Abstract
The KRAB-type zinc-finger protein Apak was recently identified as a negative regulator of p53-mediated apoptosis. However, the mechanism of this selective regulation is not fully understood. Here, we show that Apak recognizes the TCTTN2−30TTGT consensus sequence through its zinc-fingers. This sequence is specifically found in intron 1 of the proapoptotic p53 target gene p53AIP1 and largely overlaps with the p53-binding sequence. Apak competes with p53 for binding to this site to inhibit p53AIP1 expression. Upon DNA damage, Apak dissociates from the DNA, which abolishes its inhibitory effect on p53-mediated apoptosis.
Keywords: p53, p53AIP1, transcriptional regulation, apoptosis, Apak
Introduction
The tumour suppressor p53 functions as a node for organizing the cellular response to various types and levels of stress [1]. The p53-mediated transactivation of specific target genes is an essential feature of each stress response pathway, although some effects of p53 might be independent of transcription [2]. The question of how the activation of p53 leads to either cell-cycle arrest or apoptosis has received much attention. Different proteins can bind to the DNA-binding core, the tetramerization domain or the basic domain of p53 to induce different cellular outcomes. Hzf, Brn3A, YB1, c-Abl and p18/Hamlet selectively induce the p53 activation of genes encoding cell-cycle regulators, such as p21, 14-3-3σ and Gadd45, to facilitate cell-cycle arrest. In contrast, ASPP1/2, hCAS, Brn3b, p53β, NFκB/p52 and Muc1 selectively activate the proapoptotic genes such as Puma, Bax and p53AIP1, to promote cell death [3]. Whether a given promoter of a p53 downstream target is activated or repressed depends on the abundance of p53 protein, its modifications, the proteins that directly interact with p53 and the proteins that interact independently of p53 with the target promoters.
We recently reported that the KRAB-type zinc-finger (KZNF) protein Apak (ATM and p53-associated KZNF protein, also known as ZNF420) functions as a specific negative regulator of p53-mediated apoptosis. Apak preferentially inhibits the activation of the proapoptotic targets of p53 but has no significant effects on the proarrest targets [4]. This selectivity is achieved partially through the direct interaction of Apak with p53 and the recruitment of the corepressor KAP-1 and the histone deacetylase HDAC1 to attenuate p53 acetylation. Notably, Apak harbours 19 consecutive C2H2 zinc-finger repeats in its carboxy-terminus [4]. Typically, KZNF proteins bind to their corresponding target genes through these zinc-fingers. For example, ZBRK1 binds to a specific sequence, GGGN3CAGN3TTT (N represents an arbitrary nucleotide), within the Gadd45 intron 3 [5]. ZNF333 recognizes the consensus sequence ATAAT [6], whereas ZNF746 binds to TATTTT (T/G) [7]. At present, the cognate DNA site recognized by Apak remains unidentified, and the possible role of such recognition in p53 regulation has not been characterized.
Here, we first performed a target screening and identified the sequence TCTTN2−30TTGT as a specific recognition site of Apak. Then, we searched for possible Apak target genes among the p53 targets and found that the proapoptotic p53AIP1 gene uniquely contains the Apak-binding site. Intriguingly, the Apak-binding site largely overlapped with the p53-binding site within the first intron of p53AIP1. Apak competes with p53 to bind to the p53AIP1 gene. This study provides a new molecular mechanism to explain why Apak selectively inhibits p53-mediated apoptosis.
Results And Discussion
Identification of the Apak DNA-binding sequence
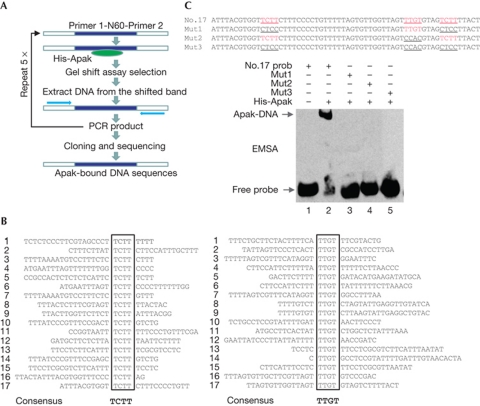
To identify the DNA-binding sequence for Apak, a cyclic amplification and selection of targets assay was performed (Fig 1A). Purified His-Apak (supplementary Fig S1 online) was incubated with a pool of double-stranded 100-mer oligonucleotides, each of which carried 20-base fixed-end sequences flanking 60 central bases of random sequences. After five rounds of selection and purification, the PCR product was cloned, amplified and sequenced. The alignment analysis revealed a consensus sequence with a core composed of two elements, TCTT and TTGT, together with a spacer of 2–30 oligonucleotides (Fig 1B). In all 18 out of 20 sequences contained the core and 1 out of 18 sequence tags were identified as duplicated, indicating that this is the primary DNA-binding sequence for Apak.
