Abstract
Mutations in phosphatase and tensin homologue-induced kinase 1 (PINK1) cause recessively inherited Parkinson's disease (PD), a neurodegenerative disorder linked to mitochondrial dysfunction. In healthy mitochondria, PINK1 is rapidly degraded in a process involving both mitochondrial proteases and the proteasome. However, when mitochondrial import is compromised by depolarization, PINK1 accumulates on the mitochondrial surface where it recruits the PD-linked E3 ubiquitin ligase Parkin from the cytosol, which in turn mediates the autophagic destruction of the dysfunctional organelles. Using an unbiased RNA-mediated interference (RNAi)-based screen, we identified four mitochondrial proteases, mitochondrial processing peptidase (MPP), presenilin-associated rhomboid-like protease (PARL), m-AAA and ClpXP, involved in PINK1 degradation. We find that PINK1 turnover is particularly sensitive to even modest reductions in MPP levels. Moreover, PINK1 cleavage by MPP is coupled to import such that reducing MPP activity induces PINK1 accumulation at the mitochondrial surface, leading to Parkin recruitment and mitophagy. These results highlight a new role for MPP in PINK1 import and mitochondrial quality control via the PINK1–Parkin pathway.
Keywords: mitochondria, mitophagy, Parkinson's disease, PINK1, proteases
Introduction
Parkinson's disease (PD) is a common neurodegenerative disorder characterized by both motor and non-motor symptoms [1]. Several lines of evidence implicate mitochondrial dysfunction in PD [2], including the role of two recessive PD genes, Parkin and phosphatase and tensin homologue-induced putative kinase 1 (PINK1), in a new mitochondrial quality control pathway [3]. A landmark study in 2008 showed that on disruption of the electrochemical potential (Δψm) across the inner mitochondrial membrane with the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP), the E3 ubiquitin ligase Parkin translocates from the cytosol to mitochondria [4]. Parkin subsequently mediates the destruction of such defective mitochondria by autophagy (termed mitophagy). A series of follow-up studies showed that the mitochondrial kinase PINK1 is required for Parkin-mediated mitophagy [5]. Under basal conditions PINK1 is rapidly degraded in a process involving Δψm-driven mitochondrial import and cleavage by mitochondrial proteases. Disrupting Δψm with CCCP blocks import and stabilizes PINK1 on the mitochondrial outer membrane, which in turn promotes Parkin recruitment [6].
Given the importance of PINK1 in mitochondrial Parkin recruitment, understanding how PINK1 is cleaved and degraded is crucial for gaining insight into mitochondrial quality control in PD. Here we identify a key step in PINK1 turnover, mediated by the mitochondrial processing peptidase (MPP). Remarkably, MPP links PINK1 proteolysis with its import into mitochondria. MPP knockdown stabilizes PINK1 at the mitochondrial surface, induces Parkin recruitment and leads to mitochondrial clearance, much like CCCP. In contrast, silencing ClpXP, m-AAA or presenilin-associated rhomboid-like protease (PARL), which were also found to affect PINK1 turnover, did not stabilize PINK1 on the surface of mitochondria and failed to promote Parkin recruitment. Thus, MPP seems to have a crucial role in coupling PINK1 import with proteolysis at a proximal step where it is uniquely poised to regulate mitochondrial quality control.
