Peritumoral lymphocytic reaction | <0.0001 | ||||
| Absent/minimal | 178 (16%) | 36 (9.7%) | 95 (16%) | 47 (30%) | |
| Mild | 782 (69%) | 265 (71%) | 418 (69%) | 99 (63%) | |
| Moderate/marked | 175 (15%) | 70 (19%) | 94 (15%) | 11 (7.0%) | |
| MSI status | 0.0001 | ||||
| MSI-low/MSS | 872 (84%) | 268 (78%) | 467 (84%) | 137 (94%) | |
| MSI-high | 171 (16%) | 74 (22%) | 88 (16%) | 9 (6.2%) | |
| CIMP status | 0.58 | ||||
| CIMP-low/0 | 818 (82%) | 261 (80%) | 446 (83%) | 111 (83%) | |
| CIMP-high | 181 (18%) | 65 (20%) | 93 (17%) | 23 (17%) | |
| KRAS mutation | 0.78 | ||||
| (−) | 671 (64%) | 221 (64%) | 360 (64%) | 90 (61%) | |
| (+) | 380 (36%) | 123 (36%) | 200 (36%) | 57 (39%) | |
| BRAF mutation | 0.076 | ||||
| (−) | 890 (85%) | 291 (84%) | 484 (87%) | 115 (79%) | |
| (+) | 159 (15%) | 55 (16%) | 74 (13%) | 301 (21%) | |
| PIK3CA mutation | 0.93 | ||||
| (−) | 809 (83%) | 265 (83%) | 431 (83%) | 113 (82%) | |
| (+) | 167 (17%) | 53 (17%) | 89 (17%) | 25 (18%) | |
| LINE-1 methylation level (Mean ± SD) |
62.7 ± 9.6 | 62.7 ± 9.5 | 62.4 ± 9.7 | 63.6 ± 9.5 | 0.44 |
(%) indicates the proportion of cases with a specific clinical, pathologic or molecular feature among each expression group. CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable; SD, standard deviation.
Histopathologic evaluations
Hematoxylin and eosin stained tissue sections from all colorectal cancer cases were reviewed by a single study pathologist (S.O.) unaware of other clinical or molecular data, to eliminate the effect of interobserver variability. Tumor differentiation was categorized as well-moderate vs. poor (>50% vs. ≤50% gland formation). The type of tumor growth pattern at the tumor margin was examined at low-power magnification and categorized as expansile, intermediate, or infiltrative (Figure 1). Tumor margins were considered expansile when the invasive margin was pushing or reasonably well circumscribed.3 They were considered intermediate when large to medium-sized glands invaded and the tumor border was not distinct. They were considered infiltrative when small glands or irregular clusters or cords of cells invaded in a diffuse manner with widespread penetration of normal tissues without distinct border.9 Tumors with a small microscopic focus of an infiltrative growth pattern were considered intermediate. The presence and extent of mucinous and/or signet ring cell component were recorded. Peritumoral lymphocytic reaction was examined as previously described.13 A subset of cases (N>100) were reviewed by a second pathologist (T.M.) and concordance was as follows (all p<0.0001): 0.77 (weighted κ=0.62) for tumor growth pattern (trichotomized); 0.96 (κ=0.73) for tumor growth pattern (dichotomized as expansile-intermediate vs. infiltrative); 0.96 (κ=0.72) for tumor differentiation (dichotomized as well-moderate vs. poor); 0.85 (κ=0.70) for presence of mucinous component; 0.93 (κ=0.60) for presence of signet ring cell component; Spearman r=0.65 for peritumoral lymphocytic reaction.13 Since our current study utilized a large sample size, an agreement study on a fraction of cases was a reasonable method to assess the interobserver reproducibility of each histopathologic feature.4, 10
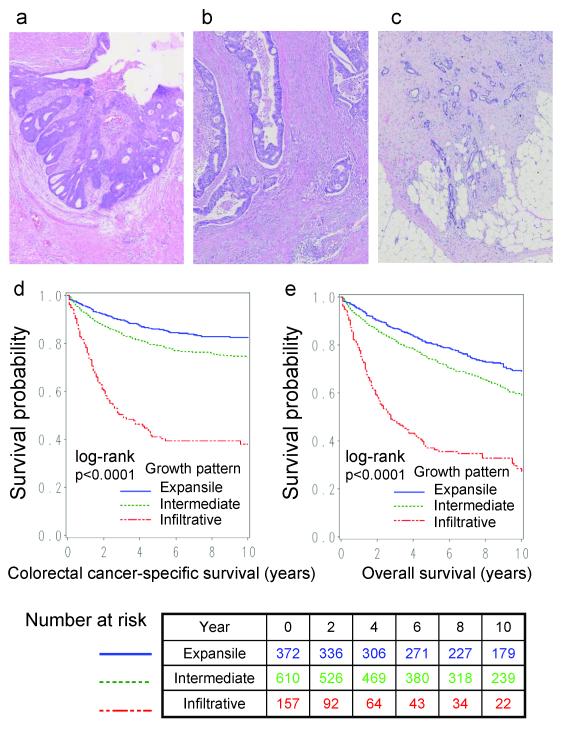
Figure 1.
