Abstract
Pulsed Q collision-induced dissociation (PQD) was developed in part to facilitate detection of low-mass reporter ions using labeling reagents (e.g. iTRAQ) on LTQ platforms. It has generally been recognized that the scan speed and sensitivity of an LTQ are superior than those of an Orbitrap using the higher-energy collisional dissociation (HCD). However, the use of PQD in quantitative proteomics is limited, primarily due to the meager reproducibility of reporter ion ratios. Optimizations of PQD for iTRAQ quantification using LTQ have been reported, but a universally applicable strategy for quantifying the less abundant proteins has not been fully established. Adjustments of the AGC target, µscan, or scan speed offer only incremental improvements in reproducibility. From our experience, however, satisfactory coefficients of variation (CVs) of reporter ion ratios were difficult to achieve using the discovery-based approach. As an alternative, we implemented a target-based approach that obviates data dependency to allow repetitive data acquisitions across chromatographic peaks. Such a strategy generates enough data points for more reliable quantification. Using cAMP treatment in S49 cell lysates and this target-based approach, we were able to validate differentially expressed proteins, which were initially identified as potential candidates using the discovery-based PQD. The target-based strategy also yielded results comparable to those obtained from HCD in an Orbitrap. Our findings should aid LTQ users who desire to explore iTRAQ quantitative proteomics but have limited access to the more costly Orbitrap or other instruments.
Keywords: Pulsed Q collision-induced dissociation (PQD), linear ion trap, triple quadrupole (QqQ), higher energy collisional dissociation (HCD), iTRAQ (Isobaric Tag for Relative and Absolute Quantification)
INTRODUCTION
Mass spectrometry-based proteomic quantification using multiplex stable-isotope labeling reagents has become increasingly popular, thanks to current efforts in systems biology that embrace such topics as multi-factorial physiological phenomena, temporal and spatial changes of protein expression, or interactomes. Among stable isotope-labeling reagents commercially available, the highest degree of multiplexing is offered by iTRAQ (isobaric tags for relative and absolute quantification, up to 8 plex), followed by tandem mass tags (TMT, up to 6 plex), stable isotope labeling with amino acids in cell culture (SILAC, typically up to 3 plex), difference gel electrophoresis (DIGE, up to 3 plex), cleavable isotope-coded affinity tags (cICAT, 2 plex), and 18O labeling (2 plex) [1–3]. Amongst these methods, iTRAQ and TMT offer particular advantages in that labeled peptides of the same sequence from different samples are isobaric (mass/charge, m/z) and nearly indistinguishable in chemical properties and therefore the retention times. While the MS signal is enhanced by summed intensities of isobaric labeled peptides, the intensities of MS2 reporter ions provide quantitative information [4–6].
Quadrupole ion trap (or more recently linear ion trap [LTQ]) is one of the most extensively utilized mass spectrometer platforms in proteomic research [7,8]. LIT employs resonance excitation in MS2 fragmentation which is subject to the low mass cut off (LMCO) effect. LMCO exists because only ions above certain m/z are trapped with stable trajectories at a particular qeject value in a quadrupole ion trap [7]. As a result, reporter ions of iTRAQ- or TMT-labeled peptides cannot be detected by a conventional LIT unless the precursor ions are no more than ~3 times larger than the corresponding reporters (e.g., to detect iTRAQ 114.1 reporter ion, the precursor needs to be < ~350 m/z). The pulsed Q collision-induced dissociation (PQD) largely eliminates the LMCO effect, thereby enabling iTRAQ/TMT quantification by LIT mass spectrometry [9]. However the use of PQD on LTQ linear ion traps in quantitative proteomic studies is limited, in part, due to the meager reproducibility of reporter ion ratios. Optimizations of PQD in iTRAQ quantitative analysis have recently been reported in both discovery-based [10–15] and target-based [16] studies. Most PQD experiments using the iTRAQ reagent were carried out from the discovery-based approach, employing data-dependent acquisition (DDA) in which the most intense precursor ions were subjected to MS/MS sequencing. Measures aimed at improving quantification accuracy such as increasing AGC (automatic gain control) target [12], increasing µscan [10,12], or adjusting activation/collision parameters [12,13] had been attempted. Quantifications with CV% < 30% on as low as 100-attomol analyte using optimized PQD conditions had been reported [12]; but CV% as aberrant as 300–500% or > 3000% have also been shown in studies using similar optimization schemes [10,14]. Thus the concern regarding whether PQD, upon optimization, is adequate for robust iTRAQ quantification remains.
In this report, we conducted systematic studies using PQD in discovery-based approach to determine methods that could improve iTRAQ ratio reproducibility. We further developed a new target-based strategy using PQD to achieve results comparable to those obtained using the higher-energy collisional dissociation (HCD) on an Orbitrap.