Results And Discussion
PINK1 is regulated by MPP, PARL, m-AAA and ClpXP
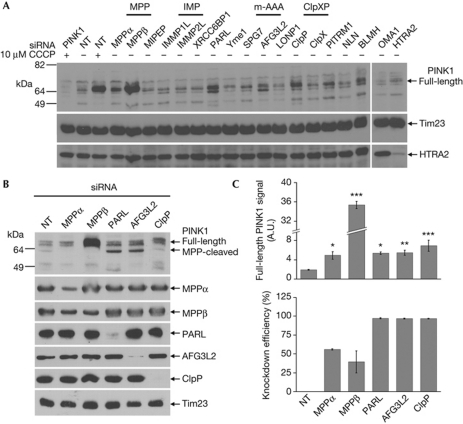
To better understand the regulation of PINK1/Parkin-mediated mitochondrial quality control, we sought to identify the mitochondrial protease(s) involved in PINK1 cleavage. We reasoned that interfering with a PINK1-cleaving protease should induce full-length PINK1 accumulation even in the absence of CCCP. Using an short interfering RNA (siRNA)-based screen of known mitochondrial proteases, we found that knockdown of MPPβ induced a large increase of full-length PINK1 levels in the mitochondrial fraction of HEK293T cells, similar to the increase produced by treatment with 10 μM CCCP for 3 h (Fig 1A). MPPβ is the catalytic subunit of the dimeric matrix protease MPP, responsible for cleaving the mitochondrial targeting sequence off of newly imported precursor proteins [7]. Knockdown of MPPα, the MPP subunit thought to be involved in substrate recognition [7], also increased PINK1 levels, albeit more modestly. Our screen identified three more mitochondrial proteases that, upon knockdown, induced the accumulation of full-length PINK1: ClpP, the catalytic subunit of the heterodimeric matrix protease ClpXP, AFG3L2, a subunit of the m-AAA protease and PARL, which mediates intramembrane cleavage at the inner membrane [8]. Knockdown of PITRM1 and BLMH also seemed to modestly increase PINK1 levels, but this was not replicated in an independent repeat of the screen (data not shown), and neither protease was pursued further. Similarly, neither HTRA2 nor Yme1 increased PINK1 levels despite their efficient knockdown (Fig 1A, supplementary Fig S1A online), supporting the specificity of our findings.
Figure 1.
PINK1 processing is regulated by MPP, PARL, m-AAA and ClpXP. (A) Immunoblots of mitochondrial fractions from siRNA-transfected HEK293T cells±CCCP. (B) Immunoblots against indicated proteases in siRNA-transfected cells. (C) Quantification of full-length PINK1 (n=4) and knockdown efficiency (n=2) compared with non-targeting (NT) siRNA from experiments performed as in B. *P<0.05; **P<0.01; ***P<0.001. CCCP, carbonyl cyanide m-chlorophenyl hydrazone; MPP, mitochondrial processing peptidase; PARL, presenilin-associated rhomboid-like protease; PINK1, phosphatase and tensin homologue-induced kinase 1; siRNA, short interfering RNA.
Knockdown of PARL, which was reported previously to regulate PINK1 cleavage [6, 9, 10, 11], and AFG3L2 also increased the levels of a cleavage fragment running just under the full-length PINK1 band (Fig 1B, upper panel and supplementary Fig S1B online). The size of this fragment and its loss on knockdown of either MPP subunit suggests that it is the product of PINK1 cleavage by MPP. It thus seems that PINK1 is cleaved in at least two steps, first by MPP, then by PARL and m-AAA. Indeed, simultaneous knockdown of PARL and AFG3L2 further increased the accumulation of this MPP-cleaved PINK1 fragment, whereas full-length PINK1 was only slightly increased (supplementary Fig S1C online). Together, these observations indicate that PARL and m-AAA are redundant in PINK1 processing and act downstream of MPP.
Given that many mitochondrial proteases require proteolytic processing to reach their mature, active forms, it is possible that one or more of the five hits identified in our screen exerts its effect on PINK1 indirectly. A particular concern was that knockdown of MPP, which is presumed to mediate processing of the newly imported PARL precursor [12], might elevate PINK1 levels by decreasing the amount of active PARL. To address this we assessed the levels of the mature forms of each of our five proteases in the context of each of the five knockdowns. In general, knockdown of each protease had little, if any, effect on the levels of the other four (Fig 1B). The knockdown of MPPβ in particular, which was fairly modest compared with the silencing efficiencies for PARL, AFG3L2 and ClpP (Fig 1C, lower panel), had no discernable effect on the levels of the other proteases, including PARL, despite producing a large accumulation of PINK1. These results confirm the specificity of the five knockdowns and indicate that MPP affects PINK1 levels directly rather than via one of the other proteases. They also show that PINK1 turnover is particularly sensitive to even small changes in MPP levels and indicate that MPP cleavage might be a rate-limiting step in PINK1 processing.