(a) Colon cancer with expansile growth pattern. Hematoxylin and eosin (HE) stain. (b) Colon cancer with intermediate growth pattern. HE stain. (c) Colon cancer with infiltrative growth pattern. HE stain. (d,e) Kaplan-Meier curves for colorectal cancer-specific survival (d) and overall survival (e).
DNA extraction, Pyrosequencing of KRAS, BRAF and PIK3CA, and microsatellite instability (MSI) analysis
DNA was extracted from paraffin-embedded tissue, and we performed PCR and Pyrosequencing targeted for KRAS (codons 12 and 13),26 BRAF (codon 600)27 and PIK3CA (exons 9 and 20).28 MSI analysis was performed using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67 and D18S487).29 MSI-high was defined as instability in ≥30% of the markers, and MSI-low/microsatellite stability (MSS) as instability in <30% of the markers.
Methylation analyses for CpG islands and LINE-1
Using validated bisulfite DNA treatment and real-time PCR (MethyLight), we quantified DNA methylation in eight CIMP-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1).30-32 CIMP-high was defined as the presence of ≥6/8 methylated promoters, and CIMP-low/0 as 0/8-5/8 methylated promoters, according to the previously established criteria.29 In order to accurately quantify relatively high LINE-1 methylation levels, we used Pyrosequencing as previously described.33, 34
Statistical analysis
All statistical analyses were performed by SAS software (Version 9.1, SAS Institute, Cary, NC). All p values were two-sided. For categorical data, the chi-square test was performed. P values were calculated by ANOVA (analysis of variance) for age and LINE-1 methylation level. The Cicchetti-Allison weight was used for calculating the weighted κ agreement coefficients for tumor growth pattern (trichotomized variable). Kaplan-Meier method and log-rank test were used for survival analyses. For analyses of colorectal cancer-specific mortality, deaths as a result of other causes were censored. To control for confounding, we used multivariate Cox proportional hazards regression models. A multivariate model initially included sex, age at diagnosis (continuous), year of diagnosis (continuous), body mass index (BMI; <30 vs. ≥30 kg/m2), family history of colorectal cancer in any first-degree relative (present vs. absent), tumor location (proximal vs. distal), tumor differentiation (well-moderate vs. poor), mucinous component (present or absent), signet ring cells (present or absent), peritumoral lymphocytic reaction (absent/minimal vs. present), MSI (high vs. low/MSS), CIMP (high vs. low/0), LINE-1 methylation (continuous), KRAS, BRAF, and PIK3CA. Disease stage (I, II, III, IV, unknown) was used as a stratifying variable using the “strata” option in the SAS “proc phreg” command, to minimize residual confounding and overfitting. A backward elimination was performed with p=0.20 as a threshold to avoid overfitting. For cases with missing information in any of the covariates [BMI (0.2% missing), tumor location (0.5%), peritumoral lymphocytic reaction (0.4%), MSI (8.4%), CIMP (12%), KRAS (7.7%), BRAF (7.9%), and PIK3CA (14%)], we included those cases in a majority category of a given covariate to avoid overfitting. We confirmed that excluding cases with missing information in any of the covariates did not substantially alter results (data not shown). The proportionality of hazards assumption was satisfied by evaluating time-dependent variables, which were the cross-product of the growth pattern variable and survival time (p>0.10). An interaction was assessed by the Wald test on the cross product of the growth pattern variable and another variable of interest (without data-missing cases) in a multivariate Cox model. To assess an interaction of tumor growth pattern and stage, we dichotomized disease stage (I-III vs. IV).
