Abstract
The transcription factor Eomesodermin (Eomes) is involved in early embryonic patterning, but the range of cell fates that it controls as well as its mechanisms of action remain unclear. Here we show that transient expression of Eomes promotes cardiovascular fate during embryonic stem cell differentiation. Eomes also rapidly induces the expression of Mesp1, a key regulator of cardiovascular differentiation, and directly binds to regulatory sequences of Mesp1. Eomes effects are strikingly modulated by Activin signalling: high levels of Activin inhibit the promotion of cardiac mesoderm by Eomes, while they enhance Eomes-dependent endodermal specification. These results place Eomes upstream of the Mesp1-dependent programme of cardiogenesis, and at the intersection of mesodermal and endodermal specification, depending on the levels of Activin/Nodal signalling.
Keywords: Eomesodermin, Mesp1, Nodal/Activin, cardiac fate, embryonic stem cell
Introduction
During early stages of embryonic development, the three germ layers (endoderm, mesoderm and ectoderm) are specified in a process called gastrulation. Gastrulation begins with the formation of the primitive streak (PS), in which epiblast cells ingress to form the endoderm and the mesoderm. Interplay between extrinsic and intrinsic cues regulates the different cell fates and patterning of the early embryo [1, 2]. A better understanding of these early events is critical for improving the differentiation of pluripotent stem cells into clinically relevant cell types [3].
Among the known extrinsic cues, Nodal/Activin signalling has a crucial role in regulating mesoderm and endoderm specification [4]. Nodal is expressed in epiblast cells and acts to promote posterior genes such as Wnt3 and Eomesodermin (Eomes), which are required for mesoderm formation [5]. In the absence of Nodal, no PS is formed, which results in ectopic neural differentiation [6]. During embryonic development and embryonic stem cell (ESC) differentiation, PS derivatives that give rise to definitive endoderm (DE) require a higher intensity of Nodal/Activin signals than posterior derivatives, which will differentiate into mesoderm [7–17].
The T-box transcription factors Brachyury and Eomes are among the well-known intrinsic cues that are critical in patterning the PS [18]. Eomes was initially identified in Xenopus to initiate mesoderm differentiation [19, 20], but subsequent studies in mouse [21, 22] and zebrafish [23] implied that Eomes was necessary for DE rather than mesoderm specification. This was further substantiated by a recent study, in which overexpression of Eomes in differentiating ESC promoted endodermal fate [7].
Here we investigated the role of Eomes in early cell fate decisions during mouse ESC differentiation, and found that in the absence of extrinsic signals, Eomes promoted the formation of cardiac mesoderm by stimulating the expression of Mesp1, a transcription factor known to have a key role in the specification of cardiovascular mesoderm [24–28]. High levels of Activin decreased the ability of Eomes to stimulate Mesp1 expression, and instead led to the induction of endodermal fate. These data illustrate how a single intrinsic cue can induce distinct fates depending on the levels of an extrinsic signal.
Results And Discussion
Eomes promotes cardiogenesis in ESC
To explore the impact of Eomes on early ESC differentiation with minimal extrinsic influence, we took advantage of a system developed recently, where ESCs are cultured as a monolayer in a chemically defined default medium (DDM) devoid of any added exogenous morphogen, except insulin [29, 30]. In these conditions, most of the cells differentiated spontaneously into a neural fate [29–32]. To study the role of Eomes, we generated a recombinant ESC line, in which the expression of a Myc-tagged version of mouse Eomes can be induced on doxycyclin (Dox) addition [33, 34] (Fig 1A and supplementary Fig S1 online).
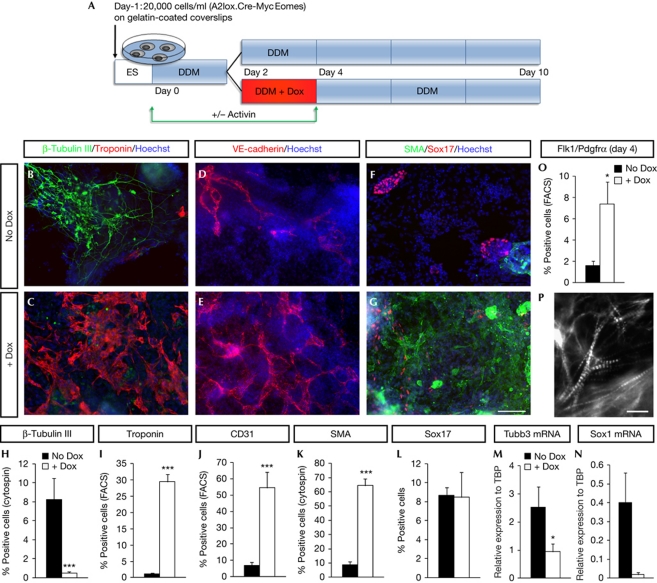
Figure 1.