One notable exception to the findings above was that siRNA directed against MPPα destabilized its dimeric partner MPPβ, yet did not lead to a comparable increase in PINK1 levels (Fig 1B,C). This suggested that the PINK1 accumulation observed using MPPβ siRNA might be due to an off-target effect. However, of the four distinct siRNAs from the MPPβ pool used above, the three that successfully knocked down MPPβ when transfected individually also caused PINK1 accumulation, while the one that failed to reduce MPPβ expression did not (supplementary Fig S2A online). This argues that the effect of MPPβ siRNA on PINK1 is in fact specific to reducing MPPβ levels. Moreover, knocking down MPPα and MPPβ together led to only a modest increase in PINK1 levels, comparable with the knockdown of MPPα alone (supplementary Fig S2B online). This also argues strongly against an off-target effect of the MPPβ knockdown and indicates that silencing MPPα can suppress the effects of knocking down MPPβ on PINK1 accumulation. While the mechanism of this suppression is unclear, one possibility might involve a requirement of the non-catalytic α-subunit of MPP to bind the PINK1 mitochondrial targeting sequence [13], in order for PINK1 to accumulate on mitochondria in the absence of the catalytic β-subunit. Finally, transfection of rat MPPβ, which is resistant to the siRNA pool directed against human MPPβ, into our knockdown cells eliminated the PINK1 accumulation observed with MPPβ siRNA alone (supplementary Fig S2C online). This ‘rescue’ experiment firmly rules out that the accumulation of PINK1 was due to an off-target effect of the MPPβ siRNAs.
Protease siRNAs do not affect mitochondrial integrity
The most straightforward interpretation of our findings so far is that MPPβ, PARL, AFG3L2 and ClpP are directly involved in PINK1 processing. However, these proteases mediate the processing and maturation of many proteins involved in essential mitochondrial functions. Thus, the knockdowns might profoundly disrupt mitochondrial integrity, which could in turn affect PINK1 indirectly, as it is known to accumulate on dysfunctional mitochondria. To survey mitochondrial protein processing and maturation, we carried out immunoblots against a panel of proteins present across all mitochondrial subcompartments. We found little effect on the levels and processing of most mitochondrial proteins (supplementary Fig S3A online), consistent with a good overall preservation of mitochondrial integrity, at least for the degree of silencing and time scale (96 h) of the experiment. In particular, even the processing of established substrates of MPP such as MnSOD, OPA1 and Rieske was affected only marginally or not at all by the MPPβ siRNA, presumably because the knockdown was incomplete (Fig 1C). This further highlights the sensitivity of PINK1 to changes in MPPβ levels, and suggests that very modest degrees of MPP inhibition might induce PINK1 accumulation without otherwise broadly affecting mitochondrial protein levels.
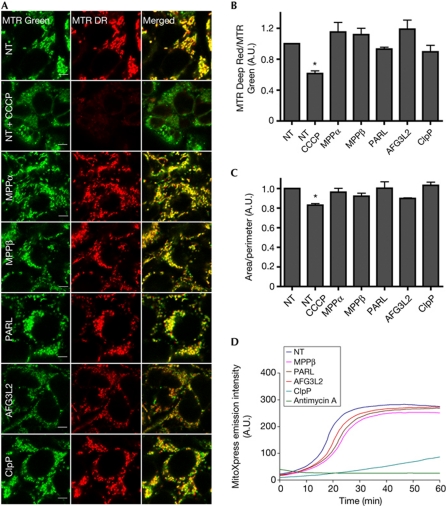
Given that PINK1 is known to accumulate on depolarized mitochondria, it was also critical to determine the effects of the knockdowns on Δψm directly. The siRNA-treated cells were imaged live by confocal microscopy after staining with the potentiometric dye MitoTracker Deep Red, as well as with MitoTracker Green, which accumulates independently of membrane potential (Fig 2A). Using the red to green ratio as a measure of mitochondrial polarization, we found that the protease knockdowns had no significant effect on mitochondrial membrane potential (Fig 2B). These results were confirmed using flow cytometry with the fluorescent potentiometric mitochondrial probe JC1 (supplementary Fig S3B online). In contrast, treatment with 10 μM CCCP for 3 h as a positive control reduced Δψm as expected (Fig 2B, supplementary Fig S3B online), but had no effect on protease levels (supplementary Fig S3C online). Moreover, mitochondrial network interconnectivity, a measure of the net effect of mitochondrial fusion and fission that is often altered with mitochondrial stress and cell death [14], was not appreciably affected by the protease knockdowns (Fig 2C). However, mitochondria in AFG3L2 and PARL knockdowns did appear slightly more fragmented and aggregated, respectively (Fig 2A). In contrast, MPPα, MPPβ and ClpP knockdowns displayed a mix of elongated and fragmented mitochondria, as seen with non-targeting siRNA, although ClpP silencing was nonetheless associated with a high level of cell death (data not shown).