Figure 1.
Identification of Apak DNA-binding sequence. (A) Strategy for the selection of the DNA consensus-binding site of Apak. (B) Alignment of individual DNA sequences selected using a CAST assay. The deduced consensus Apak DNA-binding sequence is indicated below the individually aligned sequences. (C) EMSA of His-Apak with the DNA sequence of clone no. 17 and its mutants. CAST, cyclic amplification and selection of targets; EMSA, electrophoretic mobility shift assay; Mut, mutant.
To confirm the binding of Apak to the selected sequences, we performed electrophoretic mobility shift assay (EMSA) using the randomly selected clone no. 17 as a probe. This probe contains two TCTT elements and one TTGT element (Fig 1C). TCTT, TTGT or both were mutated to generate the mutant (Mut) probes (Fig 1C). Apak bound to the wild-type (WT) probe but not to any of the Mut probes, suggesting that both elements are indispensable for Apak binding.
Apak selectively binds to the intron of p53AIP1
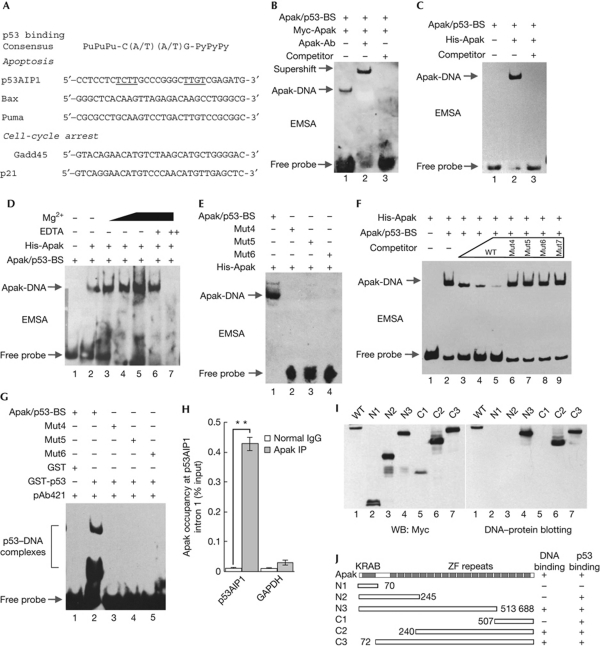
We tested whether the downstream targets of p53 contain any Apak-binding sites in their promoters or introns. A careful survey of the proapoptotic targets, including p53AIP1, Puma and Bax, and the proarrest targets, such as p21 and Gadd45, revealed that p53AIP1 contains a potential Apak-binding sequence (Fig 2A). This sequence is located within the first intron and, strikingly, overlapped with the well-defined p53-binding sequence, which is composed of two PuPuPu–C(A/T)(A/T)G–PyPyPy elements (Fig 2A). EMSA showed that the Apak-binding sequence (Apak/p53-BS) bound to the nuclear extract expressing Myc-tagged Apak, whereas incubation with an Apak-specific antibody resulted in a clear supershift (Fig 2B), confirming the binding specificity. This probe also supported the formation of a complex with the purified His-Apak protein (Fig 2C). Mg2+ was required for efficient binding (Fig 2D). The mutation of TCTT (Mut4), TTGT (Mut5) or both (Mut6) resulted in loss of the Apak/DNA complex (Fig 2E; supplementary Table S1 online). Meanwhile, the binding of Apak to the Apak/p53-BS probe was attenuated gradually by the addition of increasing amounts of the unlabelled WT probe but not the mutated probes (Fig 2F). In addition, these mutations also abolished the binding of the probes to p53 (Fig 2G). Chromatin immunoprecipitation (ChIP) assays showed that Apak interacted with the endogenous p53AIP1 intron 1 in p53−/− HCT116 cells (Fig 2H), suggesting that the binding is independent of the p53 status.