Additionally, multivariate logistic regression analysis was performed to assess relations with infiltrative tumor growth (as a binary outcome variable). Odds ratio (OR) was adjusted for sex, age (continuous), year of diagnosis (continuous), BMI (<30 vs. ≥30 kg/m2), family history of colorectal cancer in any first degree relative (present vs. absent), tumor location (proximal vs. distal), tumor differentiation (well-moderate vs. poor), mucinous component (present or absent), signet ring cells (present or absent), peritumoral lymphocytic reaction (absent/minimal vs. present), MSI (high vs. low/MSS), CIMP (high vs. low/0), LINE-1 methylation (continuous), KRAS, BRAF, and PIK3CA. A backward elimination with a threshold of p=0.20 was used to select variables in the final model.
RESULTS
Tumor growth pattern in colorectal cancer
Among 1139 colorectal cancers, 372 tumors (33%) exhibited an expansile pattern, 610 tumors (54%) showed an intermediate pattern, and 157 tumors (14%) showed an infiltrative pattern (Figure 1). Table 1 shows the relations between tumor growth pattern and various clinical, pathologic and molecular features. Infiltrative growth pattern was associated with advanced stage (p<0.0001), and inversely associated with peritumoral lymphocytic reaction (p<0.0001) and MSI-high (p<0.0001).
In multivariate logistic regression analysis, infiltrative growth pattern was inversely associated with MSI-high [multivariate odds ratio (OR)=0.15; 95% CI, 0.07-0.34; p<0.0001] and peritumoral lymphocytic reaction (multivariate OR=0.36; 95% CI, 0.24-0.54; p<0.0001), and positively with BRAF mutation (multivariate OR=2.89; 95% CI, 1.68-4.98; p=0.0001) (Table 2).
Table 2.
Multivariate logistic regression analysis to assess relations with infiltrative growth pattern in colorectal cancer
| Variables in the final model for tumor growth pattern as a binary outcome variable (infiltrative vs. expansile-intermediate) |
Multivariate OR (95% CI) |
P value |
|---|---|---|
| MSI-high (vs. MSI-low/MSS) | 0.15 (0.07-0.34) | <0.0001 |
| Peritumoral lymphocytic reaction (present vs. absent) | 0.36 (0.24-0.54) | <0.0001 |
| BRAF mutation (vs. wild-type) | 2.89 (1.68-4.98) | 0.0001 |
| Poor differentiation (vs. well-moderate) | 2.27 (1.31-3.92) | 0.0035 |
| LINE-1 hypomethylation (for a 30% decrease) | 0.55 (0.30-1.00) | 0.049 |
| Year of diagnosis (for a 10-year increase) | 0.75 (0.55-1.02) | 0.065 |
| KRAS mutation (vs. wild-type) | 1.31 (0.90-1.92) | 0.16 |
Multivariate logistic regression analysis assessing the relationship with infiltrative growth pattern (as an outcome variable) initially included age, sex, year of diagnosis, body mass index, tumor location, family history of colorectal cancer, microsatellite instability, CpG island methylator phenotype, LINE-1 methylation, KRAS, BRAF, PIK3CA, tumor differentiation, mucinous component, signet ring cell component, and peritumoral lymphocytic reaction. A backward elimination with a threshold of p=0.20 was used to select variables in the final models. Because of multiple hypothesis testing, a p value for significance was adjusted by Bonferroni correction to p=0.0029.
CI, confidence interval; MSI, microsatellite instability; MSS, microsatellite stable; OR, odds ratio.
Tumor growth pattern and patient survival in colorectal cancer
Among the 1139 patients, there were 528 deaths, including 308 colorectal cancer-specific deaths, during a median follow-up of 137 months (interquartile range, 94-189 months) for those who were censored. In Kaplan-Meier analysis, infiltrative growth pattern was significantly associated with shorter colorectal cancer-specific and overall survival (both log rank p<0.0001) (Figure 1).