MATERIALS AND METHODS
Proteins, peptides, and cell lysates
Bovine serum albumin, β-galactosidase, α-lactalbumin, β-lactoglobulin, lysozyme, apotransferrin, and glu1-fibrinopeptide B (Glu-Fib) were purchased from Sigma (St. Louis, MO). A protein mixture from the first 6 proteins described above was prepared as a 6-protein mixture (1 µg/µl each). Wild-type (WT) and kinase A-negative (kin-) murine S49 lymphoma cell lines were cultured in Dulbecco's modified Eagle’s medium supplemented with 10% heat-inactivated horse serum and 10 mM HEPES (pH 7.4) in a humidified incubator at 37°C with 5% CO2. For protein extraction, cells were cultured to a density of ~3 × 106 cells/mL and harvested by centrifugation at 1,200 rpm (306× g), and the cell pellets were then washed 6 times with PBS. The cells were sonicated five times (10 seconds each) in the presence of a 1X protease and phosphatase inhibitor cocktail solution (Pierce, Rockford, IL), followed by centrifugation at 100,000× g for 60 min. The supernatant was saved as the cell lysate. The stimulation of cultured cells (WT and kin-) with a diffusible cAMP mimic (8CPT-cAMP, Sigma) was carried out by adding the cAMP mimic to a final concentration of 100 µM for 4 hrs prior to cell harvest.
iTRAQ labeling, liquid chromatography, and ion trap mass spectrometry
The reduction, alkylation, and tryptic digestion of proteins, and the iTRAQ labeling were performed as described previously [17]. The S49 cell lysates were labeled as follows: WT (tag 114); kin- (tag 115); WT treated with the cAMP mimic (tag 116); and kin- treated with the cAMP mimic (tag 117). Samples were analyzed using Thermo Fisher Scientific’s linear ion trap (LTQ/Orbitrap XL). For iTRAQ labeled Glu-Fib (1 pmol/µL), the infusion was performed at 0.5 µL/min using PQDdefault (activation Q of 0.7; activation time of 0.1 ms), PQDmodified (activation Q of 0.55; activation time of 0.4 ms [12]), and HCD (resolution 7.5K, respectively) for 100 scans. For samples from the six protein mixture and S49 cell lysates, the analyses were performed by reversed-phase chromatography (250 nL/min) under the above PQD and HCD conditions. Briefly, ~1 µg peptides were first loaded onto a trap cartridge (Agilent Zorbax 300SB-C18, 5 µm, 300 µm i.d. × 5 mm at 5 µl/ml), then eluted onto a reversed phase PicoFrit column (New Objective, Woburn, MA, Betabasic C18, 5 µm, 150 A, 360 µm o.d. × 75 µm i.d. × 10 cm) using a linear 60-min gradient of acetonitrile (2–62%) containing 0.1% formic acid at 250 nL/min flowrate. For discovery-based data-dependent acquisitions, each MS scan was followed by 4 PQDmodified MS2 scans on LTQ; or each FT MS scan (resolution 30K) was followed by 2 HCD MS2 scans (resolution 7.5 K with collision energy of 45%) on Orbitrap, with all MS scan ranging from 300 up to 2,000 m/z; MS2 scan ranging from 100 (HCD) or 50 (PQD) up to 2,000 m/z. For target-based acquisitions using infusion, LTQ was set up using selected reaction monitoring (SRM), as exemplified in Suppl. Fig. 1. For target-based acquisitions using chromatography, LTQ was set up to enable SRM monitoring on designated precursor masses [18] (Suppl. Fig. 2). The spray voltage and ion transfer tube temperature were set at 1.9 kV and 160°C, respectively. The settings of scan speed, AGC target, and numbers of µscan were varied and indicated in respective sections.
Triple quadrupole mass spectrometry
Selected experiments were conducted on a triple quadruple (QqQ) type MS (AB Sciex Qtrap 4000, Foster City, CA) at a scan rate of 4000 (m/z per sec) under conditions previously described [17] and shown in Suppl. Fig. 3. Low and unit resolutions refer to 1 and 0.7 Da, respectively, on Q1.