Induction of Eomesodermin (Eomes) during embryonic stem cell (ESC) differentiation promotes development of cardiovascular mesoderm. (A) Schematic representation of the differentiation protocol. (B–G) ESCs were cultured in defined default medium (DDM) in the presence or absence of doxycyclin (Dox) at days 2–3, and immunostained for β-tubulin III and a cardiac-specific isoform of Troponin T (cTnT; B,C), VE-cadherin (D,E) or Sox17 and smooth muscle actin (SMA; F,G). Nuclei are stained with Hoechst dye. Scale bar, 100 μm. (H–L) Quantification of the cells expressing β-tubulin III (H), cTnT (I), CD31 (J), SMA (K) or Sox17 (L). Quantification was performed by FACS (I,J), counting after cytospin (H,K) or counting of cells cultivated on the coverslip (L). Data are presented as mean percentage of all cells+s.e.m. (M,N) Quantitative reverse transcription–PCR analysis for Tubb3 (M) and Sox1 (N) at day 10 with or without Dox. Data are presented as mean expression normalized to TBP+s.e.m. (O) Flow cytometric analysis of Flk1–Pdgfrα double-positive cells at day 4 (48 h after addition of Dox). Data are presented as mean percentage of all cells+s.e.m. (P) Higher magnification shows cTnT-positive striated myofibrils. Scale bar, 10 μm.
In the absence of Dox administration, most cells generated after 10 days corresponded to neural cells and very few cells showed expression of mesodermal or endodermal markers (Fig 1, supplementary Fig S2 online), as previously reported [30]. Transient induction of Eomes by administration of Dox at day 2 of ESC differentiation promoted a completely distinct differentiation outcome, as suggested by the appearance of beating areas (data not shown). Immunofluorescence, quantitative reverse transcription PCR (qRT–PCR) and FACS analyses performed 8 days after Eomes induction revealed a strong induction of the cardiac-specific isoform of TroponinT (cTNT) and a converse reduction of β-tubulin III, Sox1- and Pax6-expressing neural cells (Fig 1, supplementary Figs S2A,C,D and S4A online). cTnT staining revealed a striated pattern characteristic of cardiomyocyte myofibrils (Fig 1P). Eomes overexpression also promoted the appearance of endothelial cells (expressing VE-cadherin and CD31; Fig 1D,E,J) and smooth muscle cells (expressing smooth muscle actin; Fig 1F,G,K and supplementary Fig S2B online). Altogether, the three types of cells derived from multipotent cardiovascular progenitors (MCPs) [27, 35–37] were massively increased following Eomes expression in DDM, although we cannot rule out that some endothelial and smooth muscle cells could also derive from other progenitors (such as the hemangioblasts [38] that could also be induced following Eomes expression). We and others have recently demonstrated that a combination of monoclonal antibodies (Flk1/Pdgfrα) can be used to isolate the earliest Mesp1 expressing MCPs arising during ESC differentiation [16, 27]. FACS analysis of ESC 48 h after Eomes overexpression in DDM revealed a significant enrichment for Flk1/Pdgfrα-positive MCPs in Eomes overexpressing cells (Fig 1O, supplementary Fig S2E online), consistent with the increase of Mesp1 expressing MCPs by Eomes in these conditions. Interestingly, the number of Sox17-expressing endodermal cells at day 10 remained low despite high levels of Eomes expression (Fig 1F,G,L). Overall, these data indicate that Eomes induction leads to robust promotion of cardiovascular fate from ESC in the absence of added morphogens, while neural fate is strongly inhibited, and endodermal fate seems essentially unchanged.
