Abstract
Micro RNAs (miRNAs), small and labile ~22 nucleotide-sized fragments of single stranded RNA, are important regulators of messenger (mRNA) complexity and in shaping the transcriptome of a cell. In this communication, we utilized amyloid beta 42 (Aβ42) peptides and interleukin-1beta (IL1β) as a combinatorial, physiologically-relevant stress to induce miRNAs in human primary neural (HNG) cells (a co-culture of neurons and astroglia). Specific miRNA up-regulation was monitored using miRNA arrays, Northern micro-dot blots and RT-PCR. Selective NF-κB translocation and DNA binding inhibitors including the chelator and anti-oxidant pyrollidine dithiocarbamate (PDTC) and the polyphenolic resveratrol analog CAY10512 (trans-3,5,4′-trihydroxystilbene) indicated the NF-κB sensitivity of several brain miRNAs, including miRNA-9, miRNA-125b and miRNA-146a. The inducible miRNA-125b and miRNA-146a, and their verified mRNA targets, including 15-lipoxygenase (15-LOX), synapsin-2 (SYN-2), complement factor H (CFH) and tetraspanin-12 (TSPAN12), suggests complex and highly interactive roles for NF-κB, miRNA-125b and miRNA-146a. These data further indicate that just two NF-κB-mediated miRNAs have tremendous potential to contribute to the regulation of neurotrophic support, synaptogenesis, neuroinflammation, innate immune signaling and amyloidogenesis in stressed primary neural cells of the human brain.
Keywords: 15-lipoxygenase (15-LOX), Alzheimer s disease (AD), amyloidogenesis, complement factor H (CFH), evolution, micro RNA 125b (miRNA-125b), miRNA-146a, synapsin-2 (SYN-2), tetraspanin-12 (TSPAN12)
Introduction
Micro RNAs (miRNAs) are ~22 nucleotide-sized fragments of endogenously expressed single stranded RNA that are important regulators of messenger RNA (mRNA) speciation and complexity. In their typical actions miRNAs regulate gene expression through imperfect base-pairing with the 3′ un-translated region (3′-UTR) of target mRNAs, and depending on sequence complementarity within an RNA-induced silencing complex (RISC), results in either reduction or inhibition in the translational efficiency of the target mRNA (Ambros 2004; Taft et al., 2010). It is generally accepted that up-regulated mammalian miRNAs predominantly act to decrease their target mRNA levels (Guo et al., 2010). While the potential contribution of small RNA to brain genetic function has been known for at least 20 years (Lukiw et al., 1992), more recently there been an explosion into molecular-genetic research involving the neurobiological function of these non-coding RNAs in brain development, injury, aging, health and disease (Perron and Provost, 2009; Tsitsiou and Lindsay 2009; Taft et al., 2010; Lukiw et al., 2010; Madathil et al., 2011).
Pathogenically up-regulated miRNAs can be considered an epigenetic mechanism to down-regulate specific mRNAs and their expression, and up-regulated miRNA in neurodegenerative disorders such as Alzheimer s disease (AD) may help explain the large number of brain gene messages observed to be progressively down-regulated in AD affected anatomical regions (Loring et al., 2001; Colangelo et al., 2002; Lukiw et al., 2005). Interestingly, bioinformatics and sequence analysis indicates that a 22 nucleotide single stranded RNA composed of 4 different ribonucleotides can have over 1013 possible sequence combinations, so the fact that there typically only ~103 miRNAs in any single cell type suggests a very high developmental and evolutionary selection pressure to utilize only specific miRNA oligonucleotide sequences that will yield biologically useful miRNA-mRNA interactions. Further, miRNAs are highly developmental stage-, tissue- and cell-specific, even in adjacent cell types, and in human brain cells high abundance miRNAs number probably less than 102 individual species (Lukiw and Pogue, 2007; Burmistrova et al., 2007; Yuva-Aydemit et al., 2011; unpublished observations). The small size of miRNAs and recent identification of miRNA-protective protein and miRNA-containing vesicles suggests that miRNAs may be a novel means for paracrine and related forms of inter-cellular and inter-tissue communication (Wang et al., 2010; Arroyo et al., 2011). The expression of a cell s miRNA repertoire is regulated by multiple transcription factors, are transcribed as pre-miRNAs, and are not only under the transcriptional control of DNA binding proteins, transcription factors and RNAPII and RNAPIII enzymes but further by miRNA-modifying enzymes in the nucleus and cytoplasm that include DGCR8, Exportin 5, Drosha, Dicer, Argonaute and others (Perron and Provost, 2009; Guo et al., 2010). As many human neurodegenerative brain conditions such as AD appear to be associated with a disorder in the innate immune and inflammatory response, immune- and stress-induced transcription factors such as NF-κB have been shown to play determinant roles in the regulation of stress-related miRNAs, and their mRNA targets involved in the innate immune and inflammatory response.
In these studies we used a cocktail of amyloid beta 42 peptides + interleukin 1β (Aβ42+IL1β) as a AD-relevant pathogenic stressor to induce NF-κ B in human primary neural (HNG) cells (a co-culture of neurons and astroglia), and specific miRNA up-regulation were analyzed using miRNA arrays, Northern micro-dot blots and RT-PCR. Both Aβ42 peptides and the pro-inflammatory IL-1β, as well as NF-κB, have been shown to be significantly up-regulated in AD-affected brain regions (Lukiw and Bazan, 1998; Lukiw et al., 2005; Lee et al., 2010; Zhao et al., 2011; Eikelenboom et al., 2011). As required, selective NF-κB translocation and DNA binding inhibitors including the chelator and anti-oxidant pyrollidine dithiocarbamate (PDTC; pyrrolidine dithiocarbamic acid) and the polyphenolic resveratrol analog CAY10512 (trans-3,5,4′-trihydroxystilbene) indicated the NF-κB sensitivity of miRNA-9, miRNA-125b and miRNA-146a up-regulation. We provide further evidence that miRNA-125b and miRNA-146a, and several of their verified mRNA targets including complement factor H (CFH; also known as FH, CFH, HUS, β-1H), the synaptic vesicle-associated phosphoprotein synapsin-2 (SYN-2; also known as SYNII), tetraspanin-12 (TSPAN12; also known as NET-2 or transmembrane 4 superfamily member 12) and 15-lipoxygenase (15-LOX) provides a complex interactive regulatory role for NF-κB and specific miRNA species in innate and immune signaling, synaptogenesis, amyloidogenesis and neurotrophic support in stressed human brain cells.