Ratio calculation and database searches
Under the normal scan rate, the LTQ’s centroiding algorithm worked well in assigning a single centroid peak with centroiding error of ~2% due to peak splitting [19]. To correct for the signal splitting, the intensities of split peaks within a tolerance of ± 0.4, ± 0.3, and ± 0.2 Da for the normal, enhanced, and zoom scan rates, respectively, were summed for ratio calculation using an in-house written C++ program. For data from the QqQ instrument, a tolerance of ± 0.4 Da was used for summing peaks. The coefficient of variation (CV%) was defined as the standard deviation divided by the average of each data group. For infusion experiments, CV% was calculated using all the 100 consecutive scans (Figs. 1 and 3). For the least favorable conditions of CV% in infusion experiments, the 100 scans were sorted in a most dispersive way so that the data therefore represented the most dispersive CV% at the number of scans indicated (Fig. 2). Mascot 2.3 was used to match MS/MS spectra to peptides using the SwissProt databases with criteria previously described [17]. Quantification of iTRAQ labeled peptides is based on the relative intensities of the 114–117 reporter ions. Proteins having ratios ≥ 1.5 (an increase of 50% or more relative to WT) or ≤ 0.5 (a decrease of 50% or more relative to WT) are considered differentially expressed [20].
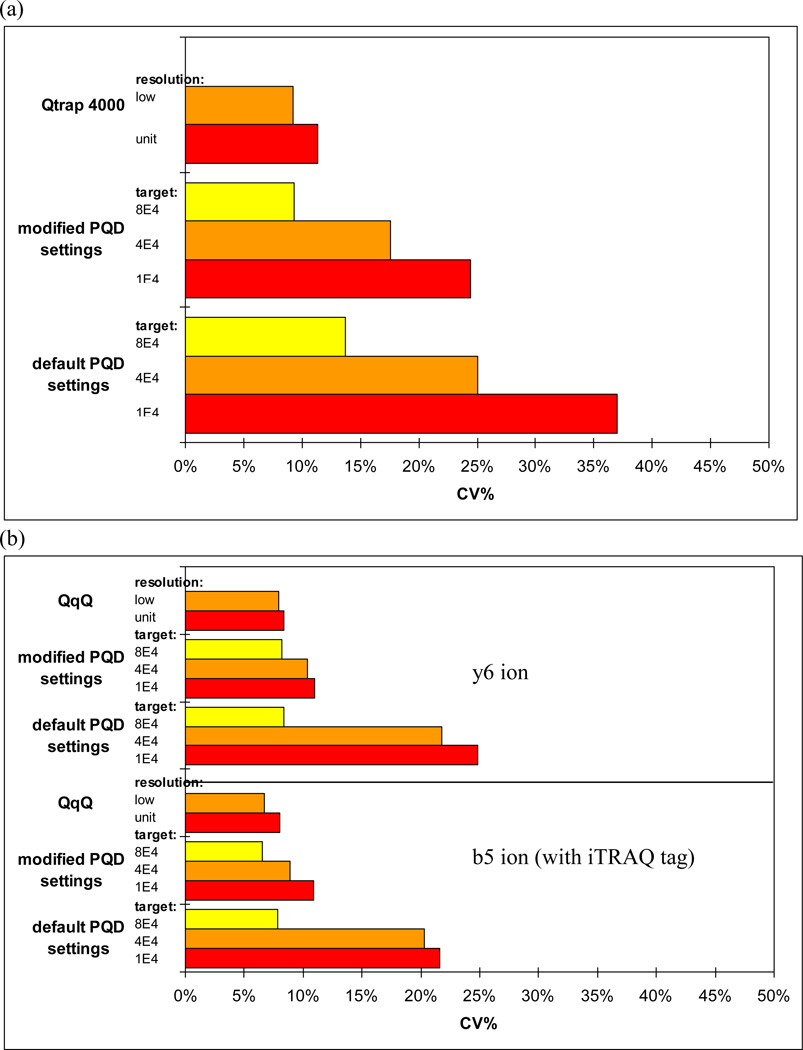
Figure 1.
Variability of iTRAQ labeled glu1-fibrinopeptide B using infusion. (a) 114 reporter ion using PQDdefault settings. (b) 114 reporter ion using PQDmodified settings. (c) y6 and b5 sequence ions from CID or PQD.
Figure 3.
Variability of iTRAQ labeled glu1-fibrinopeptide B using SRM and infusion. (a) 114.1 reporter ion. (b) y6 and b5 sequence ions.
Figure 2.
CV% of the 114 reporter ion out of the 100 scans under the least favorable conditions. iTRAQ labeled glu1-fibrinopeptide B was infused using PQDmodified settings with (a) AGC target of 1E+4. (b) AGC target of 4E+4.
RESULTS AND DISCUSSION
Discovery-based approach
We investigated how varying the scan speed, a parameter not previously tested, in conjunction with changing the AGC target, µscan, or activation parameters (PQDdefault and PQDmodified described in “Material and Methods”) would affect the CV% in iTRAQ quantification. The iTRAQ-labeled Glu-Fib was infused and the data were collected from 100 consecutive scans. To assess reproducibility, the intensities of iTRAQ reporter ions and selected b or y ions were monitored and normalized to that of the y9 ion (m/z = 1056.5, base peak ion in CID).