Infiltrative growth pattern was significantly associated with shorter colorectal cancer-specific and overall survival in univariate Cox regression analysis and in the multivariate Cox model adjusting for clinical, pathologic and molecular features (Table 3). Adjusted HR for infiltrative growth pattern (expansile pattern as a referent) was 1.74 (95% CI, 1.22-2.47) for colorectal cancer-specific survival, and 1.78 (95% CI, 1.33-2.39) for overall survival. The attenuation in the effect of infiltrative growth pattern in the multivariate analysis was principally the result of adjusting for disease stage. When we simply adjusted for disease stage, HR for infiltrative growth pattern was 1.95 (95% CI, 1.38-2.75) for colorectal cancer-specific survival, and 1.91 (95% CI, 1.44-2.54) for overall survival (Table 3). No other major confounder was present. Patients with infiltrative growth pattern experienced shorter survival compared to those with intermediate growth pattern [univariate HR=3.54 (95% CI, 2.73-4.59), multivariate HR=1.42 (95% CI, 1.06-1.91) for colorectal cancer-specific survival, and univariate HR=2.57 (95% CI, 2.06-3.21), multivariate HR=1.39 (95% CI, 1.08-1.80) for overall survival].
Table 3.
Tumor growth pattern in colorectal cancer and patient mortality
| Colorectal cancer-specific mortality |
Overall mortality |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor growth pattern |
Total N (%) |
No. of events |
Univariate HR (95% CI) |
Stage- stratified HR (95% CI) |
Multivariate stage- stratified HR (95% CI) |
No. of events |
Univariate HR (95% CI) |
Stage- stratified HR (95% CI) |
Multivariate stage- stratified HR (95% CI) |
| Expansile | 372 (33%) | 64 | 1 (referent) | 1 (referent) | 1 (referent) | 134 | 1 (referent) | 1 (referent) | 1 (referent) |
| Intermediate | 610 (54%) | 149 | 1.51 (1.13-2.03) |
1.22 (0.90-1.64) |
1.23 (0.91-1.67) |
282 | 1.41 (1.14-1.73) |
1.27 (1.03-1.58) |
1.30 (1.04-1.62) |
| Infiltrative | 157 (14%) | 95 | 5.37 (3.90-7.40) |
1.95 (1.38-2.75) |
1.74 (1.22-2.47) |
112 | 3.64 (2.82-4.69) |
1.91 (1.44-2.54) |
1.78 (1.33-2.39) |
| p for trend | <0.0001 | 0.0002 | 0.0021 | <0.0001 | <0.0001 | 0.0002 | |||
The multivariate, stage-matched (stratified) Cox regression model initially included the tumor growth pattern variable (expansile, intermediate, or infiltrative), age, sex, year of diagnosis, body mass index, tumor location, tumor differentiation, family history of colorectal cancer in any first degree relative, microsatellite instability, CpG island methylator phenotype, LINE-1 methylation, KRAS, BRAF, PIK3CA, tumor differentiation, mucinous component, signet ring cell component, and peritumoral lymphocytic reaction. A backward elimination with threshold of p=0.20 was used to select variables in the final models.
CI, confidence interval; HR, hazard ratio.
Stage-specific analysis of tumor growth pattern and survival
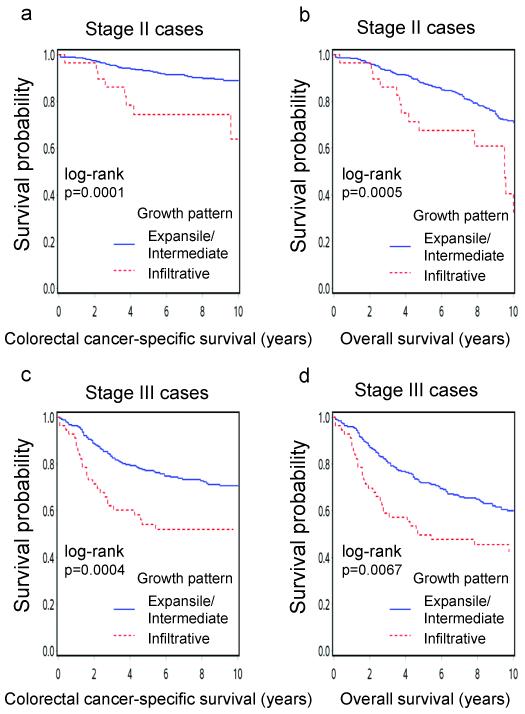
To evaluate a prognostic effect of tumor growth pattern in each disease stage, we examined stage-specific HR for tumors with infiltrative growth pattern compared with those with expansile or intermediate growth pattern (Table 4). The significant prognostic association of infiltrative growth pattern was confined to stage I-III patients. Among stage IV colorectal cancer patients, infiltrative growth pattern was not associated with survival (Pinteraction with stage=0.0001 for colorectal cancer-specific survival). The adverse effect of infiltrative growth pattern on survival in stage II or III patients was also observed in Kaplan-Meier analyses (Figure 2).