Eomes' effects are modulated by Activin signalling
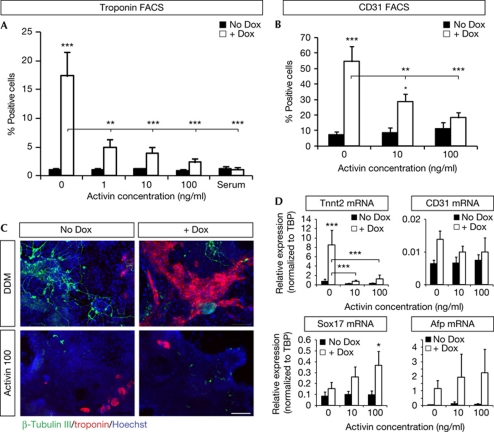
Promotion of mesodermal fates, and absence of induction of DE fates following Eomes gain of function during ESC differentiation were surprising, given the well-known effect of Eomes in promoting DE specification during embryonic development [22] and ESC differentiation [7, 39]. As those results were obtained in the presence of serum and/or various extrinsic cues such as Activin/Nodal, we next examined the effect of Eomes in our reductionist system, but in the presence of either serum or with increasing levels of Activin. Most strikingly, the presence of serum or Activin inhibited the promoting effect of Eomes on cardiomyocyte differentiation, in a dose-dependent manner. This was reflected by a sharp decrease of cTnT and CD31-positive cells as quantified by FACS (Fig 2A,B, supplementary Fig S4B online), immunofluorescence (Fig 2C, supplementary Fig S2A online) and qRT–PCR (Fig 2D), as well as by the disappearance of beating zones in differentiated ESC (data not shown). Conversely, endodermal markers Sox17 and α-fetoprotein were induced by Eomes mostly in the presence of high doses of Activin (Fig 2D).
Figure 2.
High concentrations of Activin prevent Eomesodermin (Eomes)-stimulated cells from differentiating into cardiovascular mesoderm. Embryonic stem cells (ESCs) were cultured in defined default medium (DDM) for 10 days, with or without doxycyclin (Dox) at days 2–3 and increasing concentrations of Activin (0, 1, 10 and 100 ng/ml) from day 0 to day 4, or 2% serum from day 0 to day 10. (A,B) The percentage of cells positive for cardiac-specific isoform of Troponin T (cTnT; A) or CD31 (B) was determined by FACS. Data are presented as mean percentage of positive cells+s.e.m. There is a statistically significant interaction between effects of Activin and Eomes induction on the percentage of cTnT and CD31-expressing cells; P=0.013 and 0.002, respectively. (C) Immunostaining for cTnT and β-tubulin III at day 10 with or without Activin (100 ng/ml) and/or Dox. Scale bar, 100 μm. (D) Quantitative reverse transcription–PCR analysis for Tnnt2, CD31, Sox17 and Afp at day 10 with or without Dox and with increasing concentrations of Activin (0, 10 and 100 ng/ml). Data are presented as mean expression normalized to TBP+s.e.m.
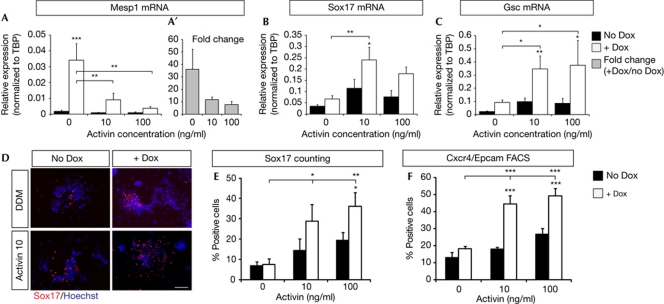
To dissect the molecular mechanism underlying the induction of cardiac mesoderm by Eomes in the absence or low dose of Activin, we next analysed the expression of Mesp1, a transcription factor known to induce cardiac mesoderm specification during ESC differentiation [24–28]. We found that in DDM and in the absence of Activin, Eomes gain of function during ESC differentiation resulted in a strong and rapid induction of Mesp1 expression, which was significantly reduced as the concentration of Activin in the culture medium was progressively increased (Fig 3A,A′). Conversely, qRT–PCR and immunofluorescence at day 3, 24 h following Dox addition, revealed that endodermal fate markers Gsc and Sox17 were induced following Eomes expression in the presence of high levels of Activin (Fig 3B–E), in accordance with previous reports [7, 39]. Consistent with the increase of endodermal differentiation, we found that Eomes in the presence of high levels of Activin promotes the appearance of cells coexpressing Cxcr4 and Epcam (Fig 3F, supplementary Fig S2F online), two markers associated with endodermal fate [15, 40].