Material and methods
Procedures for the primary culture of human neuronal-glial (HNG) cells (also referred to as human neural cells; Figure 1A) and their staining with glial-specific glial fibrillary acidic protein (GFAP), neuron-specific beta-tubulin III (βTUBIII), and Hoescht 33258 nuclear staining have been previously described in detail (Lukiw et al. 1998; Hill et al., 2001; Higaki et al., 2002; Pogue and Lukiw 2004; Cui et al., 2004, Lukiw et al., 2005; Alexandrov et al., 2005; Lukiw et al., 2008; Sethi and Lukiw, 2009; Hill et al., 2009; Pogue et al., 2009; Cui et al., 2010; Zhao et al., 2011; Li et al., 2011). Note that human primary neuronal cells do not grow well in the absence of glia; after 1 week of culture brain cells to be stressed received at each change of medium Aβ42 peptide (5 μM; Sigma-Aldrich) plus human recombinant interleukin-1β (IL-1β; 10 nM; I4019, Sigma-Aldrich Chemical, St. Louis, MO). Aβ42 peptides were prepared using the hexafluoroisopropanol (HFIP) evaporation-dimethyl sulfoxide-solubilization method as previously described (Lukiw et al., 2005; Zhao et al., 2011). Control primary cells received cell culture grade human serum albumin (HSA; Sigma-Aldrich). As required, HNG cells were treated with either the metal chelator, anti-oxidant and NF-κB translocation inhibitor pyrollidine dithiocarbamate (PDTC; pyrrolidine dithiocarbamic acid; P8765; Sigma, St Louis, MO) or the antioxidant CAY10512 (10009536; Cayman Chemical, Ann Arbor, MI) at 10 or 5 uM as previously described (Figure 1B,C) (Lukiw et al., 2008; Cui et al., 2010). After Aβ42 peptide+IL-1β addition cells were cultured for 1 additional week after which total RNA and protein fractions were analyzed using equivalent numbers of brain cells (Lukiw et al., 2005; Zhao et al., 2011). A guanidine isothiocyanate- and silica gel-based membrane total RNA purification system and miRNA isolation kit (PureLink™ Invitrogen, Carlsbad, CA) were used to isolate total RNA; total RNA concentrations were quantified using RNA 6000 Nano LabChips and a 2100 Bioanalyzer (Caliper Technologies, Mountainview, CA; Agilent Technologies, Palo Alto, CA). Specific small RNAs including 5S RNA, miRNA-9, miRNA-125b, miRNA-146a and miRNA-183 were initially analyzed and quantified using miRNA arrays (N=3 control, N=3 stressed; LC Sciences, Houston TX; for raw data see http://www.medschool.lsuhsc.edu/neuroscience/faculty_detail.aspx?name=lukiw_walter) or Northern dot blots as previously described (Lukiw et al., 2005; Cui et al., 2010; Zhao et al., 2011). For real-time quantitative PCR (qPCR) analysis of miRNAs, total RNA fractions were prepared from HNG cells using an miRNeasy Mini Kit (Qiagen, Valencia, CA), and qPCR analysis was performed using individual TaqMan miRNA assays using a TaqMan MiRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix, and an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems; Carlsbad, California) as described by our lab (Lukiw et al., 2005; Cui et al., 2010; Zhao et al., 2011). Altered small RNA levels of interest were further verified using a quantitative Northern dot blot focusing assay that utilizes a T4 PNK kinase radiolabel system employing [α-32P]-dATP (6000 Ci/m mol; Invitrogen, Carlsbad, CA) that significantly concentrates small RNA and miRNA signals (Lukiw et al., 1998; Lukiw et al., 2005; Cui et al., 2010). All 3 techniques yielded quantitatively similar results; Northern dot blot and qPCR data for 5SRNA and miRNA-9, miRNA-125b, miRNA-146a and miRNA-183 are shown in Figure 2. 5S RNA and miRNA-183 were used as endogenous controls as their levels have been previously shown not to change under control or stressed physiological conditions in any human brain cell type studied (Lukiw et al., 2005; Cui et al., 2010; Zhao et al., 2011). Gel shift assay for NF-κB binding activity, and analysis of the expression of 15-LOX, SYN-2, CFH, TSPAN12 and β-actin (as an internal control) at the level of mRNA and protein were performed as previously described (Lukiw and Bazan, 1998; Lukiw et al., 2005; Cui et al., 2010; Zhao et al., 2011). TSPAN12 protein was analyzed using a rabbit polyclonal antibody to TSPAN12 (ab90091; Abcam, Cambridge, MA) on TGSDS mini-gels (Lukiw et al., 2005; Xu et al., 2009; Li et al., 2011). Relative 5S RNA, miRNA-132 and miRNA-146a, 15-LOX, SYN-2, CFH or TSPAN12 mRNA and protein signal strengths were quantified against 5SRNA (for miRNA) or β-actin (for protein) in each sample using data-acquisition software provided with a GS250 molecular imager (Bio-Rad, Hercules, CA; Cui et al., 2010; Zhao et al., 2011). Graphic presentations were performed using Excel algorithms (Microsoft, Seattle, WA) and Adobe Photoshop 6.0 (Adobe Systems, San Jose CA). Statistical significance was analyzed using a two-way factorial analysis of variance (p, ANOVA; SAS Institute, Cary, NC). A ‘p’ value of <0.05 was deemed as statistically significant; experimental values in the Figures are expressed as means +/− one standard deviation (SD) of that mean. Bioinformatics analysis that yielded miRNA-mRNA complementarity maps was performed using www.mirbase.org and/or www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/b (University of Manchester, UK) algorithms; additional molecular and genetic details on CFH, SYN-2, TSPAN12 and 15-LOX were accessed using GeneCards Version 3 at www.genecards.org (Weizmann Institute of Science, Rehovot, Israel).