Table 4.
Stage-specific analysis of patient mortality in colorectal cancer with infiltrative growth pattern compared with colorectal cancer with expansile/intermediate growth pattern
| Colorectal cancer-specific mortality in tumors with infiltrative growth pattern (vs. tumors with expansile- intermediate growth pattern as a referent) |
Overall mortality in tumors with infiltrative growth pattern (vs. tumors with expansile-intermediate growth pattern as a referent) |
|||||
|---|---|---|---|---|---|---|
| Number of deaths/cases (expansile-intermediate vs. infiltrative ) |
Univariate HR (95% CI) |
Multivariate HR (95% CI) |
Number of deaths/cases (expansile-intermediate vs. infiltrative ) |
Univariate HR (95% CI) |
Multivariate HR (95% CI) |
|
| Stage I | 20/254 vs. 1/3 | 5.02 (0.67-37.6) |
7.70 (1.01-58.6) |
71/254 vs. 2/3 | 3.67 (0.89-15.1) |
5.24 (1.26-21.8) |
| Stage II | 33/319 vs. 9/29 | 3.56 (1.70-7.45) |
2.81 (1.33-5.94) |
118/319 vs. 16/29 | 2.16 (1.28-3.66) |
1.97 (1.16-3.35) |
| Stage III | 70/255 vs. 27/56 | 2.20 (1.41-3.43) |
1.82 (1.16-2.86) |
116/255 vs. 33/56 | 1.69 (1.15-2.49) |
1.63 (1.10-2.42) |
| Stage IV | 75/85 vs. 51/57 | 1.09 (0.76-1.57) |
0.99 (0.68-1.45) |
76/85 vs. 51/57 | 1.09 (0.76-1.56) |
0.94 (0.65-1.37) |
| P for interaction (tumor growth pattern and stage) |
<0.0001 | 0.0001 | 0.0022 | 0.0007 | ||
The multivariate Cox regression model included the same set of covariates selected as in Table 3.
CI, confidence interval; HR, hazard ratio.
Figure 2.
Kaplan-Meier curves for colorectal cancer-specific survival (a,c) and overall survival (b,d) in stage II (a,b) and stage III (c,d) patients.
Stratified analysis of tumor growth pattern and mortality
We examined whether the prognostic association of tumor growth pattern was modified by any of the other variables including sex, age, year of diagnosis, BMI, family history of colorectal cancer, tumor location, tumor differentiation, mucinous or signet ring cell component, peritumoral lymphocytic reaction, MSI, CIMP, LINE-1 methylation, BRAF, KRAS, and PIK3CA. We did not observe a significant modifying effect by any of the variables (all Pinteraction>0.05). Notably, the effect of tumor growth pattern did not significantly differ between the two cohort studies (Pinteraction=0.83).
DISCUSSION
We examined the prognostic significance of tumor growth pattern in a population of stage I to IV colorectal cancer patients who were concurrently assessed for other clinical, pathologic and molecular predictors of patient outcome. Advancing tumor margin is considered to be a tumor area which manifests tumor aggressiveness.1 We found that an infiltrative growth pattern was associated with worse prognosis among stage I-III colorectal cancer patients, independent of other clinical, pathologic, and molecular characteristics including MSI, CIMP, LINE-1 methylation, and KRAS, BRAF, and PIK3CA mutations. Our results support the role of tumor growth pattern as an independent prognostic factor among stage I-III colorectal cancer patients.
Tumor growth pattern can be assessed at low magnification with ease on routine histopathologic examination of resected colorectal cancer, and an evaluation of this feature can be implemented in clinical practice. Tumor growth pattern has generated considerable interest as an additional prognostic factor.35 An evaluation of tumor growth pattern has also been integrated into standard practice in Japan.36 Previous studies4, 10, 37 as well as the present study demonstrated that tumor growth pattern can be reliably assessed with acceptable levels of intraobserver and/or interobserver agreement. Moreover, assessment is not significantly affected by the site of sampling of the invasive tumor margin.37 Beyond the previous reports,2-10 the present study has shown that this parameter is a reliable prognostic factor independent of tumor molecular variables by multivariate analysis.