Figure 2.
Modest effects of transient MPPβ, PARL and AFG3L2 knockdown on mitochondrial integrity. (A) Live confocal micrographs of siRNA-transfected HEK293T cells±CCCP stained with MitoTracker (MTR) Green and Deep Red (DR); Scale bar, 5 μm. (B) MTR DR/Green ratio from images as in A. N=3; *P<0.05 compared with NT siRNA. (C) Mitochondrial interconnectivity from images as in A, N=3. (D) Oxygen consumption in state 3 respiration in mitochondrial fractions from siRNA-transfected cells. Antimycin A with NT siRNA was used as a control. A.U., arbitrary unit; CCCP, carbonyl cyanide m-chlorophenyl hydrazone; MPP, mitochondrial processing peptidase; NT, non-targeting; PARL, presenilin-associated rhomboid-like protease; siRNA, short interfering RNA.
To further assess mitochondrial function, we monitored ADP-stimulated (state 3) oxygen consumption using the MitoXpress probe and succinate as a substrate. As shown in Fig 2D, mitochondria from the knockdowns consumed oxygen at a reduced rate compared with those from cells transfected with non-targeting siRNA. However, the defect was minor except for ClpP, which displayed a major oxygen consumption deficit, almost to the degree observed with the complex III inhibitor antimycin A. Similar results were found for unstimulated (state 2) respiration, although in this case the ClpP knockdown deficit was less pronounced (data not shown), which might explain the lack of depolarization found above, under basal conditions (Fig 2B, supplementary Fig S3B online).
Given the respiration deficit and cell death observed with ClpP knockdown, results obtained for PINK1 with the ClpP siRNA should be interpreted with caution. However, the fact that neither Δψm (Fig 2B, supplementary Fig S3B online) nor the levels of mitochondrial proteins (Fig 1B, supplementary Fig S3A online) were significantly affected with ClpP siRNA suggests that several key mitochondrial functions were preserved at steady state, arguing against an indirect effect of ClpP on PINK1. More importantly, our findings indicate that, overall, the transient knockdowns of MPPβ, PARL and AFG3L2 achieved in our experiments do not lead to a profound disruption of mitochondrial integrity and function, consistent with the proteases participating in PINK1 processing directly. Moreover, they support the observation that, possibly because of PINK1's rapid turnover, it is unusually sensitive to changes in the levels of mitochondrial proteases, particularly MPPβ.
PARL and AFG3L2 generate a 52kDa PINK fragment
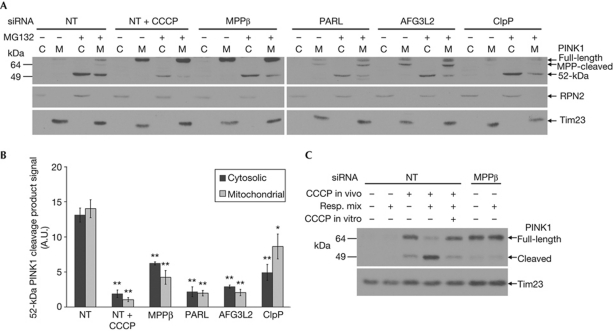
PINK1 processing has previously been shown to generate a 52-kDa cleavage product that is rapidly degraded by the proteasome, as evidenced by its accumulation with the proteasomal inhibitors MG132 or epoxomycin [3, 15]. To examine which proteases are required for 52-kDa PINK1 formation, we induced the fragment's accumulation by treating with MG132. As expected, prior and concomitant treatment of cells with CCCP dramatically reduced the 52-kDa PINK1 accumulation in both the cytosolic and mitochondrial fractions (Fig 3A,B). This effect was mimicked by knockdown of PARL or AFG3L2, and to a lesser extent by knockdown of MPPβ or ClpP, indicating that these proteases are involved in the formation of the 52-kDa PINK1 fragment. Moreover, knockdown of PARL or AFG3L2 significantly increased the levels of the putative MPP-processed band, which runs just below the full-length fragment (Figs 1B,3A, supplementary Fig S1B online). Conversely, silencing either MPP subunit significantly decreased the levels of this band. Together, these findings indicate that this band is an intermediate PINK1 species, produced by MPP-mediated cleavage and available for further processing by PARL and AFG3L2 into the mature 52-kDa form.