Effects of the scan speeds
The scan rates (m/z per second) used for LTQ were as follow: turbo scan (125,000); normal scan (16,700); enhanced scan (5,000); and zoom scan (1,100). Our initial observation indicated a poor resolution of the turbo scan on peaks around m/z 114–117. Other scans were able to resolve the reporter peaks (data not shown). Some iTRAQ reporter ions in profile mode showed peak-splitting (M shape with a dip around the apex), likely due to the ion statistics. A slower scan (enhanced or zoom) tends to turn the dip around the apex, deep enough that Xcalibur assigns more than one peak for a given iTRAQ reporter, thus resulting in peak splitting (data not shown). The observed peak splitting in enhanced or zoom scan is probably intrinsic to the higher resolution or poorer ion statistics due to fewer AGC targets. Split peaks were summed for the purpose of ratio calculation described in “Material and Methods”.
The variability of iTRAQ reporter ion (m/z 114.1) from different scan speeds is shown in Fig. 1a and b. Using either PQDdefault or PQDmodified, the normal scan (target: 1E+4 ions) consistently yielded better CV% than those of zoom or enhanced scan. A similar trend was found for other reporter ions (data not shown). The better CV% from normal scan might be ascribed to the relatively higher AGC target (default, 1E+4 ions), which is 2 to 3 times more than the enhanced (5E+3) or zoom scan (3E+3). In addition, a slower scan rate requires a smaller excitation amplitude [21] which apparently negates the proper settings required for PQD which employs higher excitation amplitude than CID [9].
Effects of AGC target
Increasing the AGC target improves the reproducibility of iTRAQ quantification ratio under the normal scan rate (Fig. 1a and 1b). As more ions are injected, the improvement in reproducibility is ascribed to the better ion statistics. However, several features unique to PQD were not readily explainable by the ion statistics alone. On two selected sequencing (b5 and y6) ions, poorer CV% were observed for PQD, relative to CID, all under the same AGC target (1E+4) (Fig. 1c). The reason is not apparent, as these sequencing ions are abundant and therefore, in theory, their CV% would not be affected by the ion statistics. Unlike CID, which undergoes a slower activation process with milder kinetic energy imparted at a longer duration [22], PQD is bestowed with higher kinetic energy imparted in a much shorter duration. This may cause aberrant fragmentation of the precursor ions, leading to the poorer CV% under PQD. Since PQD has a lower fragmentation efficiency (overall ~49% of that of CID [17]), we expect MS2 AGC target at 2E+4 for PQD should produce similar levels of fragment ions as those at 1E+4 for the default CID. Because the ratio variability is greater with PQD (Fig. 1c), an AGC target of > 2E+4 is likely needed to compensate for aberrant fragmentations. We used an AGC target up to 4E+4 (Fig. 1a and b) and found no space charging effect on the iTRAQ reporter ions (data not shown). This is in contrast with the notion that iTRAQ ratio variability from PQD was likely caused by local space charging [10].
Activation parameters and µscans
A slight improvement in CV% was seen with PQDmodified than with PQDdefault (Fig. 1a and b). At all scan rates investigated increasing the number of µscans improved the ratio reproducibility of the reporter ions (Fig. 1a and b) as well as other sequencing ions (Fig. 1c). However, due to the slow speed of the zoom scan of LTQ and that of QqQ and HCD, acquiring > 2 µscans under the chromatographic time scale is not practical and hence not attempted. Our findings corroborate previous reports showing that the CV% of iTRAQ reporter ions using PQD could be improved by summing 3–5 µscans [8] or 2–10 µscans [12].
Comparison with the quadrupole-type activation
Parallel studies were also conducted on instruments using the quadrupole-type (QqQ and HCD) activation. QqQ, at the same numbers of µscans, showed comparable CV% as to that from the normal scan (AGC 4E+4) under PQDdefault (Fig. 1a). In the QqQ mass spectra, the intensity ratios of the reporter ions to the y9 ion were much higher (~50%) than those from PQD (< 10%) (data not shown). This might explain, at least in part, the better CV% of QqQ than that of PQDdefault under the same µscan. However, the number of proteins identified by the QqQ is typically much fewer than by LTQ, presumably due to the slower scan rate of QqQ (4,000 Da/s) and thus fewer searchable MS2 spectra. HCD, at the same numbers of µscans, showed better CV% than all others. Similar to QqQ, the intensities of the reporter ions from HCD were higher (≥ 50% of the base peak) and the duty cycles were lower than the counterparts from PQD. The lower duty cycles are mainly due to the longer fill time as a result of the 20X AGC target (2E+5) for the Orbitrap.