Figure 2.
Apak selectively binds to the p53AIP1 intron via its zinc-fingers. (A) The p53-binding sequence in the promoter or intron of the indicated target genes is shown. For p53AIP1 intron 1, the Apak-binding core sequence is underlined. (B) Apak binds to the Apak/p53-BS of p53AIP1 in vitro. Supershift EMSA using nuclear extracts derived from p53−/− HCT116 cells harbouring Myc-Apak. The Apak antibody and an unlabelled competitor probe were added as indicated. (C) EMSA was performed with 100 ng of His-Apak and biotin-labelled Apak/p53-BS oligonucleotides. (D) Mg2+ affects the binding of His-Apak with Apak/p53-BS. The indicated amounts of Mg2+ and EDTA were added into the EMSA reactions. (E) The mutant Apak/p53-BS probes (for sequences, see supplementary Table S1 online) were used in the EMSA with Apak protein. (F) The binding of Apak with Apak/p53-BS was abolished by the addition of a molar excess of unlabelled cold probe, but not the mutated probes. (G) The mutant probes were used in the EMSA with the p53 protein and p53 antibody. (H) ChIP assays in p53−/− HCT116 cells. The p53AIP1 intron was amplified from immunoprecipitates of anti-Apak or IgG control. Graph shows the mean±s.d. (n=3). s.d., standard deviation. **P<0.01. (I) p53−/− HCT116 cells were transfected with various Apak deletion mutants. The cell lysates were analysed by western blotting using anti-Myc antibody (left). The same samples were hybridized to the Apak/p53-BS probe (right). (J) A schematic diagram of the DNA-binding and p53-binding regions in Apak. ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Mut, mutant; ZF, zinc-finger.
We used various Apak deletion Muts to test their binding ability to the p53AIP1 intron. DNA–protein blotting analysis showed that the zinc-fingers of Apak, but not its KRAB domain, were required for DNA binding (Fig 2I). Apak also uses the zinc-fingers to bind to the p53 protein [4]. Interestingly, a careful comparison showed that the four zinc-fingers of Apak are sufficient for p53 binding [4] but not for DNA binding, and that 14 zinc-fingers can mediate the DNA binding efficiently (Fig 2J). A similar phenomenon has been previously observed in the repressor NRSF [8].
Apak is a transcriptional repressor of p53AIP1
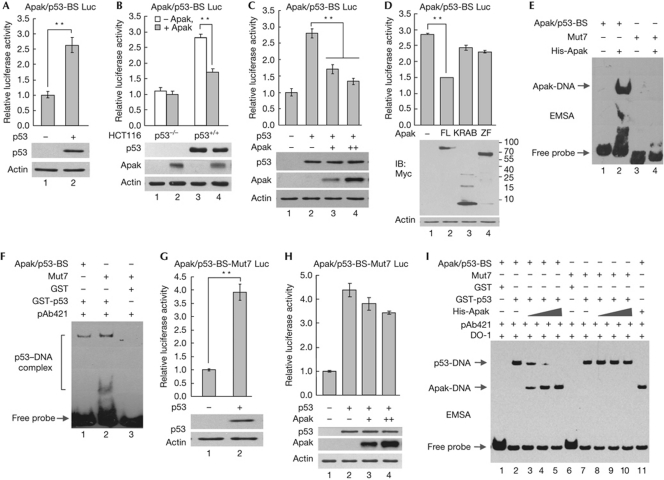
To evaluate the functional relevance of the DNA-binding activity of Apak, we performed a reporter gene assay with the heterologous luciferase gene fused to one copy of Apak/p53-BS upstream in the pGL3 vector. Co-transfection of Apak/p53-BS-luc with p53 into p53−/− HCT116 cells resulted in a 2.5-fold increase of luciferase activity (Fig 3A). Overexpression of Apak repressed the p53-mediated Apak/p53-BS-luc activity (Fig 3B), and the repression was dose dependent (Fig 3C). Individual expression of either the KRAB domain or the zinc-fingers of Apak resulted in moderate effects on Apak/p53-BS activity compared with full-length Apak (Fig 3D), suggesting that both KRAB and the zinc-fingers contribute to the p53AIP1 repression. The insufficiency of zinc-fingers for the repression might be due to the mislocalization of the ZF truncate to the nucleolus during overexpression in HCT116 cells (supplementary Fig S2 online).