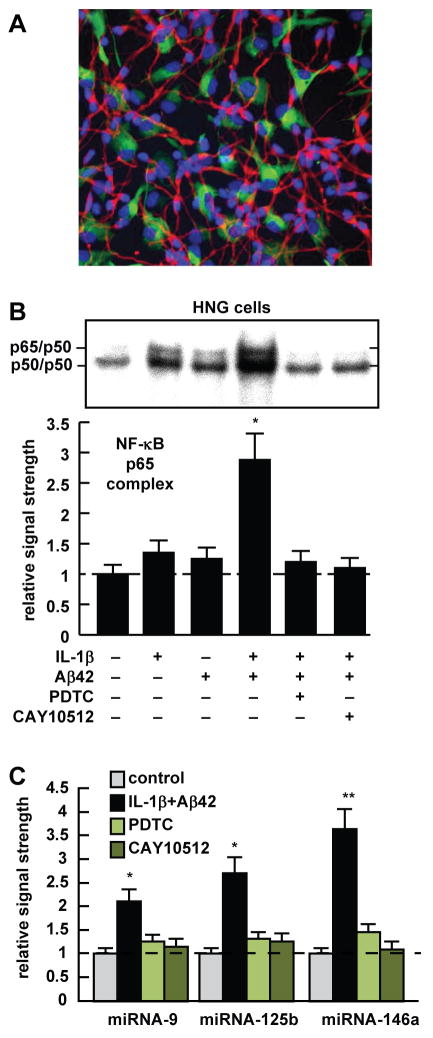
Figure 1.
(A) control human neuronal-glial (HNG) cells in primary culture stained with antibody to glial fibrillary acidic protein (GFAP), a glial-specific cytoplasmic marker (green fluorescence; λmax=556 nm); with antibody to βTUBIII, a neuron-specific cytoplasmic marker (red; λmax=702 nm), and with Hoescht 33258 to highlight the morphological features of both glial- and neuronal-cell nuclei (blue; λmax=461 nm; 2 weeks in culture; 20x magnification) (Zhao et al., 2011); note large nuclear area, relative to both glial or neuronal cytoplasmic area, indicative of high levels of transcriptional activity (Cui et al., 2005); (B) relative induction of NF-κB in HNG cells by IL-1β and Aβ42, either alone or in combination, and effects of PDTC and CAY10512, as analyzed by human NF-κB gel-shift assay (Lukiw and Bazan, 1998; Pogue et al., 2009); IL-1β and Aβ42 together show synergistic effects as previously described in older HNG cell cultures (Lukiw et al., 2008); (C) Aβ42+IL-1β-induced HNG cells in primary culture show significant up-regulation of miRNA-9, miRNA-125b and miRNA-146a, and inhibition by the metal ion chelator, anti-oxidant and NF-κB inhibitor PDTC, and CAY10512 (Lukiw et al., 2008; Cui et al., 2010). Aβ42+IL-1β has been previously shown to induce NF-κB and selective miRNA expression in several different human brain cell types (Li et al., 2011); other classes of NF-κB inhibitors, including the polyphenolic free radical scavenger curcumin are also known to significantly quench the up-regulation of brain-enriched miRNAs (Lukiw et al., 2008; Cui et al., 2010).
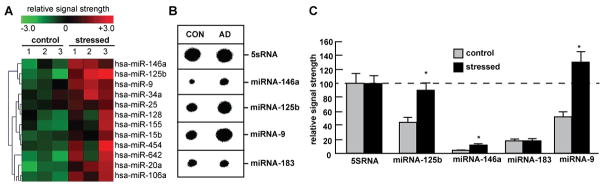
Figure 2.
(A) Cluster diagram of NF-κB-up-regulated miRNAs in Aβ42+IL1β-stressed HNG primary cells (N=3) compared to untreated controls (N=3); hsa miR = homo sapiens micro RNA; (B) dot blot confirmation of up-regulated miRNA-9, miRNA-125b and miRNA-146a abundance in stressed HNG cells compared to control 5SRNA and miRNA-183 (N=2 control; N=2 Aβ42+IL1β-stressed); and (C) RT-PCR confirmation of up-regulated miRNA-146a, miRNA-125b and miRNA-9 (N=5 control; N=5 Aβ42+IL1β-stressed); note in parts (B) and (C) 5SRNA was loaded at 1/20 the amount of all other miRNAs; as miRNAs have been shown to possess relatively short half-lives, down-regulated miRNA abundance values are suspect to degradation interference and were not considered in these experiments (Sethi and Lukiw, 2009). Members of the Let7 miRNA family were also found to be up-regulated by stress in these studies cells but did not reach statistical significance (data not shown). Genbank-based DNA sequence analysis indicates miRNA-9, miRNA-125b and miRNA-146a all contain canonical, and often multiple, NF-κB binding sites in their respective pre-miRNA promoters, a feature absent within the 15-LOX, SYN-2, CFH and TSPAN12 immediate promoters (Lukiw et al., 2008; Pogue et al., 2010, Pogue et al., 2011; see also Fig. 1; unpublished observations). In (C) a dashed horizontal line at 100 indicates levels of the 5SRNA control for ease of comparison; *p<0.05, ANOVA).
Results
Figure 1A shows a typical culture of 2 week old HNG cells studied in this report and Figure 1B shows NF-κB induction by Aβ42 peptide and/or IL-1β, and the inhibitory effects by both PDTC and CAY10512 in this human brain cell type. Figure 1C shows in bar graph format the results of Aβ42+IL-1β-mediated induction of miRNA-9, miRNA-125b and miRNA-146a and inhibition using 2 different classes of NF-κB inhibitors. Untreated (control) or Aβ42+IL-1β-treated HNG cells exhibited no overt differences in neuronal or glial cell morphology over the 1 week treatment period. While the basal abundance of miRNA-9, miRNA-125b and miRNA-146a was miRNA-125b≫miRNA-9⋙miRNA-146a, the rank order of induction was miRNA-146a⋙miRNA-9≫miRNA-125b. We observe significant quenching of at least 85% of induced miRNA-9, miRNA-125b and miRNA-146a signals by both PDTC and CAY10512. The induction of miRNA-9, miRNA-125b and miRNA-146a by NF-κB, and agents known to induce NF-κB signaling (LPS, Aβ42 peptides, IL-1β, TNFα, oxidative stress, neurotoxic metal sulfates), has been further supported and discussed in multiple studies and reviews (Taganov et al., 2006; Lukiw an Pogue 2007; Lukiw et al., 2008; Pogue et al., 2009; Ma et al., 2011; Smale et al., 2011).
Figure 2A shows a green-red color coded cluster diagram of NF-κB up-regulated miRNAs in Aβ42+IL1β-stressed HNG cells; miRNA-9, miRNA-125b and miRNA-146a were found to be the most consistently up-regulated (Sethi and Lukiw 2009; Cui et al., 2010). Figure 2B shows verification of miRNA-9, miRNA-125b and miRNA-146a up-regulation using a Northern dot blot concentration technique (Lukiw et al., 2008) and Figure 2C shows the relative up-regulation of miRNA-9, miRNA-125b and miRNA-146a in comparison to 5SRNA in the same sample.