Inferences from the infusion data
Although a CV% of < 30% using PQD is achievable (Fig. 1a and b), the results are based on 100 consecutive scans through infusion. A CV% of 25% (114 tag) has been reported for 50 consecutive scans in a previous study [12]. Paradoxically with the DDA, if a protein has its peptides sequenced 50–100 times, that protein is likely to be highly abundant and therefore may represent a ‘house-keeping’ function, with probably low levels of experimental expression alteration. In contrast, peptides from the less abundant proteins are subjected to limited sequencing events (typically ≤ 5 times) in DDA, despite the fact that some are likely to be experimentally differentially expressed. As an illustration, CV% of the 114 reporter ion out of the 100 scans using PQDmodified under the least favorable conditions (described in “Material and Methods”) is shown in Fig. 2. At 1 µscan and default AGC target (1E+4) under the least favorable conditions, it takes at least 100 scans to reach a value of ≤ 25% CV.
Based on calculations using individual dta files of the iTRAQ labeled Glu-Fib and 6-protein mixture (data not shown), the 4 iTRAQ reporter ions account for, at best, 1/50 of the total fragment ions covering the 50–2000 m/z range, equivalent to < 50 ions per reporter ion. The reproducibility of iTRAQ quantification is thus compromised by the insufficient ion statistics, which is particularly problematic for the low abundant peptides. Increasing the AGC target and/or µscan offers incremental improvement in reproducibility (Fig. 1a and b), but at the expense of numbers of sequencing events under chromatographic time scale which in turn favors the identification of more abundant proteins. A new software pipeline (LTQ-iQuant) [23] and Mascot improve the ratio reliability at the protein level by assigning ratios based on intensity-dependent weighting of peptide data. Although optimization based on method parameters and/or software is amenable for improved iTRAQ quantification [10,12,13,23], uncertainty remains for achieving a satisfactory quantification of medium- to low- abundant proteins using the discovery-based approach.
Target-based approach
Depending on the sample complexity, low abundant peptides may, or may not, trigger MS2 fragmentation in a typical discovery-based scheme. Even when triggered, the reproducibility of iTRAQ reporter ion ratios is likely to be marginal, given the large CV% discussed above. We therefore investigated a target-based approach in which ions of interest are constantly monitored. We first assessed the reproducibility by infusing iTRAQ labeled Glu-Fib. The LTQ SRM setting used and the spectrum of the product and reporter ions on the precursor (858.3 m/z) are shown in Suppl. Fig. 1.
The CV% of iTRAQ 114.1 reporter ion and two sequencing ions of Glu-Fib are shown in Fig. 3. Better CV% was consistently obtained from QqQ (the benchmark instrument for SRM) or LTQ PQDmodified at higher targets. QqQ at low Q1 resolution showed better CV% than unit resolution, likely due to better ion statistics. Although better CV% was obtained with a target of 8E+4, there was a significant mass shift (≥ 0.3) due to space charging (Suppl. Fig. 4), and thus not investigated further. Since generally acceptable CV% (≤ 25%) using default AGC and PQDmodified settings were obtained, this suggests that a reliable quantification could be achieved on LTQ as long as sufficient data points are collected (100 scans in the case shown in Fig. 2a). We used the default MSn target of 1E+4 and PQDmodified setting for subsequent studies. More than 1 µscan or higher MSn target was not required, as a higher target necessitates a longer fill time and our sole goal was to maximize the number of data points across chromatographic peaks to yield better statistics.
The 4-plex iTRAQ labeled peptides from lysates of control and kin- S49 cells, cultured in the presence or absence of a diffusible cAMP analog, were analyzed using the discovery-based as well as the target-based approaches using LTQ/PQDmodified. Several candidate proteins identified with the discovery approach yielded only a few peptides, providing only meager power of statistics in iTRAQ ratios. Based on parent masses and retention times of identified peptides of the candidate proteins, target-based analysis was conducted. The results are summarized in Table 1. For comparison, results obtained from a discovery-based approach using Orbitrap/HCD were also listed. A total of 26 proteins were identified as being differentially regulated (ratios ≥ 1.5 or ≤ 0.5) using PQD, as compared to 16 such proteins using HCD. Among those identified by both platforms, the trend of changes is generally consistent. However, it is apparent that some of the ratios can only be determined using the target-based approach.
Table 1.