Figure 3.
Eomesodermin (Eomes) promotes Mesp1 expression in the absence of Activin. (A–C) Quantitative reverse transcription (qRT)–PCR for Mesp1 (A), Sox17 (B) and Gsc (C) at day 3 in defined default medium (DDM) with increasing concentrations of Activin (0, 10 and 100 ng/ml), 24 h after addition of doxycyclin (+Dox). (D) Immunofluorescence for Sox17 at day 3 of differentiation, with or without Dox or Activin (10 ng/ml). Scale bar, 100 μm. (E) Quantification of the percentage of Sox17-positive cells at day 3. (F) FACS quantification of Cxcr4–Epcam double-positive cells at day 4 with increasing concentrations of Activin (0, 10 and 100 ng/ml), 48 h after addition of Dox. Data from quantifications are presented as mean percentage+s.e.m. qRT–PCR data are presented as mean expression normalized to TBP+s.e.m. (A–C), or as mean fold change+s.e.m. of Dox-treated cells compared with non-treated cells at the same time (A′). For Mesp1 qRT–PCR and Cxcr4–Epcam FACS, there is a statistically significant interaction between effects of Activin and Eomes induction; P=0.042 and 0.016, respectively.
These results indicate that Eomes can induce either endodermal or cardiac mesodermal fate during ESC differentiation, depending on the levels of Activin/Nodal signalling. This is reminiscent of the in vivo situation, where a gradient of Nodal signalling across the PS is proposed to pattern differentially the germ layers towards an endodermal or mesodermal fate [11–13].
Eomes binds directly to the Mesp1-promoter
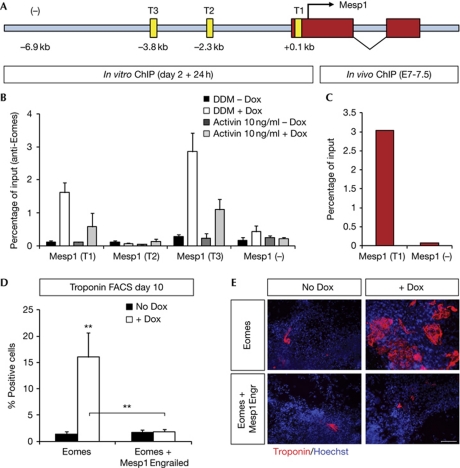
As Mesp1 is already expressed 24 h after overexpression of Eomes, we reasoned that Eomes might directly control the transcription of Mesp1. To address this possibility we analysed the genomic region flanking the Mesp1 locus and found two conserved T-box core motifs [41, 42] (T1 and T3, Fig 4A), and a third one (T2) which is not conserved across mammalian species. T3 is located in a highly conserved enhancer region, previously shown to drive Mesp1 expression in the early mesoderm [42, 43]. We performed chromatin immunoprecipitation (ChIP) using an anti-Eomes antibody or control immunoglobulin-G coupled to qPCR. Amplification of the promoter regions containing T1 and T3 was strongly enriched following ChIP with the anti-Eomes antibody following Eomes overexpression, as compared with ChIP in the absence of Eomes overexpression or using control immunoglobulin-G. The T2 region and a negative-control region containing no T-box-binding site (−) showed no enrichment (Fig 4B,C, and data not shown). As a positive control for these ChIP experiments, we tested a recently described Eomes-binding site within the Mixl1 promoter [7], and found similar enrichment for this genomic region (Mixl1(T1); supplementary Fig S3A,B online). To test for the in vivo relevance of these findings, we performed ChIP experiments on E7 mouse embryos, and found significant enrichments for the identified regulatory elements in the Mesp1 locus, as well as on the Mixl1 locus (Fig 4C and supplementary Fig S3C online). Consistent with our observations, a recent study identified the same Eomes-binding sites (T1 and T3) in the Mesp1 promoter during the differentiation of P19 cells, an embryonic carcinoma cell line [44].
Figure 4.