Figure 3.
The levels of mature 52-kDa PINK1 are regulated by MPPβ, PARL, AFG3L2, ClpP and the proteasome. (A) Immunoblots of mitochondrial (M) and cytosolic (C) fractions from siRNA-transfected HEK293T cells±CCCP for 3.5 h, ±MG132 for the final hour. (B) Quantification of 52-kDa PINK1 from A (with MG132), n=3; *P<0.05; **P<0.001 compared with NT siRNA. (C) Mitochondria from siRNA-transfected cells treated with 10 μM CCCP or dimethylsulphoxide were incubated with or without respiration substrate (Resp. mix) for 30 min at 37°C. CCCP, carbonyl cyanide m-chlorophenyl hydrazone; MPP, mitochondrial processing peptidase; NT, non-targeting; PARL, presenilin-associated rhomboid-like protease; PINK1, phosphatase and tensin homologue-induced kinase 1; siRNA, short interfering RNA.
To more directly establish the importance of MPPβ in PINK1 processing, we used an in organello PINK1 cleavage assay. Mitochondria from HEK293T cells that had been treated with or without CCCP were isolated and incubated at 37 °C with or without succinate as a respiration substrate. Including succinate led to rapid conversion of full-length PINK1 accumulated by CCCP treatment into the 52-kDa form (Fig 3C), presumably via reestablishment of Δψm and PINK1 import, thus permitting cleavage. Indeed, when CCCP was included in the reaction buffer the formation of 52-kDa PINK1 was completely blocked. When MPPβ knockdown was used instead of CCCP treatment to accumulate full-length PINK1, incubation with succinate did not induce the production of the 52-kDa cleavage form, confirming the requirement for MPPβ in PINK1 processing.
MPPβ knockdown blocks PINK1 import
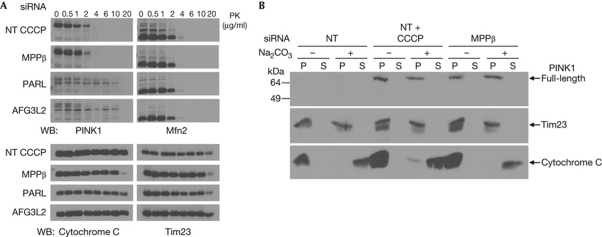
To determine the localization of the various PINK1 forms discussed so far, we added increasing amounts of proteinase K externally to mitochondria isolated from cells with protease knockdowns or treated with CCCP. Fig 4A shows that the MPP-cleaved form of PINK1 that accumulates upon knockdown of PARL or AFG3L2 is more resistant to proteinase K in comparison with full-length PINK1 accumulated with either CCCP treatment or MPPβ knockdown. This is consistent with the former being protected within mitochondria, while the latter is exposed at the mitochondrial surface. The findings are also consistent with previous work showing that depolarizing mitochondria with CCCP blocks PINK1 import, and indicate that reducing MPPβ levels mimics this effect. MPP-mediated cleavage of newly imported precursor proteins is thought to be tightly coupled to import itself [16], and there is long-standing evidence to indicate that the protease is actually required for import [17], although this is controversial [18]. Our data support a role for MPP in coupling proteolysis of PINK1 with mitochondrial import.
Figure 4.
Knockdown of MPPβ leads to full-length PINK1 accumulation at the outer mitochondrial membrane. (A) Mitochondria from CCCP-treated or siRNA-transfected HEK293T cells were incubated with increasing concentrations of proteinase K (PK). (B) Mitochondria from CCCP-treated or MPPβ knockdown cells were incubated ±0.1 M Na2CO3 before supernatant (S) and pellet (P) were separated by centrifugation. CCCP, carbonyl cyanide m-chlorophenyl hydrazone; MPP, mitochondrial processing peptidase; NT, non-targeting; PARL, presenilin-associated rhomboid-like protease; PINK1, phosphatase and tensin homologue-induced kinase 1; siRNA, short interfering RNA; WB, western blot.