Proteins differentially expressed in cultured S49 and its kin- mutant cells in the presence or absence of a diffusible cAMP analog. Both target-based (PQD/LTQ) and discovery-based (HCD/Orbitrap) acquisitions were conducted with the same tryptic peptides prepared from the lysates. The iTRAQ labeling schemes are described in Materials and Methods. Suppl. Table 1 details the number of peptides and peptide sequences identified.
| Target-based acquisition using PQD | Discovery-based acquisition using HCD | ||||||
|---|---|---|---|---|---|---|---|
| Mutational effect | Ratio115/114 | Ratio116/114 | Ratio117/114 | Ratio115/114 | Ratio116/114 | Ratio117/114 | |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P_MOUSE | 1.44 | 1.18 | 1.76 | 1.53 | 1.15 | 1.79 |
| Asparagine synthetase [glutamine-hydrolyzing] | ASNS_MOUSE | 1.41 | 0.92 | 1.30 | 1.58 | 0.92 | 1.58 |
| Staphylococcal nuclease domain-containing protein 1 | SND1_MOUSE | 1.43 | 1.00 | 1.64 | 1.55 | 1.00 | 1.47 |
| Galectin-1 | LEG1_MOUSE | 3.17 | 1.42 | 3.57 | 2.42 | 1.31 | 2.27 |
| Protein disulfide-isomerase A4 | PDIA4_MOUSE | 1.62 | 1.01 | 1.57 | 1.63 | 1.01 | 1.77 |
| Rho GDP-dissociation inhibitor 2 | GDIR2_MOUSE | 1.70 | 0.96 | 1.89 | 2.02 | 0.83 | 1.96 |
| Metallothionein-2 | MT2_MOUSE | 0.33 | 0.80 | 0.41 | --- | --- | --- |
| Serine-threonine kinase receptor-associated protein | STRAP_MOUSE | 1.50 | 0.83 | 1.43 | 1.39 | 0.89 | 1.17 |
| Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase | MTDC_MOUSE | 1.59 | 1.05 | 1.74 | 1.52 | 0.92 | 1.39 |
| RuvB-like 1 | RUVB1_MOUSE | 1.41 | 1.12 | 1.52 | 1.38 | 1.09 | 1.31 |
| Adenine phosphoribosyltransferase | APT_MUSPA | 0.48 | 0.93 | 0.54 | 0.57 | 0.89 | 0.58 |
| Glutathione reductase, mitochondrial | GSHR_MOUSE | 1.43 | 0.91 | 1.52 | 1.06 | 0.83 | 1.01 |
| Phosphoenolpyruvate carboxykinase (GTP), mitochondrial | POKGM_MOUSE | 1.87 | 0.85 | 1.81 | 1.75 | 0.88 | 1.60 |
| Response gene to complement 32 protein | RGC32_MOUSE | 0.46 | 1.37 | 0.70 | --- | --- | --- |
| Thymosin beta-10. | TYB10_MOUSE | 3.45 | 1.08 | 4.01 | 7.07 | 0.92 | 9.06 |
| Mitotic spindle assembly checkpoint protein MAD2A | MD2L1_MOUSE | 1.50 | 1.20 | 1.55 | --- | --- | --- |
| Glycyl-tRNA synthetase | SYG_MOUSE | 1.43 | 1.00 | 1.33 | 1.50 | 1.01 | 1.54 |
| Chromobox protein homolog 3 | OBX3_MOUSE | 1.46 | 0.98 | 1.39 | 1.60 | 1.07 | 1.66 |
| cAMP effect | |||||||
| BTB/POZ domain-containing protein TNFAIP1 | TNAP1_MOUSE | 1.14 | 1.70 | 1.74 | --- | --- | --- |
| UPF0565 protein | U566_MOUSE | 1.06 | 1.71 | 1.79 | --- | --- | --- |
| T-complex protein 1 subunit delta | TCPD_MOUSE | 1.09 | 1.50 | 1.73 | 1.03 | 1.16 | 1.21 |
| combined (cAMP and mutational) effect | |||||||
| Histone H1.1 | H11_MOUSE | 1.29 | 1.37 | 1.54 | --- | --- | --- |
| Histone H1.2 | H12_MOUSE | 1.39 | 1.43 | 1.65 | 1.26 | 1.30 | 1.63 |
| Histone H1.3 | H13_MOUSE | 1.38 | 1.45 | 1.66 | 1.30 | 1.37 | 1.71 |
| Thioredoxin | THIO_MOUSE | 1.03 | 0.97 | 1.39 | 1.32 | 1.17 | 1.62 |
| Proteasome subunit alpha type-6 | PSA6_MOUSE | 1.14 | 1.24 | 1.54 | 1.21 | 1.39 | 1.63 |
| Proteasome subunit alpha type-4 | PSA4_MOUSE | 1.12 | 1.38 | 2.12 | --- | --- | --- |
| Ab1 interactor 1 | ABI1_MOUSE | 0.72 | 0.73 | 0.46 | --- | --- | --- |
| Neurabin-2 | NEB2_MOUSE | 0.78 | 0.96 | 0.48 | --- | --- | --- |
| Actin-related protein 2 | ARP2_MOUSE | 1.31 | 1.27 | 1.47 | 1.42 | 1.33 | 1.63 |
Note: - - - indicates not detected.