Eomesodermin (Eomes) directly binds to the Mesp1 promoter in vitro and in vivo. (A) Representation of the genomic region of Mesp1, showing the exons (red), putative Eomes-binding sites (yellow), and a control negative region (−) located 6.9 kb upstream of the transcription start site. (B,C) Quantification as measured by quantitative PCR of DNA fragment enrichment by chromatin immunoprecipitation (ChIP) on differentiating embryonic stem cells (ESCs) at day 3, with or without Activin 10 ng/ml from day 0 to 3 and/or doxycyclin (Dox) for 24 h (B) or on E7 embryos (C) using anti-Eomes antibody and primers encompassing the indicated regions of the Mesp1 promoter (T1, T2, T3 or (−)). Data are presented as percentage of the input. In vitro data are presented as mean+s.e.m. (B). (D,E) FACS quantification of the percentage of cTnT expression (D) and immunostaining for cTnT (E) at day 10 following induction at days 2–3 of either MycEomes alone or MycEomes combined with Flag–Mesp1–Engrailed in defined default medium without Activin. Scale bar, 100 μm.
Given the striking modulatory effects of Activin on Eomes-dependent induction of Mesp1, we next compared the binding of Eomes to the Mesp1 promoter in the presence of Activin (Fig 4B). This revealed a strong decrease in the binding of Eomes to the Mesp1 promoter, suggesting that during endoderm differentiation, Activin may act in part by regulating the recruitment of Eomes to the Mesp1 promoter. Indeed, mice lacking two negative regulators of the transforming growth factor-β pathway (Tgif1 and Tgif2), which present enhanced Nodal signalling, do not express Mesp1 in vivo [45]. Decreasing Nodal signalling in this context rescues Mesp1 expression and gastrulation, suggesting that Nodal signalling restricts Mesp1 expression in vivo as well [45]. Moreover, it has been shown previously that Mesp1 not only induces key cardiovascular transcription factors, but also represses endodermal transcription factors such as Sox17 or Foxa2, further ensuring the specificity in the promotion of cardiovascular cell lineages induced by Mesp1 [25].
Despite direct binding of Eomes to the Mesp1 promoter and the strong and rapid induction of Mesp1 following Eomes overexpression, transactivation assays using different fragments of the Mesp1 promoter [27, 43] cloned into a luciferase-reporter vector did not result in a significant increase of luciferase activity following Eomes overexpression in P19 cells (supplementary Fig S3D–F online). These negative results could reflect technical issues, including weak promoter activity, inappropriate cellular context for proper transactivation, the absence of a particular cofactor that would be required with a precise stoichiometry for Eomes to work properly on the Mesp1 promoter, or the need for more epigenetic factors that would not function properly in transient transfection assays. To test further and more directly the functional link between Eomes and Mesp1, we then examined the effect of Eomes on cardiac induction in the presence of a Mesp1–Engrailed fusion protein, previously known to inhibit endogenous Mesp1 functions [27]. Concomitant expression of Eomes and Mesp1–Engrailed fusion protein (supplementary Fig S1B,C online) blocked the ability of Eomes to promote cardiac fate in these conditions, as assessed by cTnT expression (Fig 4D,E, supplementary Fig S4C online). Mesp1 thus acts genetically downstream of Eomes, possibly as a direct target gene. This could explain the similar phenotype between Mesp1/Mesp2 double-knockout mice and Eomes knockout mice that fail to undergo epithelial to mesenchymal transition and to form cardiac mesoderm [21, 22, 46, 47]. Our findings are also fully consistent with a recent report, in which Eomes was demonstrated to be required for cardiac lineage specification in vivo and in vitro, and was suggested to act directly upstream of Mesp1 [44]. It was also recently shown that Brachyury, another Tbx transcription factor, directly binds to the T2 regulatory sequence of Mesp1 and promotes its expression [48], suggesting that several T-Box genes might cooperate to induce Mesp1 expression in the cardiac mesoderm. Interestingly, the same enhancer region and Tbx-binding site (T3) are also required to promote Mesp1 expression in the presomitic mesoderm, suggesting that the same Tbx-binding site is used sequentially to promote Mesp1 expression in different mesodermal cell populations [42]. Moreover, in Ciona intestinalis, the orthologue of Tbx6 acts upstream of Mesp and directly controls its expression allowing cardiac mesoderm specification [49], suggesting that a Tbx–Mesp transcriptional circuit has been conserved throughout evolution to promote cardiac progenitor specification.
Altogether, our results indicate a model whereby Eomes acts at least in part through Mesp1 to promote MCP specification and cardiovascular differentiation, although it remains to be determined whether this interaction is mostly direct or also indirect, and which other Eomes target genes may be involved in this process. They also show that these effects are strongly influenced by Activin signalling, pointing to a mechanism by which a single intrinsic cue induces different cell fates depending on the levels of an extrinsic signal.