Our findings so far indicate that the effects of MPPβ knockdown mimic several key features of CCCP treatment on PINK1 processing, including inhibition of PINK1 import and cleavage. To further characterize the effects of MPPβ siRNA and CCCP at a biochemical level, we used sodium carbonate to release soluble and peripheral membrane proteins from organelles, allowing them to be separated from integral membrane proteins by centrifugation. In both cases, PINK1 was retained within the pellet, much like the integral membrane protein Tim23 and unlike the peripheral membrane protein Cytochrome C (Fig 4B). The most straightforward interpretation is that PINK1 is laterally released from the translocase of the outer membrane (TOM) complex into the lipid phase of the outer membrane when import is arrested, either by disrupting Δψm or by interfering with MPP cleavage. Lateral release of import substrates from the TOM complex has been shown previously [19], although the process remains poorly understood. Nonetheless, this interpretation is difficult to reconcile with the finding that PINK1 processing (and presumably therefore import) can resume on washout of CCCP and reestablishment of Δψm (Fig 3C), as the reversible movement of an integral membrane protein back into the pore of the TOM complex has not previously been described. Certain other import substrates such as the F1-ATPase subunit-β seem to remain associated with the TOM complex when import is arrested by CCCP, as manifested by their sensitivity to carbonate stripping [20]. This persistent association with the TOM complex is believed to allow import to resume once CCCP is washed out and Δψm is reestablished [21]. Thus, another possible interpretation of our findings is that, on arrest of import, PINK1 remains within or adjacent to the TOM complex but adopts a conformation that makes it resistant to high pH stripping.
MPPβ knockdown induces mitophagy
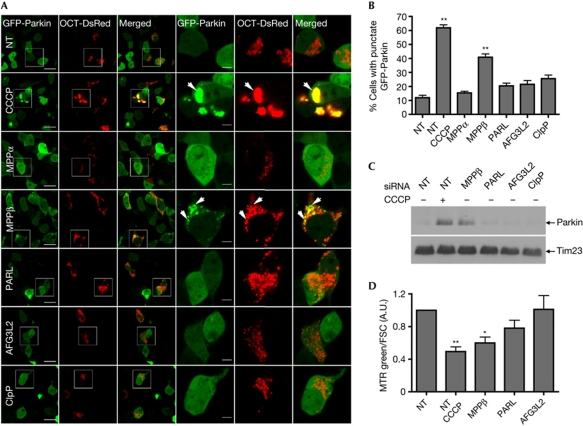
Regardless of its precise localization, our findings indicate that full-length PINK1 accumulates at the outer surface of mitochondria on MPPβ knockdown or CCCP treatment. We therefore asked whether MPPβ siRNA could mimic CCCP by inducing Parkin recruitment to mitochondria. We coexpressed green fluorescent protein (GFP)-Parkin with the mitochondrial marker OCT-DsRed in the knockdown cells and used confocal microscopy to image the cells live 24 h after transfection. Both CCCP and MPPβ siRNA induced substantial GFP-Parkin translocation to mitochondria, whereas siRNA against MPPα, PARL, AFG3L2 or ClpP did not (Fig 5A). Moreover, while both CCCP and MPPβ siRNA led to an increase in the proportion of cells showing a punctate mitochondrial pattern of GFP-Parkin distribution (Fig 5B), several differences were apparent. Indeed, MPPβ knockdown primarily induced GFP-Parkin recruitment to isolated mitochondria (Fig 5A). In contrast, CCCP induced larger perinuclear GFP-Parkin-positive mitochondrial clusters, as reported previously [22]. It is also worth noting that in MPPβ knockdown cells, GFP-Parkin puncta occasionally did not colocalize with OCT-DsRed (Fig 5A, arrows). While the identity of these structures is uncertain, they may represent mitochondria undergoing mitophagy, as might be expected after 24 h of GFP-Parkin overexpression in cells with significant mitochondrial surface accumulation of PINK1 induced by MPPβ silencing. Importantly, knocking down MPPβ also increased the levels of endogenous Parkin in the mitochondrial fraction to a similar extent as CCCP (Fig 5C). Consistent with our results above using overexpressed GFP-Parkin (Fig 5A,B) and with our PINK1 localization studies (Fig 4), endogenous Parkin recruitment was not observed on knockdown of PARL, AFG3L2 or ClpP.