Examining prognostic and predictive factors is important in cancer research.38-42 Infiltrative growth pattern has been associated with shorter survival in colorectal cancer.2-10 However, the mechanism underlying the survival disadvantage associated with infiltrative growth pattern remains speculative. Infiltrative growth pattern may be an indicator of less host immune and inflammatory responses to tumor cells,1, 4, 5 leading to shorter patient survival.11-15 Alternatively, infiltrative growth pattern may reflect specific tumor molecular alterations associated with aggressive tumor behavior. Indeed, studies16-20 have shown that infiltrative growth pattern is inversely associated with MSI, which has been associated with better patient survival.21-23 Recent studies have further shown that MSI-high in colorectal cancer is associated with CIMP, BRAF mutation,43 and high LINE-1 methylation level,33 and all of these factors (MSI, CIMP, BRAF mutation and LINE-1 methylation) have been independently related with prognosis of colorectal cancer patients.29, 34, 44 Therefore, numerous pathologic and molecular features (lymphocytic reactions, MSI, CIMP, BRAF mutation, and LINE-1 methylation) could account for the adverse effect of infiltrative growth pattern. However, none of the previous studies on tumor growth pattern and patient survival2-10 has comprehensively examined the aforementioned molecular features in colorectal cancer beyond MSI. In our analysis, the disadvantage associated with infiltrative growth pattern remained significant after adjusting for these various pathologic and molecular features.
The frequency of infiltrative growth pattern greatly varies ranging from 17%6 to 80%45 among the previous studies that used the dichotomous classification (i.e. expansile or infiltrative). This fact suggests that there are many cases that may fall into a gray zone between expansile and infiltrative growth patterns. Therefore, to examine the effect of unequivocal infiltrative growth pattern, we used the trichotomous categorization (i.e. expansile, intermediate or infiltrative), which is similar to the Japanese classification.36 It is likely that tumors with intermediate growth pattern are included in the infiltrative category in the classical dichotomous classification. Results of the survival analysis showed that patients with infiltrative growth pattern experienced shorter survival compared to those with expansile or intermediate growth pattern, highlighting the adverse prognostic significance of unequivocal infiltrative growth pattern. In addition, inverse associations between unequivocal infiltrative growth pattern and peritumoral lymphocytic reaction or MSI were also evident (Table 1). Thus, the trichotomous categorization (expansile, intermediate or infiltrative) may be a reasonable classification when one evaluates the tumor growth pattern.
Interestingly, we found that an infiltrative growth pattern was inversely associated with MSI-high, and positively with BRAF mutation in multivariate analysis. While MSI-high is associated with better patient survival,21-23 BRAF mutation is associated with worse patient survival.44, 46, 47 Our results suggest that an infiltrative growth pattern may reflect specific molecular alterations associated with aggressive tumor behavior. Considering that the effects of adjuvant therapy may differ according to tumor molecular features,48-50 it may be interesting in future studies to determine the predictive role of tumor growth pattern for response to adjuvant therapies in colorectal cancer, since tumor growth pattern can be easily assessed on routine histopathologic examination.
There are limitations in this study. For example, data on cancer treatment were limited. Nonetheless, it is unlikely that chemotherapy use substantially differed according to tumor growth pattern, since such data were not typically used for treatment decision making. In addition, our multivariate survival analysis finely adjusted for disease stage (I, II, III, IV, unknown), on which treatment decision making was mostly based. As another limitation, beyond cause of mortality, data on cancer recurrences were not available in these cohorts. Nonetheless, colorectal cancer-specific survival is a reasonable surrogate of colorectal cancer-specific outcome.
There are advantages in using the database of the two independent prospective cohort studies, the Nurses’ Health Study and the Health Professionals Follow-up Study, to examine prognostic significance of tumor growth pattern and its interactions with tumoral and host factors. Anthropometric measurements, family history, other clinical information, pathologic and tumor staging data, and tumor molecular features were prospectively collected blinded to patient outcome. Cohort participants who developed cancer were treated at hospitals throughout the U.S., and thus more representative of colorectal cancers in the general U.S. population than patients in one to a few academic hospitals. There were no demographic difference between cases with tumor tissue analyzed and those without tumor tissue analyzed.24 Tumor growth pattern of colorectal cancer was examined by the single pathologist, and a subset of cases were reexamined by a second pathologist for the agreement study. Finally, our rich tumor database enabled us to simultaneously assess pathologic and tumor molecular features and control for confounding by a number of tumor molecular alterations or histological features. None of the previous studies on tumor growth pattern and patient outcome has examined as many molecular variables as we did in this study.