Figure 3 shows highly complementary targets for miRNA-125b and miRNA-146a with mRNA-3 -UTR sequences showing free energy of associations (EA) of −21.7 kcal/mol or less; this is indicative of a very high and physiologically favorable potential to form double stranded RNA structures (Fabian et al., 2010; Witkos et al., 2011). Indeed high free energies of association (≤−21 kcal/mol) in duplex formation between miRNAs and their mRNA 3′UTR targets may be predictive for down-regulated gene expression. Several additional studies and comprehensive reviews on regulatory pathways involving NF-κB, miRNA-125b and miRNA-146a mRNA targets, including the ones studied here, have been recently published (Li et al., 2011a; Li et al 2011b; Ma et al., 2011; Smale 2011).
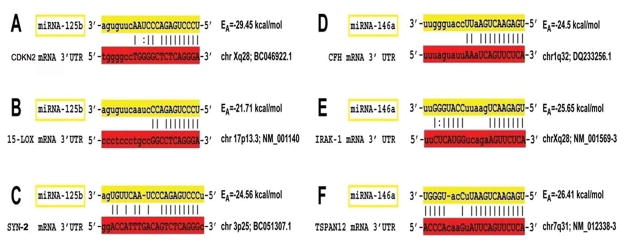
Figure 3.
In silico computation and bioinformatics analysis using www.mirbase.org and/or www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/b algorithms; predicted miRNA-mRNA complementarity maps for miRNA-125b (A–C) and miRNA-146a (D–F) of ≤ −21.71 kcal/mol (Cui et al., 2010). Specific mRNA targets for miRNA-9 are currently being investigated. miRNA-mRNA oligonucleotide complementarity yielding free energies of at least ≤ −21kcal/mol may favor selective miRNA-mRNA targeting and down-regulation of specific gene expression (unpublished observations). As indicated, miRNA-125b and miRNA-146a sequences are highlighted in yellow, and complementary sequence in the 3′ un-translated region (3′ UTR) of target mRNAs are highlighted in red; an “|” between the miRNA and mRNA indicates a hydrogen bond; an “:” between the miRNA and mRNA indicates a partial hydrogen bond. Energies of association (EA), chromosomal location and Genbank accession numbers of target mRNAs are indicated. miRNA-125b-mediated down-regulation of CDKN2A, 15-LOX and SYN II has implications for, respectively, glial cell proliferation (Pogue et al., 2010), neurotrophism (Lukiw et al., 2005; Zhao et al., 2011) and synaptic signaling (Yao et al., 2003; Lukiw 2004). Evidence for miRNA-146a targeting of CFH, IRAK-1 and TSPAN12 (also known as NET-2 or TM4SF12) mRNA, and down-regulation of CFH, IRAK-1 and TSPAN12 expression in human brain cells is further supported by recent studies (Lukiw et al., 2008; Cui et al., 2009; Li et al., 2011a; Li et al., 2011b).
Figure 4 shows protein levels for four of the miRNA-bound mRNA targets shown in Figure 3 - 15-LOX, SYN-2, CFH and TSPAN12. All Western blots generated one single prominent band and the SYN-2, CFH and TSPAN12 bands were further associated with a small faster running band (Kinders et al., 1998; Yao et al., 2003; Hennig et al., 2007; Lukiw et al., 2008; Junge et al., 2009; Cui et al., 2010; Xu et al., 2009). To eliminate the contribution of Western blot-to-blot variability, control and stressed samples were processed in the same Western blot under identical analytical conditions. Each of the 15-LOX, SYN-2, CFH and TSPAN12 RNA and protein signals were found to be significantly reduced in stressed HNG cells. These overall results suggests that in primary HNG cells that at least four of the predicted miRNA-mRNA interactions in Figure 3 likely occur, and result in the down-regulation of selective miRNA-125b- and miRNA-146a-mRNA targets including those encoding 15-LOX, SYN-2, CFH and TSPAN12.
Figure 4.
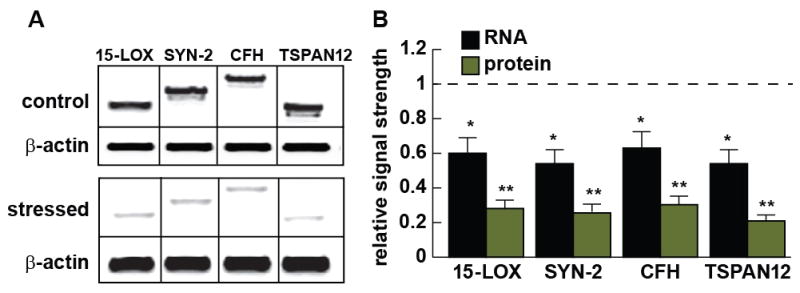
(A) Western analysis indicating down-regulation of 2 miRNA-125b targets (15-LOX and SYN-2) and 2 miRNA-146a targets (CFH and TSPAN12) in control and Aβ42+IL1β-stressed HNG primary cells, compared to a β-actin internal control within the same sample; 15-LOX, SYN-2, CFH and TSPAN12 protein levels (molecular weights ~63, 74, 150, and 35 kDa, respectively) were found to be down-regulated between 0.21 and 0.29 of controls; note that in the ‘stressed’ panel (part A, bottom) total proteins were loaded at 1.5 times the amount as in the ‘control’ panel so that the faint bands for ‘stressed’ 15-LOX, SYN-2, CFH and TSPAN12 became more clearly visible for a publication quality photo; (B) Comparison of RNA and protein signal strengths for 15-LOX, SYN-2, CFH and TSPAN12; mRNA signal strengths (gel data not shown) were found to be down-regulated between 0.54 and 0.63 of controls; relative RNA and protein control levels were both set to 1.0 (dashed horizontal line) for ease of comparison; N=3; *p<0.05, **p<0.01 (ANOVA).
Figure 5 describes in a highly schematicized format several mRNA targets for miRNA-125b and miRNA-146a, and the potential influence these human brain miRNAs has on several inter-related aspects of AD neuropathology including glial cell proliferation, synaptic failure, neurotrophic failure, neuroinflammation and amyloidogenesis.
Figure 5.