Methods
P19 and ESC culture. ICE (A2lox.Cre) mouse ESCs [33] were routinely propagated as described [50]. For differentiation, ESCs were plated at low density (20 × 103/ml) on gelatin-coated coverslips. After 1 day, medium was switched to DDM [50] and changed every 2 days. When specified, Activin A (R&D) was added from day 0 to 4 at 1, 10 or 100 ng/ml, 1 μg/ml doxycyclin (Sigma) was added from days 2–4 and 2% fetal bovine serum was added to DDM from day 0 to day 10.
P19 cells were routinely cultured as monolayer in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamin and 1% penicillin/streptomycin.
Generation of tetracycline-inducible ESC lines. To generate the tetracycline-inducible Myc–Eomes ESC line, the N-terminal Myc-tagged murine Eomes open reading frame was amplified by PCR, sequence verified and cloned into the p2Lox backbone [33]. To generate a cell line allowing inducible and combined expression of Myc–Eomes and Flag–Mesp1–Engrailed [27], we introduced both constructs downstream of two separate Tet-O in tandem in the p2Lox backbone [27]. After electroporation into A2Lox.Cre cells and cassette exchange recombination [33], neomycin-resistant clones were screened for their expression of Myc–Eomes or Flag–Mesp1–Engrailed by immunofluorescence (supplementary Fig S1 online) and qRT–PCR (data not shown) 24 h after induction. Results obtained with the Myc–Eomes ESC line were confirmed in three independent clones.
Mesp1-promoter plasmids and transactivation assay. Fragments of the Mesp1 promoter containing one or both T-box sequences T1 and T3 were cloned into a pGL3 basic, a pGL3 enhancer or a pGL4.23 vector backbone (Scheme of cloning depicted in Supplementary Fig S3D online) starting from the pMesp1 plasmid [27] containing a 5.6-kb fragment upstream of the Mesp1 translation site [43]. For transfection, P19 cells were seeded at 25 × 103 cells/24-well and 24 h later transfected using Lipofectamin 2000 reagent with 10 ng pRL-TK, 500 ng of one of the pMesp1 vectors or pGL3 basic alone (negative control) and 200 ng pCIG or pCIG–Eomes. pCIG was used as control vector up to a final DNA amount of 1 μg. Positive control was a validated Hes5–promoter construct co-transfected with Notch1ΔE [51]. Renilla and Firefly Luciferase activity were measured after 48 h using Dual Luciferase kit (Promega). Data are presented as Firefly Luciferase activity relative to Renilla activity.
RNA isolation and qRT–PCR. RNA extraction, DNAse treatment and RT–PCR were performed as previously described [25, 30]. All qRT–PCR were performed in duplicate using the Power SybrGreen Mix (Applied Biosystems) and a 7500 Real-Time PCR System (Applied Biosystems). Results were normalized to the housekeeping gene TBP; primers used are summarized in supplementary Table S1 online.
Chromatin immunoprecipitation and ChIP–qPCR. ChIP was performed as described previously [52] on ESC after 10 days of differentiation or on dissected E7–7.5 embryos using either the rabbit anti-Eomes antibody (ab2345; Abcam) or the rabbit anti-haemagglutinin isotype antibody (sc-805; Santa Cruz) as control. Primers for qPCR analysis are listed in supplementary Table S1 online. For each primer set, qPCR was performed in duplicate. Results were analysed using the 2−Δ Ct method, comparing anti-Eomes to anti-haemagglutinin and to the input. Data are presented as percentage of the input.
Immunofluorescence staining and FACS analysis. Immunofluorescence was performed as previously described [30]. For cytospin analysis, cells were dissociated by trypsinization, cytospun on Superfrost Plus glass slides and fixed for 5 min in 2% paraformaldehyde. Primary antibodies used were the following: Mouse anti-cardiac isoform of TroponinT Ab1 (1/100; NeoMarkers), Rabbit anti-β-tubulin III (1/2,000; Covance), Goat anti-Sox17 (1/1,000; R&D), Mouse anti-c-Myc (1/1,000; Roche), Mouse anti-Smooth muscle actin (1/200; Sigma), Rabbit anti-Pax6 (1/1,000; Covance), Rat anti-CD144 (VE-cadherin; 1/100; BD Biosciences) and Mouse anti-Flag (M2; 1/500; Sigma).