In summary, our large cohort study suggests that an infiltrative growth pattern is associated with shorter survival of colorectal cancer patients, independent of other clinical, pathologic, and tumor molecular characteristics. Our data suggest a possible role of tumor-stromal interaction as an independent prognostic determinant of behavior of colorectal cancer cells. Future studies are needed to confirm these results as well as to elucidate exact mechanisms by which tumor-stromal interaction affects behavior of colorectal cancer.
Acknowledgments
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study, for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Funding
This work was supported by U.S. National Institute of Health (NIH) grants P01 CA87969 (to S. Hankinson), P01 CA55075 (to W. Willett), P50 CA127003 (to C.S.F.), and R01 CA151993 (to S.O.) and by grants from the Bennett Family Fund and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. T.M. was supported by a fellowship grant from the Japan Society for Promotion of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
(TM, AK and ZRQ contributed equally)
No conflict of interest is present.
References
- 1.Zlobec I, Lugli A. Invasive front of colorectal cancer: dynamic interface of pro-/anti-tumor factors. World J Gastroenterol. 2009;15:5898–906. doi: 10.3748/wjg.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jass JR, Atkin WS, Cuzick J, Bussey HJ, Morson BC, Northover JM, Todd IP. The grading of rectal cancer: historical perspectives and a multivariate analysis of 447 cases. Histopathology. 1986;10:437–59. doi: 10.1111/j.1365-2559.1986.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR, Love SB, Northover JM. A new prognostic classification of rectal cancer. Lancet. 1987;1:1303–6. doi: 10.1016/s0140-6736(87)90552-6. [DOI] [PubMed] [Google Scholar]
- 4.Halvorsen TB, Seim E. Association between invasiveness, inflammatory reaction, desmoplasia and survival in colorectal cancer. J Clin Pathol. 1989;42:162–6. doi: 10.1136/jcp.42.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepherd NA, Saraga EP, Love SB, Jass JR. Prognostic factors in colonic cancer. Histopathology. 1989;14:613–20. doi: 10.1111/j.1365-2559.1989.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 6.Kubota Y, Petras RE, Easley KA, Bauer TW, Tubbs RR, Fazio VW. Ki-67-determined growth fraction versus standard staging and grading parameters in colorectal carcinoma. A multivariate analysis. Cancer. 1992;70:2602–9. doi: 10.1002/1097-0142(19921201)70:11<2602::aid-cncr2820701106>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 7.Roncucci L, Fante R, Losi L, et al. Survival for colon and rectal cancer in a population-based cancer registry. Eur J Cancer. 1996;32A:295–302. doi: 10.1016/0959-8049(95)00532-3. [DOI] [PubMed] [Google Scholar]
- 8.Cianchi F, Messerini L, Palomba A, et al. Character of the invasive margin in colorectal cancer: does it improve prognostic information of Dukes staging? Dis Colon Rectum. 1997;40:1170–5. doi: 10.1007/BF02055162. discussion 5-6. [DOI] [PubMed] [Google Scholar]
- 9.Cianchi F, Messerini L, Comin CE, Boddi V, Perna F, Perigli G, Cortesini C. Pathologic determinants of survival after resection of T3N0 (Stage IIA) colorectal cancer: proposal for a new prognostic model. Dis Colon Rectum. 2007;50:1332–41. doi: 10.1007/s10350-007-0222-9. [DOI] [PubMed] [Google Scholar]
- 10.Zlobec I, Baker K, Minoo P, Hayashi S, Terracciano L, Lugli A. Tumor border configuration added to TNM staging better stratifies stage II colorectal cancer patients into prognostic subgroups. Cancer. 2009;115:4021–9. doi: 10.1002/cncr.24450. [DOI] [PubMed] [Google Scholar]
- 11.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 12.Pages F, Kirilovsky A, Mlecnik B, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. 2009;27:5944–51. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- 13.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–20. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–66. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mlecnik B, Tosolini M, Kirilovsky A, et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29:610–8. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 16.Greenson JK, Bonner JD, Ben-Yzhak O, et al. Phenotype of microsatellite unstable colorectal carcinomas: Well-differentiated and focally mucinous tumors and the absence of dirty necrosis correlate with microsatellite instability. Am J Surg Pathol. 2003;27:563–70. doi: 10.1097/00000478-200305000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Colomer A, Erill N, Vidal A, et al. A novel logistic model based on clinicopathological features predicts microsatellite instability in colorectal carcinomas. Diagn Mol Pathol. 2005;14:213–23. doi: 10.1097/01.pas.0000177800.65959.48. [DOI] [PubMed] [Google Scholar]