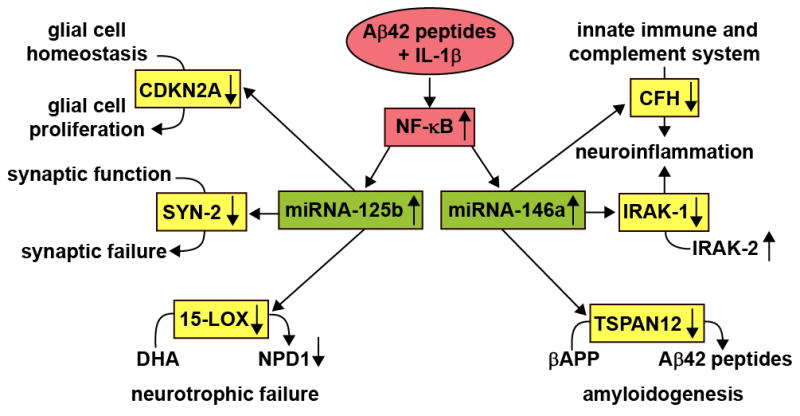
Several key pathological features of AD - glial cell proliferation, synaptic failure, neurotrophic failure, neuroinflammation and amyloidogenesis - can be explained in part by the actions of just two up-regulated miRNAs - miRNA-125b and miRNA-146a. Initially, in this pathogenic cascade, the combination of Aβ42 peptide+IL-1β up-regulates the pro-inflammatory transcription factor NF-κB which drives the transcription of miRNA-125b and miRNA-146a (Lukiw et al., 2007; Pogue et al., 2009). Up-regulated miRNA-125b results in down-regulation in the expression of CDKN2A, a negative regulator of glial cell proliferation (Pogue et al., 2010), down-regulation in the abundance of SYN-2, an essential neuronal phosphoprotein implicated in synaptogenesis and the modulation of neurotransmitter release (Figure 4), and down-regulation in the abundance of 15-LOX, a key enzyme in the biosynthesis of neuroprotectin D1 (NPD1) from the essential omega-3 fatty acid docosahexaneoic acid (DHA; Lukiw et al., 2005; Lukiw and Bazan 2008; Zhao et al., 2011). Similarly, an up-regulated miRNA-146a targets the mRNAs for CFH, IRAK-1 and TSPAN12, down-regulates CFH, IRAK-1 and TSPAN12 gene expression, and this has implications for up-regulated neuroinflammation and amyloidogenesis (notably a down-regulated IRAK-1 is associated with a compensatory surge in the abundance of IRAK-2; Cui et al., 2010). At this time we cannot exclude the participation of other misegulated miRNAs that may contribute epigenetically to the initiation or advancement of the AD process.
Discussion
Micro-RNAs and small RNAs are acquiring increasingly important roles in modulating the pathogenesis of human neurologic disorders including inflammatory neurodegeneration, AD and murine transgenic models of AD (Tg-AD) (Lukiw et al., 1992; Lukiw 2007; Coolen and Bally-Cuif, 2009; Hebert and De Strooper, 2009; Taft et al., 2010; Fabian et al., 2010; Baltimore 2011; Li et al., 2011a, 2011b). Recent interrelated and independent studies further suggest the sensitivity of human miRNA-9, miRNA-125b and miRNA-146a to central nervous system (CNS)-relevant stress and neuropathology as NF-κB-mediated miRNAs (Lukiw et al., 2008; Cui et al., 2010; Ma et al., 2011; Smale 2011; Sen 2011). Only up-regulated miRNAs were analyzed in these studies as down-regulation of miRNA abundance may be, in part, a consequence of their relatively short half-life, and uncontrolled and rapid degradation, especially in human post-mortem tissues (Sethi and Lukiw 2009). Moreover it has been recently shown that the primary mode of (up-regulated) miRNA action in mammalian cells is to down-regulate their target mRNA levels (Guo et al., 2010). The neurological activities of miRNA-9, miRNA-125b and miRNA-146a and several of their mRNA targets, and implications, are further discussed in the sections below:
miRNA-9
miRNA-9 is a human brain abundant miRNA with a half-life of 0.5–1 hr in both cultured human primary brain cells and tissues, possessing the shortest half-life of any human brain miRNA studied to date (Sethi and Lukiw., 2009; Bazzoni et al., 2011). Interestingly, miRNA-9 exerts significant control of neural progenitor cell proliferation and differentiation in the developing telencephalon by regulating the expression of multiple transcription factors including Foxg1, Nr2e1 and Pax6 (Shibata et al., 2011). We note that miRNA-9 decreases in expression as human brain cells age in primary culture, in accordance with its established role as a developmentally regulated miRNA (Yuva-Aydemir et al., 2011). Knockdown of miRNA-9 in neural progenitor cells, results in an inhibition of neurogenesis along the anterior-posterior axis of the CNS (Bonev et al., 2011), and miRNA-9 is significantly down-regulated in the tissues of fetuses with severe congenital abnormalities such as anencephaly (Zhang et al., 2010). MiRNA-9 has been further reported to be up-regulated in glioma cell lines and tissues and appears to be the target the chromobox protein homolog CBX7 Involved in maintaining the transcriptionally repressive state of genes and regulator of cellular lifespan by maintaining the repression of the cyclin-dependent kinase inhibitor 2A (CDKN2A) thus linking miRNA-9 with CDKN2A-mediated glial cell proliferation (Chao et al., 2009; Pogue et al., 2010).
miRNA-125b
One of the most human brain abundant, if not the most abundant CNS miRNAs and intensively studied miRNAs, is the inducible miRNA-125b, first shown to be up-regulated in differentiating mouse and human neurons, and since implicated in mammalian neuronal development and function (Sempere et al., 2004). miRNA-125b has been shown to be induced by neurotoxic metal sulfates that generate robust oxidative stress, and is also up-regulated in brain cancers where it apparently targets CDKN2A, a negative regulator of cell growth (Lukiw 2007; Lukiw and Pogue, 2007; Sethi and Lukiw 2009; Pogue et al., 2010; Feng et al., 2011). Up-regulated miRNA-125b thus associates with glial cell proliferation and is further associated with astrogliosis in neurodegenerative conditions such as AD and Down s syndrome, as well as in glioma and glioblastoma multiforme (Pogue et al., 2010). Interestingly miRNA-125b and miRNA-146a have tandem binding sites in the human CFH mRNA 3′-UTR (Cui et al., 2010; unpublished observations).