Figure 3.
Apak is a transcriptional repressor of p53AIP1. (A) p53−/− HCT116 cells were co-transfected with Apak/p53-BS-Luc and p53. After 48 h, the luciferase activity was measured. (B) p53−/− and p53+/+ HCT116 cells were co-transfected with Apak/p53-BS-Luc and Apak, and the luciferase activity was measured. (C) p53−/− HCT116 cells were co-transfected with Apak/p53-BS-Luc, p53 and Apak as indicated and the luciferase activity was measured. (D) Both KRAB domain and the zinc-fingers are required for Apak to repress luciferase activity efficiently. (E) EMSA was performed with 100 ng His-Apak and biotin-labelled mutant Apak/p53-BS. (F) EMSA of the p53 protein with Apak/p53-BS and mutants. (G) p53−/− HCT116 cells were co-transfected with Apak/p53-BS-Mut7-Luc (for Mut7 sequence, see supplementary Table S1 online) and p53. After 48 h, the luciferase activity was measured. (H) Apak cannot significantly repress the Apak/p53-BS-Mut7-Luc activity increased by p53. (I) Apak competes with and displaces the binding of p53 to Apak/p53-BS, but not to Mut7. In vitro EMSAs were performed with GST-p53 or His-Apak proteins. To distinguish between the p53–DNA and the Apak-DNA migration bands, two anti-p53 antibodies (pAb421 and DO-1) were included in the reaction. Data in (A–D), (G) and (H) are means±s.d. (n=3). **P<0.01. EMSA, electrophoretic mobility shift assay; GST, glutathione S-transferase; IB, immunoblotting; Mut, mutant.
As Apak can interact with p53 directly and inhibit p53 acetylation by recruiting the KAP1–HDAC1 complex via the KRAB domain, the inhibitory effect of Apak on p53AIP1 might be caused by the regulation of p53 activity, direct binding to the p53AIP1 intron or a combined effect. To clarify this issue, we generated a mutated p53AIP1 intron sequence that loses the ability to bind to Apak but retains the ability to bind to p53. The Apak-binding TCTT and TTGT elements were therefore mutated to GCTT and TTGC, respectively. EMSA showed that the produced Mut (Mut7) indeed failed to interact with Apak (Fig 3E) but retained the ability to bind to p53 (Fig 3F). Through a reporter assay with the Mut7-driven luciferase, we found that ectopic p53 expression resulted in a 4-fold increase of luciferase activity (Fig 3G). The higher sensitivity of Apak/p53-BS-Mut7 compared with Apak/p53-BS-WT (Fig 3B) might be caused by the prevention of endogenous Apak binding. Indeed, Apak overexpression had only weak repression effects on Mut7 activity (Fig 3H), indicating that the direct binding of Apak to the p53AIP1 intron has a critical role in Apak's repression of p53AIP1 transcription.
To obtain further proof that Apak competes with p53 to bind directly to the p53AIP1 intron, we set up an in vitro competition experiment. EMSA showed that increasing amounts of Apak competed and displaced the binding of p53 to Apak/p53-BS but had no significant effect on p53's binding to the Mut7 probe (Fig 3I), strongly indicating that Apak competes with p53 through direct DNA binding. The above data suggest that Apak functions as a transcriptional repressor of p53AIP1 through direct binding to the p53AIP1 intron.