Fig. 4a illustrates the feature of repetitive data acquisitions of 164 individual SRMs (in “stick” mode) across the 20-second chromatographic peak on peptide SFVLNLGK (LEG1_MOUSE). The figure insets (Fig. 4a) show variations of the intensities of iTRAQ reporter ions at randomly selected sampling points. The mass spectrum around iTRAQ reporter ions and the calculated reporter ion ratios (median ratio±CV%), relative to the peak intensity of the WT control, are shown in Fig 4b. The CV% of ≤ 27% (n = 164 sampling points) obtained suggests that this target-based approach provides sufficient power of statistics to confirm data obtained from the discovery-based approach. Such levels of CV% and sampling events (n ≥ 100) are not readily achievable from the discovery-based approach over the chromatographic time scale.
Figure 4.
(a) Variability in ratios of iTRAQ labeled SFVLNLGK using SRM of precursor 583.4 m/z → product ions (110–120 m/z) across the chromatographic peak (n = 164 between the red lines); (b) the median ratio±CV% and the spectrum showing the m/z of reporter ions.
The 164 data points (Fig. 4a) across a 20-second peak is equivalent to ~100 msec triple quadrupole-like “dwell time” for each SRM transition. As an illustration, the target-based method (Suppl. Fig. 2) consists of scan events within several segments of retention time windows, which could be designed to accommodate a long list of selected parent masses. Each segment performs MS2 for selected parent ions whose retention times fall within the window. To maximize the data points within a given segment, the scan events could be limited to no more than three. A targeted data acquisition method involving PQD has recently been reported [16]. It differs from our target-based approach in two ways. Firstly, the MS1 scan was performed on an Orbitrap [16], our approach is based on MS1 scan on LTQ, which is a more widely available machine platform. Secondly, the referenced method was subjected to data dependency. This is unlike our strategy which obviates data dependency to allow repeated data acquisitions across chromatographic peaks, thus generating many more data points for averaging.
CONCLUSIONS
In contrast to the finding that optimized PQD is amenable for iTRAQ quantification, we found that there is a limitation for PQD to accurately quantify medium- to low- abundant proteins using the discovery-based approach. At 1 µscan under the default AGC target, it may require at least 100 scans to reach CV ≤ 25%. The reproducibility of iTRAQ quantification seems to be compromised by inadequate ion statistics at ~50 ions per reporter ion, which is particularly problematic for low abundance peptides. Increasing AGC target and/or µscan may offer incremental improvements in reproducibility, but only at the expense of the numbers of sequencing events. We implemented a target-based approach that avoids data dependency to allow repeated data acquisitions across chromatographic peaks, thus greatly improving the number of data points for averaging and statistics. We successfully validated this platform on differential protein expression of control and stimulated cultured cells using PQD in both discovery- and target-based approaches and showed comparable results to those obtained using HCD in an Orbitrap. Our described approach can facilitate the ability of the greater number of MS users that have access to LTQ machines, rather than Orbitraps, to effectively use iTRAQ-based quantitative proteomics.
Highlights.