miRNA-146a
miRNA-146a was first described as an NF-κB-regulated pro-inflammatory miRNA that was found to target signaling proteins of innate immune responses, and more specifically the 3′-UTR of complement factor H (CFH) in human monocytes (Taganov et al., 2006). Elevated miRNA-146a in AD brain was previously shown to also target CFH and the interleukin-1 associated kinase 1 (IRAK-1), and is believed to contribute to altered innate immune responses and neuro-inflammation in degenerating human brain cells and tissues (Lukiw et al., 2008; Cui et al., 2010). CFH is a highly abundant human serum protein of hepatic origin, abundant CFH presence in brain and retinal tissues suggests CFH involvement in the innate immune response and inflammatory regulation within the privileged immunology of these tissues (Lukiw et al., 2008). Although miRNA-146a is the least basally abundant miRNA when compared to miRNA-9 and miRNA-125b, it is the most inducible and up-regulated miRNA in AD brain compared to all other NF-κB-regulated species so far indentified (Fig. 1). Interestingly, miRNA-146a may be the most induced of the miRNAs studied here due to the presence of 3 cannonical tandem NF-κB binding sites in the pre-miRNA-146a promoter (Taganov et al., 2006; Lukiw et al., 2008).
miRNA-9, miRNA-125b and miRNA-146a mRNA targets in the brain
Up-regulation in the brain-abundant miRNA-125b is associated with down-regulation of both the 15-lipoxygenase 15-LOX and the synaptic vesicle-associated phosphoprotein synapsin-2 (SYN-2). The 15-LOX enzyme is essential in the conversion of the essential omega-3 fatty acid docosahexaenoic acid (DHA) into the potent neuroprotectin D1 (NPD1), and deficits in 15-LOX correlate with NPD1 deficits in AD brain (Hennig et al., 2007; Zhao et al., 2011). The neuronal-enriched phosphoprotein SYN-2 that associates with the cytoplasmic surface of synaptic vesicles is also a miRNA-125b target, and miRNA-125b up-regulation is associated with SYN-2 down-regulation (Fig. 4) (Yao et al., 2003). Similarly, CFH is a key negative regulator of the innate immune system, and miRNA-146a up-regulation associates with decreased CFH in AD, prion disease and temporal lobe epilepsy (Kinders et al., 1998; Lukiw et al., 2008; Saba et al., 2008; Hebert and De Strooper 2009; Tsitsiou and Lindsay 2009; Aronica et al., 2010). The mRNA encoding a 4-time membrane spanning integral membrane protein TSPAN12 is also a target for miRNA-146a; and up-regulated miRNA might be expected to contribute to the down-regulation of TSPAN12 as is observed in AD brain and in stressed human brain cells (Fig. 4) (Xu et al., 2009; Li et al., 2011). Interestingly, sufficient TSPAN12 appears to be required for the neurotrophic cleavage of the beta-amyloid precursor protein (βAPP); insufficient TSPAN12 is associated with the induction of amyloidogenesis (Xu et al., 2009; Junge et al., 2009). Hence the integrated miRNA-mRNA interactions of as few as two human brain miRNAs – miRNA-125b and miRNA-146a – may in part explain not only the observed down-regulation of 15-LOX, SYN-2, CFH and TSPAN12 but also pathogenic deficiencies in innate and immune signaling, synaptogenesis, amyloidogenesis and neurotrophic support in the pathogenic brain.
Conclusions
The five main conclusions of this study are: (a) that normally aging, and stressed HNG cells in primary culture are a proven and human-relevant brain cell model to study the mechanism of transcription factor mediated miRNA activation and speciation, including the actions of specific transcription factor inhibitors, under normal aging and Aβ42 peptide- and cytokine-induced stress conditions; (b) that the down-regulated expression of several bioinformatics- and computationally-predicted mRNAs and proteins are targeted by increases in specific human brain-relevant miRNA abundances; (c) that high free energies of association (≤−21 kcal/mol) between miRNAs and their mRNA 3′UTR targets may be predictive for selective down-regulation of gene expression; (d) that stressors known to induce NF-κB activate select and pathologically relevant human brain cell miRNAs, and NF-κB inhibitors selectively negate this induction; and (e) that single miRNAs (such as miRNA-125b and miRNA-146a) appear to have the capability to regulate multiple mRNA nodes within neurobiological and neuro-immunological pathways. Indeed, some of these mRNA targets are known to associate with neurodegenerative disease, and participate in complex positive or negative NF-κB-mediated feedback and signaling loops (Lukiw et al., 2008; Cui et al., 2010; Ma et al. 2011). The targeted over-abundance of NF-κB in specific anatomical regions in AD neocortex and hippocampus further implicates an NF-κB-mediated, miRNA-regulated disease mechanism that appears to selectively down-regulate different pathology-associated brain gene transcripts during the AD-process, including those encoding 15-LOX, SYN-2, CFH and TSPAN12 (Lukiw and Bazan, 1998; Loring et al., 2001; Colangelo et al., 2002; Lukiw et al., 2004; Alexandrov et al., 2005; Li et al., 2011a; 2011b). NF-κB may be singularly important in regulating genetic responses to brain stress through the innate immune response because it belongs to the category of ‘pre-formed’ primary transcription factors that are already present in cells in an ‘inactive-sensory’ state, and do not require new protein synthesis to be activated. The more chronic and persistent activation of NF-κB via the interleukin-1 receptor-associated kinase-2 (IRAK-2) signaling pathway in AD has recently been described (Taganov et al., 2006; Cui et al., 2010; Baltimore, 2011; Sen 2011; Smale 2011). It will certainly be interesting to further analyze the role of NF-κB and other transcription factors, chromatin-mediated mechanisms and other epigenetic influences on specific miRNA-mRNA activation pathways to further understand their surprisingly dynamic interactive roles, and their contribution to the neurogenetics of brain cell aging in both health and degenerative disease.
Supplementary Material
Acknowledgments
The studies were presented in part at the 11th annual Alzheimer s Association International Conference on Alzheimer s disease (AAICAD 11) conference in Versailles, France, 16–20 July 2011; thanks are extended to Drs. Yuhai Zhao, Evgeny I. Rogaev, Surjyadipta Bhattacharjee, and Darlene Guillot for expert technical assistance, unpublished data, recent publications in this research area and helpful interpretative discussions. Research on the structure and function of NF-κB and miRNA expression in the Lukiw laboratory were supported through Grant Number P20RR016456 from the National Center for Research Resources (NCRR), a Translational Research Initiative (TRI) Grant from LSU Health Sciences Center New Orleans (WJL), an Alzheimer Association Investigator-Initiated Research Grant IIRG-09-131729 (WJL), and NIH NIA Grants AG18031 and AG038834 (WJL). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging, National Center for Research Resources, or the National Institutes of Health.