Apak specifically regulates p53AIP1
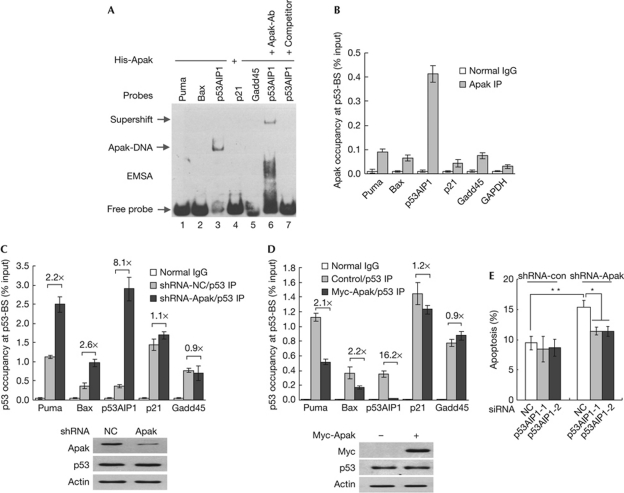
We next examined whether Apak binds to other proapoptotic genes in addition to p53AIP1. EMSA showed that Apak did not bind to the p53-responsive elements of other p53 targets we examined in vitro, including the proapoptotic genes Puma and Bax and the proarrest genes p21 and Gadd45 (Fig 4A). ChIP assays revealed that Apak was co-immunoprecipitated with the p53AIP1 intron with a higher affinity in p53−/− HCT116 cells, while it showed very weak affinity to the promoter of Puma, Bax, p21 or intron 1 of Gadd45 (Fig 4B). Moreover, in p53+/+ HCT116 cells, Apak depletion by short hairpin RNA resulted in a marked increase in the binding of p53 to p53AIP1 gene and a moderate increase in binding to Puma and Bax, but had no significant effect on p21 and Gadd45, as indicated by quantitative ChIP assays (Fig 4C). Conversely, Apak overexpression inhibited the binding of p53 to p53AIP1 gene and, to a lesser extent, to Puma and Bax, but had no significant effect on p21 and Gadd45 (Fig 4D). The observed moderate effects of Apak depletion or overexpression on Puma and Bax should be caused by the altered interaction between Apak and p53 proteins, followed by changes in p53 acetylation, as we reported previously [4]. Consistent with this hypothesis, we previously showed that a decrease in Apak levels caused a 5–17-fold increase in mRNA expression and significant increases in the protein expression of proapoptotic p53 targets, including Puma, p53AIP1, Noxa and FAS, but had only weak effects on proarrest targets, including p21, 14-3-3σ and Gadd45 (Fig 1e in reference 4). Notably, the regulatory effect of Apak on p53AIP1 gene was more significant than that on other proapoptotic genes (Fig 4C,D; reference 4, Fig 1e,g). This should be caused by the unique ‘direct DNA competition’ mechanism on p53AIP1.
Figure 4.
Apak specifically regulates p53AIP1 depending on its DNA-binding activity to regulate apoptosis. (A) The binding activity of Apak to oligonucleotides containing p53-binding sites in the promoter of Puma, Bax and p21, the intron 1 of p53AIP1, and the intron 3 of Gadd45 was determined by EMSA. (B) ChIP assays were performed in p53−/− HCT116 cells using control IgG or anti-Apak antibody, and PCR was performed. Data are means±s.d. (n=3). (C) ChIP assays were performed in Apak-depleted or control p53+/+ HCT116 cells using control IgG or anti-p53 antibody. Data are means±s.d. (n=3). (D) ChIP assays were performed in p53+/+ HCT116 cells harbouring overexpressed Myc-Apak using IgG or anti-p53 antibody. Data are means±s.d. (n=3). (E) HCT116 cells were transfected with Apak shRNA, p53AIP1 siRNA (no. 1 and 2) alone or in combination, and cell apoptosis was measured. Data are means±s.d. (n=3). *P<0.05, **P<0.01. ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IP, immunoprecipitation; NC, non-targeting control; shRNA, short hairpin RNA; siRNA, short interfering RNA.
p53AIP1 was originally identified as the p53-inducible apoptotic target [9]. However, the inhibitory mechanism of the p53AIP1 gene remains poorly understood. These results indicate that Apak specifically regulates p53AIP1 through its DNA-binding activity. As far as we know, the current findings provide the first evidence to identify the transcriptional repressor of p53AIP1.
We then asked whether the inhibition of p53AIP1 is critical for the antiapoptotic effect of Apak. As seen in Fig 4E, Apak depletion alone promoted apoptosis, whereas the simultaneous depletion of p53AIP1 relieved such effects, indicating that Apak regulates apoptosis through the regulation of p53AIP1, at least partially.