Figure 5.
MPPβ knockdown induces Parkin recruitment and mitophagy. (A) Live confocal micrographs of siRNA-, GFP-Parkin- and OCT-DsRed-transfected HEK293T cells ±CCCP; Scale bars, 20 μm (low magnification) and 5 μm (high magnification). (B) Percentage of transfected cells displaying a punctate mitochondrial pattern of GFP-Parkin distribution, n=3; *P<0.05; **P<0.01 compared with NT siRNA. (C) Immunoblots of endogenous Parkin in mitochondrial fractions from siRNA-transfected cells ±CCCP. (D) Mitochondrial mass measured by flow cytometry of MTR Green fluorescence in siRNA-transfected cells treated ±CCCP for 24 h, n=3. A.U., arbitrary unit; CCCP, carbonyl cyanide m-chlorophenyl hydrazone; GFP, green fluorescent protein; MPP, mitochondrial processing peptidase; MTR, MitoTracker; NT, non-targeting; PARL, presenilin-associated rhomboid-like protease; PINK1, phosphatase and tensin homologue-induced kinase 1; siRNA, short interfering RNA.
Finally, we examined whether Parkin recruitment induced by MPPβ knockdown could trigger mitophagy, as has been shown for CCCP-induced Parkin recruitment. We used the non-potentiometric dye MitoTracker Green to stain cells treated with CCCP or siRNA, and found that both CCCP and MPPβ knockdown significantly decreased mitochondrial mass (Fig 5D). This shows that knocking down MPPβ is sufficient to clear mitochondria from cells even without overexpressing Parkin. Presumably, the accumulation of PINK1 on the mitochondrial surface induced by silencing MPPβ leads to the recruitment of endogenous parkin, which targets the mitochondria for degradation via the autophagy pathway.
Conclusions
Our study adds weight to the notion that PINK1 import into, and cleavage within, mitochondria are responsible for keeping PINK1 levels low under basal conditions so that they may be specifically and rapidly elevated when mitochondria become dysfunctional. Our findings support the previously identified role for PARL in PINK1 cleavage, and implicate three more proteases, MPP, m-AAA and ClpXP, in the cleavage of this PD-linked protein (see supplementary Fig S4 online for model). Mutations in the AFG3L2 subunit of m-AAA have been linked to the neurodegenerative disorder dominant hereditary ataxia SCA28 [23], and it will be interesting to examine whether altered PINK1 cleavage has a role in this disease. As a considerable proportion of endogenous PINK1 is found in the 52-kDa form (Fig 1A), it will also be interesting to determine whether this cleavage product exerts a physiological function, rather than simply being a degradation intermediate.
Importantly, we have shown that a relatively modest knockdown of MPPβ, which has little overall effect on mitochondrial integrity, is sufficient to induce Parkin recruitment to mitochondria and promote mitochondrial clearance. MPPβ inhibition may therefore represent a useful model for future studies of the mitophagy pathway. In addition, based on our findings it is possible that tonic, low-level inhibition of MPPβ activity could induce a moderate increase in PINK1 levels, Parkin recruitment and mitophagy of defective organelles, without otherwise broadly affecting mitochondrial function. This could represent an exciting therapeutic opportunity for slowing the progression of PD by reducing the accumulation of mitochondrial defects.
Methods
Confocal microscopy. Cells were imaged live with or without MitoTracker Green and MitoTracker Deep Red (Invitrogen) staining on MatTek glass-bottom dishes on a Zeiss LSM710 microscope using a × 63 Plan Apo objective (Zeiss). Mitochondrial interconnectivity and MitoTracker Deep Red fluorescence/Total MitoTracker Green fluorescence ratio were analysed using ImageJ (National Institute of Health).