Abbreviations
- 15-LOX
15-lipoxygenase
- AD
Alzheimer s disease
- CFH
complement factor H
- HNG
human neuronal-glial
- LPS
lipopolysaccharide
- miRNA
micro RNA
- PDTC
pyrollidine dithiocarbamate
- SYN-2
SYN-II; synapsin-2
- TNFα
tumor necrtosis factor alpha
- TSPAN-12
tetraspanin 12
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov PN, Zhao Y, Pogue AI, Tarr MA, Kruck TP, Percy ME, Cui JG, Lukiw WJ. Synergistic effects of iron and aluminum on stress-related gene expression in primary human neural cells. J Alzheimers Dis. 2005;8(2):117–127. doi: 10.3233/jad-2005-8204. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431 (7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of miRNA-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci. 2010;31(16):1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. NF-κB is 25. Nat Immunol. 2011;12(8):683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106 (13):5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20(1):19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11) Biochemistry (Mosc) 2007;72(5):578–582. doi: 10.1134/s0006297907050161. [DOI] [PubMed] [Google Scholar]
- Chao TF, Zhang Y, Yan XQ, Yin B, Gong YH, Yuan JG, Qiang BQ, Peng XZ. MiR-9 regulates the expression of CBX7 in human glioma. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2008;30(3):268–274. [PubMed] [Google Scholar]
- Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70(3):462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19(5):461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Cui JG, Kuroda HH, Chandrasekharan NV, Pelaez RP, Simmons DL, Bazan NG, Lukiw WJ. Cyclooxygenase-3 gene expression in Alzheimer hippocampus and in stressed human neural cells. Neurochem Res. 2004;29(9):1731–1737. doi: 10.1023/b:nere.0000035809.70905.8a. [DOI] [PubMed] [Google Scholar]
- Cui JG, Zhao Y, Lukiw WJ. Isolation of high spectral quality RNA using run-on gene transcription; application to gene expression profiling of human brain. Cell Mol Neurobiol. 2005;25(3–4):789–794. doi: 10.1007/s10571-005-4035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J Biol Chem. 2010;285(50):38951–38960. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikelenboom P, Veerhuis R, van Exel E, Hoozemans JJ, Rozemuller AJ, van Gool WA. The early involvement of the innate immunity in the pathogenesis of late-onset Alzheimer’s disease: neuropathological, epidemiological and genetic evidence. Curr Alzheimer Res. 2011;8(2):142–150. doi: 10.2174/156720511795256080. [DOI] [PubMed] [Google Scholar]
- Ershov AV, Lukiw WJ, Bazan NG. Selective transcription factor induction in retinal pigment epithelial cells during photoreceptor phagocytosis. J Biol Chem. 1996;271(45):28458–28462. doi: 10.1074/jbc.271.45.28458. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Feng J, Kim ST, Liu W, Kim JW, Zhang Z, Zhu Y, Berens M, Sun J, Xu J. An integrated analysis of germline and somatic, genetic and epigenetic alterations at 9p21.3 in glioblastoma. Cancer. 2011 doi: 10.1002/cncr.26250. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466 (7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends in Neuroscience. 2009;32 (4):199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hennig R, Kehl T, Noor S, Ding XZ, Rao SM, Bergmann F, Fürstenberger G, Büchler MW, Friess H, Krieg P, Adrian TE. 15-lipoxygenase-1 production is lost in pancreatic cancer and overexpression of the gene inhibits tumor cell growth. Neoplasia. 2007;9(11):917–926. doi: 10.1593/neo.07565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki S, Gebhardt BM, Lukiw WJ, Thompson HW, Hill JM. Effect of immunosuppression on gene expression in the HSV-1 latently infected mouse trigeminal ganglion. Invest Ophthalmol Vis Sci. 2002;43(6):1862–1869. [PubMed] [Google Scholar]
- Hill JM, Lukiw WJ, Gebhardt BM, Higaki S, Loutsch JM, Myles ME, Thompson HW, Kwon BS, Bazan NG, Kaufman HE. Gene expression analyzed by microarrays in HSV-1 latent mouse trigeminal ganglion following heat stress. Virus Genes. 2001;23(3):273–280. doi: 10.1023/a:1012517221937. [DOI] [PubMed] [Google Scholar]
- Hill JM, Zhao Y, Clement C, Neumann DM, Lukiw WJ. HSV-1 infection of human brain cells induces miRNA-146a and Alzheimer-type inflammatory signaling. Neuroreport. 2009;20 (16):1500–1505. doi: 10.1097/WNR.0b013e3283329c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge HJ, Yang S, Burton JB, Paes K, Shu X, French DM, Costa M, Rice DS, Ye W. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139(2):299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- Kinders R, Jones T, Root R, Bruce C, Murchison H, Corey M, Williams L, Enfield D, Hass GM. Complement factor H or a related protein is a marker for transitional cell cancer of the bladder. Clin Cancer Res. 1998;4(10):2511–2520. [PubMed] [Google Scholar]
- Lee YJ, Han SB, Nam SY, Oh KW, Hong JT. Inflammation and Alzheimer’s disease. Arch Pharm Res. 2010;33(10):1539–1556. doi: 10.1007/s12272-010-1006-7. [DOI] [PubMed] [Google Scholar]
- Li YY, Cui JG, Dua P, Pogue AI, Bhattacharjee S, Lukiw WJ. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci Lett. 2011a;499(2):109–113. doi: 10.1016/j.neulet.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. Increased expression of miRNA-146a in Alzheimer’s disease transgenic models. Neurosci Lett. 2011b;487(1):94–98. doi: 10.1016/j.neulet.2010.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring JF, Wen X, Lee JM, Seilhamer J, Somogyi R. A gene expression profile of Alzheimer’s disease. DNA Cell Biol. 2001;20(11):683–695. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Handley P, Wong L, McLachlan DRC. BC200 and other small RNAs RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD) Neurochem Res. 1992;17(6):591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, LeBlanc HJ, Carver L, McLachlan DRC, Bazan NG. Run-on gene transcription in human neocortical nuclei. Inhibition by nanomolar aluminum and implications for neurodegenerative disease. J Mol Neurosci. 1998;11(1):67–78. doi: 10.1385/JMN:11:1:67. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Bazan NG. Strong nuclear factor-kappaB-DNA binding parallels cyclooxygenase-2 gene transcription in aging and in sporadic Alzheimer’s disease superior temporal lobe neocortex. J Neurosci Res. 1998;53(5):583–592. doi: 10.1002/(SICI)1097-4547(19980901)53:5<583::AID-JNR8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Gene expression profiling in fetal, aged, and Alzheimer hippocampus: a continuum of stress-related signaling. Neurochem Res. 2004;29(6):1287–1297. doi: 10.1023/b:nere.0000023615.89699.63. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Percy ME, Kruck TPa. Nanomolar aluminum induces pro-inflammatory and pro-apoptotic gene expression in human brain cells in primary culture. J Inorg Biochem. 2005;99(9):1895–1898. doi: 10.1016/j.jinorgbio.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Bazan NG. Survival signaling in Alzheimer’s disease. Biochem Soc Trans. 2006;34(6):1277–1282. doi: 10.1042/BST0341277. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Cui JG, Marcheselli VL, Bodker M, Botkjaer A, Gotlinger K, Serhan CN, Bazan NG. A role for docosahexaenoic acid-derived neuroprotectin D1 in neural cell survival and Alzheimer disease. J Clin Invest. 2005;115(10):2774–2783. doi: 10.1172/JCI25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Cui JG, Yuan YY, Bhattacharjee PS, Corkern M, Clement C, Hill JM. Acyclovir or Aβ42 peptides attenuate HSV-1-induced miRNA-146a levels in human primary brain cells. Neuroreport. 2010;21 (14):922–927. doi: 10.1097/WNR.0b013e32833da51a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101(9):1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Bazan NG. Docosahexaenoic acid and the aging brain. J Nutr. 2008;138(12):2510–4. doi: 10.3945/jn.108.096016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Zhao Y, Cui JG. An NF-κB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283(46):31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Cui JG, Li YY, Culicchia F. Up-regulation of miRNA-221 (chr Xp11.3) and caspase-3 accompanies down-regulation of the survivin-1 homolog BIRC1 (NAIP) in glioblastoma multiforme (GBM) J Neurooncol. 2009;91(1):27–32. doi: 10.1007/s11060-008-9688-0. [DOI] [PubMed] [Google Scholar]
- Ma X, Becker Buscaglia LE, Barker JR, Li YY. MicroRNAs in NF-kB signaling. J Mol Cell Biol. 2011;3(3):159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madathil SK, Nelson PT, Saatman KE, Wilfred BR. MicroRNAs in CNS injury: potential roles and therapeutic implications. Bioessays. 2011;33(1):21–26. doi: 10.1002/bies.201000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MP, Provost P. Protein components of the microRNA pathway and human diseases. Methods Mol Biol. 2009;487(1):369–385. doi: 10.1007/978-1-60327-547-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue AI, Lukiw WJ. Angiogenic signaling in Alzheimer’s disease. Neuroreport. 2004;15(9):1507–1510. doi: 10.1097/01.wnr.0000130539.39937.1d. [DOI] [PubMed] [Google Scholar]
- Pogue AI, Li YY, Cui JG, Zhao Y, Kruck TP, Percy ME, Tarr MA, Lukiw WJ. Characterization of an NF-κB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J Inorg Biochem. 2009;103(11):1591–1595. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. miRNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476(1):18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- Pogue AI, Dua P, Eicken Hill JM, Lukiw WJ. Up-regulation of micro RNA-146a (miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler Scheinker (GSS) syndrome. Journal of Toxicology and Environmental Health (JTEH) 2011 doi: 10.1080/15287394.2011.618973. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazanskaia N, Lukiw WJ, Grigorenko A, Korovaitseva G, Dvoryanchikov G, Moliaka Y, Nicolaou M, Farrer L, Bazan NG, Rogaev E. Regulatory region variability in the human presenilin-2 (PSEN2) gene: potential contribution to the gene activity and risk for AD. Mol Psychiatry. 2002;7 (8):891–898. doi: 10.1038/sj.mp.4001101. [DOI] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana J, Hajduch M, Michalek J, Vyzula R, Slaby O. MicroRNAs and glioblastoma: roles in core signaling pathways and potential clinical implications. J Cell Mol Med. 2011;15(8):1636–1644. doi: 10.1111/j.1582-4934.2011.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R. The origins of NF-κB. Nat Immunol. 2011;12(8):686–8. doi: 10.1038/ni.2071. [DOI] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459(2):100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31(9):3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukitt-Hale B, Lau FC, Carey AN, Galli RL, Spangler EL, Ingram DK, Joseph JA. Blueberry polyphenols attenuate kainic acid-induced decrements in cognition and alter inflammatory gene expression in rat hippocampus. Nutr Neurosci. 2008;11(4):172–182. doi: 10.1179/147683008X301487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Hierarchies of NF-κB target-gene regulation. Nat Immunol. 2011;12(8):689–694. doi: 10.1038/ni.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol. 2009;9(4):514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38(20):7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos TM, Koscianska E, Krzyzosiak WJ. Practical Aspects of microRNA target prediction. Curr Mol Med. 2011;11(2):93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Sharma C, Hemler ME. Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. FASEB J. 2009;23(11):3674–3681. doi: 10.1096/fj.09-133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PJ, Zhu M, Pyun EI, Brooks AI, Therianos S, Meyers VE, Coleman PD. Defects in expression of genes related to synaptic vesicle trafficking in frontal cortex of Alzheimer’s disease. Neurobiol Dis. 2003;12(2):97–109. doi: 10.1016/s0969-9961(02)00009-8. [DOI] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. MicroRNA-9: Functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8(4) doi: 10.4161/rna.8.4.16019. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chang H, Li Y, Zhang T, Zou J, Zheng X, Wu J. MicroRNAs: potential regulators involved in human anencephaly. Int J Biochem Cell Biol. 2010;42(2):367–374. doi: 10.1016/j.biocel.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Calon F, Julien C, Winkler JW, Petasis NA, Lukiw WJ, Bazan NG. Docosahexaenoic acid-derived neuroprotectin D1 induces neuronal survival via secretase- and PPARγ-mediated mechanisms in Alzheimer’s disease models. PLoS One. 2011;6(1):e15816. doi: 10.1371/journal.pone.0015816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kB p65-dependent transactivation of miRNA genes following cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5(12):e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes. Nucleic Acids Res. 2010;38(10):3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.