Apak dissociates from p53AIP1 upon DNA damage
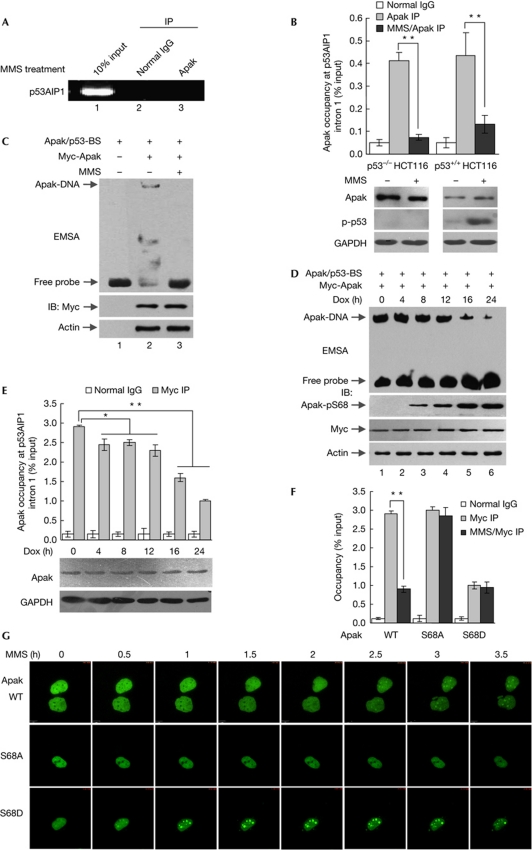
If the DNA-binding activity of Apak contributes to its negative regulation of p53 activity, Apak should dissociate from the p53AIP1 intron in response to DNA damage in order to promote the transcription of p53AIP1. To verify this hypothesis, we investigated the cooperation of Apak with the p53AIP1 gene in response to DNA damage induced by methyl methanesulphonate (MMS). To rule out the involvement of p53 during this response, p53−/− HCT116 cells were used to express ectopic Apak proteins. ChIP assays confirmed that Apak dissociated from the p53AIP1 gene after MMS treatment (Fig 5A,B, columns 1–3). Similar to the case of p53−/− HCT116 cells, Apak dissociated from the p53AIP1 intron in p53+/+ HCT116 cells (Fig 5B, columns 4–6). EMSA showed that the Apak protein/p53AIP1–intron complex could not be formed in the nuclear extract derived from MMS-treated cells, although the Apak protein levels were comparable to the untreated cells (Fig 5C). Thus, the MMS-induced Apak dissociation from the p53AIP1 gene was independent of p53.
Figure 5.
In response to DNA damage, Apak dissociates from the p53AIP1 gene. (A) ChIP assays in MMS-treated p53−/− HCT116 cells. (B) ChIP assays in p53−/− and p53+/+ HCT116 cells that were treated with or without MMS (0.02% for 4 h). Data are means±s.d. (n=3). **P<0.01. (C) Biotin-labelled Apak/p53-BS was used in an EMSA with nuclear extract derived from cultured p53−/− HCT116 cells that were transfected with Myc-Apak and treated with or without MMS. (D) EMSA was performed with Apak/p53-BS and nuclear extract derived from p53−/− HCT116 cells harbouring Myc-Apak and treated with Dox for the indicated times. (E) p53−/− HCT116 cells were transfected with Myc-Apak and treated with Dox. ChIP assays were performed to amplify the p53AIP1 gene. Data are means±s.d. (n=3). *P<0.05; **P<0.01. (F) p53−/− HCT116 cells were transfected with Myc-Apak WT, S68A and S68D, respectively. After treatment with or without MMS for 4 h, ChIP assays were performed to amplify p53AIP1. Data are means±s.d. (n=3). **P<0.01. (G) Subcellular localization analysis of GFP-Apak WT, S68A and S68D in response to MMS treatment. Selected frames from time-lapse movies of representative cells are shown. Dox, doxorubicin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IP, immunoprecipitation; MMS, methyl methanesulphonate; WT, wild type.