In organello cleavage assay. Mitochondria from cells treated with either 10 μM CCCP or dimethylsulphoxide for 3 h were enriched and resuspended in reaction buffer (100 mM sucrose, 100 mM KCl, 2 mM KH2PO4, 10 μM EGTA, 5 mM Tris, pH 7.4). The mitochondria were incubated at 0.8 μg/μl in 30 μl reaction buffer with or without respiration mix (1 mM ATP, 0.08 mM ADP, 5 mM NaSuccinate, 2 mM K2HPO4, pH 7.5) and 1 μM CCCP for 30 min at 37°C.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Luca Pellegrini for the PARL antibody, and Robert Screaton for advice about siRNA. We thank Eric Shoubridge for critical reading of the manuscript. This work was supported by an Operating Grant from the Canadian Institutes of Health Research (CIHR) and by the Brain Repair Program from Neuroscience Canada to E.A.F. and D.S.P. A.W.G. is supported by the Fonds de recherche en Santé du Québec (FRSQ). K.G. and H.M.M. are supported by CIHR. D.S.P. is supported by Heart and Stroke Foundation of Ontario (HSFO). E.A.F. is a Chercheur National of the FRSQ.
Author contributions:Experiments were performed by A.W.G., K.G., M.A.A. and S.M. A.W.G., K.G. and E.A.F. designed and analysed the experiments and wrote the article. R.F., M.E.H., H.M.M. and D.S.P. provided important advice and insights.
Footnotes
The authors declare that they have no conflict of interest.
References
- Shulman JM, De Jager PL, Feany MB (2011) Parkinson's disease: genetics and pathogenesis. Annu Rev Pathol 6: 193–222 [DOI] [PubMed] [Google Scholar]
- Abou-Sleiman PM, Muqit MM, Wood NW (2006) Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat Rev Neurosci 7: 207–219 [DOI] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Przedborski S (2011) Mitophagy: the latest problem for Parkinson's disease. Trends Mol Med 17: 158–165 [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakh O, Cavadini P, Isaya G (2002) Mitochondrial processing peptidases. Biochim Biophys Acta 1592: 63–77 [DOI] [PubMed] [Google Scholar]
- Koppen M, Langer T (2007) Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol 42: 221–242 [DOI] [PubMed] [Google Scholar]
- Deas E et al. (2010) PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 20: 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK (2011) The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem 117: 856–867 [DOI] [PubMed] [Google Scholar]
- Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, Ross OA, Farrer M, McQuibban GA, Bulman DE (2011) Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson's disease. Hum Mol Genet 20: 1966–1974 [DOI] [PubMed] [Google Scholar]
- Sik A, Passer BJ, Koonin EV, Pellegrini L (2004) Self-regulated cleavage of the mitochondrial intramembrane-cleaving protease PARL yields Pbeta, a nuclear-targeted peptide. J Biol Chem 279: 15323–15329 [DOI] [PubMed] [Google Scholar]
- Dvorakova-Hola K, Matuskova A, Kubala M, Otyepka M, Kucera T, Vecer J, Herman P, Parkhomenko N, Kutejova E, Janata J (2010) Glycine-rich loop of mitochondrial processing peptidase alpha-subunit is responsible for substrate recognition by a mechanism analogous to mitochondrial receptor Tom20. J Mol Biol 396: 1197–1210 [DOI] [PubMed] [Google Scholar]
- Campello S, Scorrano L (2010) Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep 11: 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Kang UJ (2008) Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem 106: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano P, Geoffroy S, Brandt A, Hernandez JF, Geli V (1997) Functional cooperation of the mitochondrial processing peptidase subunits. J Mol Biol 272: 213–225 [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G (1984) Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA 81: 4819–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli V, Yang MJ, Suda K, Lustig A, Schatz G (1990) The MAS-encoded processing protease of yeast mitochondria. Overproduction and characterization of its two nonidentical subunits. J Biol Chem 265: 19216–19222 [PubMed] [Google Scholar]
- Harner M, Neupert W, Deponte M (2011) Lateral release of proteins from the TOM complex into the outer membrane of mitochondria. EMBO J 30: 3232–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Hartl FU, Guiard B, Neupert W (1987) Mitochondrial precursor proteins are imported through a hydrophilic membrane environment. Eur J Biochem 169: 289–293 [DOI] [PubMed] [Google Scholar]
- Rapaport D, Mayer A, Neupert W, Lill R (1998) cis and trans sites of the TOM complex of mitochondria in unfolding and initial translocation of preproteins. J Biol Chem 273: 8806–8813 [DOI] [PubMed] [Google Scholar]
- Okatsu K et al. (2010) p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella D et al. (2010) Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat Genet 42: 313–321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.