- 18.Halvarsson B, Anderson H, Domanska K, Lindmark G, Nilbert M. Clinicopathologic factors identify sporadic mismatch repair-defective colon cancers. Am J Clin Pathol. 2008;129:238–44. doi: 10.1309/0PP5GDRTXUDVKAWJ. [DOI] [PubMed] [Google Scholar]
- 19.Greenson JK, Huang SC, Herron C, et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am J Surg Pathol. 2009;33:126–33. doi: 10.1097/PAS.0b013e31817ec2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roman R, Verdu M, Calvo M, et al. Microsatellite instability of the colorectal carcinoma can be predicted in the conventional pathologic examination. A prospective multicentric study and the statistical analysis of 615 cases consolidate our previously proposed logistic regression model. Virchows Arch. 2010;456:533–41. doi: 10.1007/s00428-010-0896-6. [DOI] [PubMed] [Google Scholar]
- 21.Gryfe R, Kim H, Hsieh ET, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 22.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 23.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87. e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–42. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 25.Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–94. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–8. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–84. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 33.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–73. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–8. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Compton C, Fenoglio-Preiser CM, Pettigrew N, Fielding LP, American Joint Committee on Cancer Prognostic Factors Consensus Conference: Colorectal Working Group Cancer; 2000; pp. 1739–57. [DOI] [PubMed] [Google Scholar]
- 36.Japanese Society for Cancer of the Colon and Rectum . Japanese Classification of Colorectal Carcinoma. 2nd ed Kanehara; Tokyo: 2009. [Google Scholar]
- 37.Dundas SA, Laing RW, O’Cathain A, Seddon I, Slater DN, Stephenson TJ, Underwood JC. Feasibility of new prognostic classification for rectal cancer. J Clin Pathol. 1988;41:1273–6. doi: 10.1136/jcp.41.12.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu D, Li Y, Wang W, et al. High level of Notch1 protein is associated with poor overall survival in colorectal cancer. Ann Surg Oncol. 2010;17:1337–42. doi: 10.1245/s10434-009-0893-7. [DOI] [PubMed] [Google Scholar]
- 39.Huh JW, Kim HR, Kim YJ. Prognostic value of perineural invasion in patients with stage II colorectal cancer. Ann Surg Oncol. 2010;17:2066–72. doi: 10.1245/s10434-010-0982-7. [DOI] [PubMed] [Google Scholar]
- 40.Muratore A, Ribero D, Zimmitti G, Mellano A, Langella S, Capussotti L. Resection margin and recurrence-free survival after liver resection of colorectal metastases. Ann Surg Oncol. 2010;17:1324–9. doi: 10.1245/s10434-009-0770-4. [DOI] [PubMed] [Google Scholar]
- 41.Kang H, Min BS, Lee KY, Kim NK, Kim SN, Choi J, Kim H. Loss of E-cadherin and MUC2 expressions correlated with poor survival in patients with stages II and III colorectal carcinoma. Ann Surg Oncol. 2011;18:711–9. doi: 10.1245/s10434-010-1338-z. [DOI] [PubMed] [Google Scholar]
- 42.Li D, Peng X, Yan D, Tang H, Huang F, Yang Y, Peng Z. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Ann Surg Oncol. 2011;18:2074–83. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 43.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–9. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 45.Harrison JC, Dean PJ, el-Zeky F, Vander Zwaag R. From Dukes through Jass: pathological prognostic indicators in rectal cancer. Hum Pathol. 1994;25:498–505. doi: 10.1016/0046-8177(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 46.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, van den Brule AJ. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 47.Yokota T, Ura T, Shibata N, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–62. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 50.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]