Next we asked whether Apak dissociates from the p53AIP1 gene in response to other damage triggers. A relatively mild DNA-damaging agent, doxorubicin, was used to treat p53−/− HCT116 cells. We previously showed that the ATM kinase phosphorylates Apak on Ser68 in response to DNA damage, and that the phosphorylation is critical for p53-induced apoptosis [4, 10]. As expected, doxorubicin treatment induced a gradual increase of Apak phosphorylation on Ser68 without altering Apak protein levels (Fig 5D). During this process, the interaction between Apak and the p53AIP1 intron was gradually decreased, as indicated by both EMSA (Fig 5D) and ChIP (Fig 5E). The phosphorylation level of Apak-Ser68 was inversely related to the binding affinity between Apak and the p53AIP1 gene. The Apak S68A mutation (preventing phosphorylation) blocked the dissociation of Apak from the p53AIP1 gene when damage occurred, whereas the S68D mutation (mimicking phosphorylation) resulted in a significant attenuation of such binding, even in the absence of damage (Fig 5F).
Then we examined whether the subcellular localization of Apak is altered upon DNA damage. GFP-tagged Apak was used to analyse the localization of Apak. GFP-Apak overexpression downregulated the protein level of p53AIP1 but not 14-3-3σ, nor p53; this ability was diminished after treatment with MMS (supplementary Fig S3 online). These results suggested that GFP-Apak was functional and inhibited p53-mediated p53AIP1 induction similar to non-fused Apak. Confocal analysis revealed that GFP-Apak was translocated from the nucleoplasm into the nucleoli at the late stage after MMS treatment (Fig 5G). The Apak-S68A Mut was retained in the nucleoplasm, and Apak-S68D was translocated rapidly into the nucleoli following stress (Fig 5G). The changes in the subcellular localization of Apak proteins were consistent with the alterations of Apak–p53AIP1 intron binding. These results indicated that in response to DNA damage, Apak was phosphorylated and dissociated from the p53AIP1 gene, which might contribute to the removal of Apak repression and the simultaneous p53 activation of the p53AIP1 gene.
Taken together, our work shows that the p53AIP1 intron 1 contains a specific Apak-binding site that overlaps with, but is not identical to, the p53-binding site. Apak directly competes with p53 to bind to the p53AIP1 gene and then inhibits p53AIP1 transcription. These findings provide a new mechanism to explain why Apak selectively inhibits p53-mediated apoptosis. Apak represses the expression of not only p53AIP1 but also Puma, Bax and Noxa, all of which belong to the proapoptotic subset of genes activated by p53 [4]. Therefore, we propose that Apak might utilize two modes to regulate p53 activity. The first is by the recruitment of the corepressor KAP-1 and the histone deacetylase HDAC1 to attenuate p53 acetylation [4], and the other is by competition with p53 binding to the target gene p53AIP1 (this study). Compared with other selective regulators of p53, including Hzf, Brn3, hCAS and ASPP1/2, Apak seems to the only regulator that directly binds to the p53-binding site in the target gene. Therefore, this ‘direct DNA competition’ mechanism provides new insight into selective p53 regulation.
We also found that, in response to DNA damage, Apak dissociates from the p53-responsive element of the p53AIP1 gene. Thus, there is a dynamic interaction between the p53AIP1 gene and Apak. Given that the critical domains of Apak are highly conserved among the approximately 400 members of the KZNF superfamily and that more members of the KZNF family than Apak itself are involved in p53 regulation [4], whether more KZNF proteins have a role in the selective regulation of the diverse downstream outcomes of p53 and whether the current findings can be expanded to other KZNF proteins to explain the selectivity is worthy of further investigation. Moreover, identification of the target sequence of each KZNF protein will surely contribute to the elucidation of its function. In support of this hypothesis, previous studies have identified the specific binding sequences of ZBRK1 [5], K-RBP [11], ZNF333 [6] and ZNF746 [7].