-
○
Difficult to quantify lower abundant proteins using PQD in discovery-based approach
-
○
Our target-based approach greatly improves reproducibility in iTRAQ quantification
-
○
The target-based approach yields results comparable to those obtained from HCD
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Programs of National Institute on Aging and National Heart, Lung, and Blood Institute, National Institutes of Health (NIH). The authors would like to acknowledge the following for technical advice: Dr. Howard Jaffe, NINDS, NIH; Drs. Jae C. Schwartz, Mike W. Senko, and John E.P. Syka of ThermoFisher Scientific Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wu WW, Wang G, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J. Proteome. Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 2.Gevaert K, Impens F, Ghesquiere B, Van DP, Lambrechts A, Vandekerckhove J. Stable isotopic labeling in proteomics. Proteomics. 2008;8:4873–4885. doi: 10.1002/pmic.200800421. [DOI] [PubMed] [Google Scholar]
- 3.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 4.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal. Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 6.Viner RI, Zhang T, Second T, Zabrouskov V. Quantification of post-translationally modified peptides of bovine alpha-crystallin using tandem mass tags and electron transfer dissociation. J. Proteomics. 2009;72:874–885. doi: 10.1016/j.jprot.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Louris JN, Cooks RG, Syka JEP, Kelley PE, Stafford GC, Todd JFJ. Instrumentation, applications, and energy deposition in quadrupole ion-trap tandem mass-spectrometry. Anal. Chem. 1987;59:1677–1685. [Google Scholar]
- 8.Schwartz JC, Senko MW, Syka JEP. A two-dimensional quadrupole ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz JC, Syka JEP, Quarmby ST. Improving the fundamentals of MSn on 2D linear ion traps: new ion activation and isolation techniques. The 53rd ASMS Conference on Mass Spectrometry and Allied Topic; June 5–9; San Antonio, TX. 2005. [Google Scholar]
- 10.Griffin TJ, Xie H, Bandhakavi S, Popko J, Mohan A, Carlis JV, Higgins L. iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J. Proteome Res. 2007;6:4200–4209. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 12.Bantscheff M, Boesche M, Eberhard D, Matthieson T, Sweetman G, Kuster B. Robust and sensitive iTRAQ quantification on an LTQ orbitrap mass spectrometer. Mol. Cell Proteomics. 2008;7:1702–1713. doi: 10.1074/mcp.M800029-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo T, Gan CS, Zhang H, Zhu Y, Kon OL, Sze SK. Hybridization of pulsed-Q dissociation and collision-activated dissociation in linear ion trap mass spectrometer for iTRAQ quantitation. J. Proteome Res. 2008;7:4831–4840. doi: 10.1021/pr800403z. [DOI] [PubMed] [Google Scholar]
- 14.Armenta JM, Hoeschele I, Lazar IM. Differential protein expression analysis using stable isotope labeling and PQD linear ion trap MS technology. J. Am. Soc. Mass Spectrom. 2009;20:1287–1302. doi: 10.1016/j.jasms.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, Wu S, Stenoien DL, Zhao R, Monroe ME, Gritsenko MA, Purvine SO, Polpitiya AD, Tolic N, Zhang Q, Norbeck AD, Orton DJ, Moore RJ, Tang K, Anderson GA, Pasa-Tolic L, Camp DG, Smith RD. Combined pulsed-Q dissociation and electron transfer dissociation for identification and quantification of iTRAQ-labeled phosphopeptides. Anal. Chem. 2009;81:4137–4143. doi: 10.1021/ac802605m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savitski MM, Fischer F, Mathieson T, Sweetman G, Lang M, Bantscheff M. Targeted data acquisition for improved reproducibility and robustness of proteomic mass spectrometry assays. J. Am. Soc. Mass Spectrom. 2010;21:1668–1679. doi: 10.1016/j.jasms.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Wu WW, Wang G, Insel PA, Hsiao CT, Zou S, Martin B, Maudsley S, Shen RF. Identification of proteins and phosphoproteins using pulsed Q collision induced dissociation (PQD) J. Am. Soc. Mass Spectrom. 2011;22:1753–1762. doi: 10.1007/s13361-011-0197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu WW, Dufresne CP, Shen RF. Target-based approaches using data dependent and independent schemes on LTQ/Orbitrap and comparison with QTrap 5500 results. The 58th ASMS Conference on Mass Spectrometry and Allied Topic; May 23–27; Salt Lake City, UT. 2010. [Google Scholar]
- 19.Li Q, Xia Q, Wang T, Meila M, Hackett M. Analysis of the stochastic variation in LTQ single scan mass spectra. Rapid Commun. Mass Spectrom. 2006;20:1551–1557. doi: 10.1002/rcm.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu WW, Wang G, Yu MJ, Knepper MA, Shen RF. Identification and quantification of basic and acidic proteins using solution-based two-dimensional protein fractionation and label-free or 18O-labeling mass spectrometry. J. Proteome. Res. 2007;6:2447–2459. doi: 10.1021/pr060621c. [DOI] [PubMed] [Google Scholar]
- 21.Jackson GP, Hyland JJ, Laskay UA. Energetics and efficiencies of collision-induced dissociation achieved during the mass acquisition scan in a quadrupole ion trap. Rapid Commun. Mass Spectrom. 2005;19:3555–3563. [Google Scholar]
- 22.Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. J. Mass Spectrom. 2004;39:1091–1012. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- 23.Onsongo G, Stone MD, Van Riper SK, Chilton J, Wu B, Higgins LA, Lund TC, Carlis JV, Griffin TJ. LTQ-iQuant: a freely available software pipeline for automated and accurate protein quantification of isobaric tagged peptide data from LTQ instruments. Proteomics. 2010;10:3533–3538. doi: 10.1002/pmic.201000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.