Methods
Cyclic amplification and selection of targets assay. A pool of double-stranded random oligomers was obtained by PCR amplification using the 100-base random oligonucleotide (5′-GTACACATAGAACCAGCAGA-N60-TCTCATACTTGTAGCAGTCG-3′) as the template (125 ng). The double-stranded DNA (∼0.5 μg) was incubated with 0.5 μg of His-Apak protein for 1 h with rotation in a final volume of 150 μl of binding buffer (25 mM HEPES, pH 7.9, 50 mM KCl, 50 mM MgCl2, 0.1% NP-40, 1 mM phenylmethyl sulphonyl fluoride, 0.16 mM dithiothreitol, 10 μM ZnSO4, 10% glycerol) containing 10 μg/ml poly(dI–dC). The resulting Apak/DNA complex was resolved by gel-shift assay selection, and the bound oligonucleotides were recovered and subjected to PCR amplification using primers corresponding to the 20 fixed bases at both ends of the 100-mer. After five rounds of selection and purification, the final PCR product was cloned directly into the pGEM-T easy vector system (Promega), amplified and sequenced. The sequences were analysed, aligned using the BioEdit analysis program and adjusted manually.
Electrophoretic mobility shift assay. The double-stranded oligonucleotides used for EMSA were end-labelled with biotin. The labelled probes were incubated with the protein(s) for 30 min in binding buffer (10 mM Tris–HCl (pH 7.5), 5 mM KCl, 5 mM MgCl2, 10 μM ZnSO4, 50 μg/ml of poly[dI–dC], 5 μg/μl bovine serum albumin, 0.67 mM dithiothreitol, 0.67 mM phenylmethyl sulphonyl fluoride, 2.5% glycerol) in the presence or absence of unlabelled probes. If an antibody was added to detect the supershift, the antibody and protein were pre-incubated for 20 min before the labelled probes were added. The protein/DNA-binding samples were loaded onto a native 6–10% polyacrylamide gel in TBE (Tris/borate/EDTA) buffer and then transferred to a Biodyne membrane. The membrane was blocked and then incubated with the HRP-conjugated streptavidin for 15 min. The membrane was washed three times, treated with SuperSignal (Pierce Biotechnology) detection reagents and exposed to Kodak Light films.
Chip assay. The ChIP assays were performed as described previously [4]. Primer sequences used for all ChIPs are listed in supplementary Table S2 online.
Luciferase reporter assay. The p53AIP1 intron sequence and the mutated sequences were each cloned into the pGL3 luciferase plasmid. The luciferase reporter assays were performed as described previously [4]. After transfection for 48 h, cells were lysed in a passive lysis buffer (Promega). Luciferase activity was measured with the Dual Luciferase Assay System (Promega) in accordance with the manufacturer's protocol.
Statistical analysis. Statistical evaluation was carried out using the Student's t-test.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We are grateful to Drs. Zhixiong Xiao and Qiang Yu for their critical comments on the manuscript. This study was supported by the National Basic Research Programs (2012CB910702, 2011CB910802, 2010CB912202, 2007CB914601), National Natural Science Foundation Projects (30871373, 31125010) and the Beijing Natural Science Foundation Project (5102034).
Author contributions: The project was conceived by L.Z. and the experiments were designed by L.Y., C.T. and L.Z. The experiments were performed by L.Y., C.T., H.W., S.S., D.L. and G.X. The data were analysed by L.Y., C.T., Y.Y., F.H. and L.Z. The manuscript was written by L.Y., C.T. and L.Z.
The authors declare that they have no conflict of interest.
03/14/2012
Unfortunately, due to a typesetting error, two mistakes were originally published in the labelling within Figs 1 and 3. Fig 1C, second line, Mut has been corrected to Mut1. Fig 3I, third line, GAT has been corrected to GST.
References
- Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev Mol Cell Biol 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F (2009) KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 11: 580–591 [DOI] [PubMed] [Google Scholar]
- Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen P, Boyer TG, Lee W (2000) Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell 6: 757–768 [DOI] [PubMed] [Google Scholar]
- Jing Z, Liu Y, Dong M, Hu S, Huang S (2004) Identification of the DNA binding element of the human ZNF333 protein. J Biochem Mol Biol 37: 663–670 [DOI] [PubMed] [Google Scholar]
- Shin J-H et al. (2011) PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's disease. Cell 144: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ et al. (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Oda K et al. (2000) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- Wang S, Tian C, Xiao T, Xing G, He F, Zhang L, Chen H (2010) Differential regulation of Apak by various DNA damage signals. Mol Cell Biochem 333: 181–187 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wen H, Minhas V, Wood C (2009) The zinc finger DNA-binding domain of K-RBP plays an important role in regulating Kaposi's sarcoma-associated herpesvirus RTA-mediated gene expression. Virology 391: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.