For flow cytometry, stainings against cTnT (Ab1-NeoMarkers), Flk1 (Vegfr2; Avas12a1; eBioscience), Pdgfrα (APA5; eBioscience), Cxcr4 (2B11; eBioscience), Epcam (G8.8; Biolegend) and CD31 (MEC13.3; BD Biosciences) were performed as described previously [25, 27].
Mice. Embryos were dissected from timed pregnant CD1 mice at embryonic day E7–E7.5. The plug date was defined as embryonic day E0.5. Animal care and procedures were in compliance with local ethical committees.
Statistical analysis. Unless stated otherwise, data are presented as mean of at least three biologically independent experiments+ standard error of the mean. qPCR data are presented as linearized Ct-values normalized to TBP (2−Δ Ct). To calculate fold increase of qPCR data, TBP-normalized Ct-values of +Dox conditions were normalized to −Dox (ΔΔCt) for each independent experiment. Interaction between the effects of Dox and Activin was tested using a two-way analysis of variance test.
For counting of cells, at least 1,000 cells were counted in three different fields, from at least three biologically independent experiments. All P-values were calculated using a two-way analysis of variance test with a post hoc Tukey test for multiple comparisons. *P<0.05; **P<0.01; ***P<0.001 in all figs.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank G. Vassart for continuous support and interest, and members of the lab and IRIBHM for helpful discussions and advice. We thank M. vander Heyden, UMC Utrecht, for providing P19 cells. This work was funded by grants from the Belgian Queen Elizabeth Medical Foundation, the Action de Recherches Concertées Programs, the Interuniversity Attraction Poles Program, Belgian State, Federal Office for Scientific, Technical and Cultural Affairs, the Fondations Pierre Clerdent and Roger de Spoelberch (to P.V.), the Belgian FNRS and FRSM, the Welbio and Programme d’Excellence CIBLES of the Walloon Region (to P.V. and C.B.). C.B. is supported by the Fund Gaston Ithier, a starting grant of the European Research Council, and the EMBO Young Investigator Program. L.T. was supported by an EMBO Long Term Fellowship. P.V. is Research Director, C.B. Chercheur Qualifié, A.B. and L.T. Chargés de Recherche, and J.v.d.A. Research Fellow of the FNRS.
Author contributions: J.v.d.A., L.T., A.B., C.P. and A.H. performed all the experiments. M.K. and M.L. provided crucial cellular tools. J.v.d.A., L.T., A.B., C.B. and P.V. designed the experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Tam PP, Loebel DA (2007) Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet 8: 368–381 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ (2009) Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10: 91–103 [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G (2008) Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132: 661–680 [DOI] [PubMed] [Google Scholar]
- Schier AF (2003) Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol 19: 589–621 [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ (2001) Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411: 965–969 [DOI] [PubMed] [Google Scholar]
- Camus A, Perea-Gomez A, Moreau A, Collignon J (2006) Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev Biol 295: 743–755 [DOI] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L (2011) Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25: 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G (2004) Development of definitive endoderm from embryonic stem cells in culture. Development 131: 1651–1662 [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE (2005) Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541 [DOI] [PubMed] [Google Scholar]
- Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA (2009) Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 4: 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ (2003) Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev 17: 1646–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn NR, Vincent SD, Oxburgh L, Robertson EJ, Bikoff EK (2004) Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development 131: 1717–1728 [DOI] [PubMed] [Google Scholar]
- Lowe LA, Yamada S, Kuehn MR (2001) Genetic dissection of nodal function in patterning the mouse embryo. Development 128: 1831–1843 [DOI] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM (2006) Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci USA 103: 16806–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, Nishikawa S, Chiba T, Era T (2005) Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol 23: 1542–1550 [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G (2011) Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8: 228–240 [DOI] [PubMed] [Google Scholar]
- Gouon-Evans V, Boussemart L, Gadue P, Nierhoff D, Koehler CI, Kubo A, Shafritz DA, Keller G (2006) BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol 24: 1402–1411 [DOI] [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL (2004) T-box genes in early embryogenesis. Dev Dyn 229: 201–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Bourillot P, Stennard F, Gurdon JB (2000) The Xenopus eomesodermin promoter and its concentration-dependent response to activin. Mech Dev 94: 133–146 [DOI] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Mitchell A, Gurdon JB (1996) Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell 87: 989–1000 [DOI] [PubMed] [Google Scholar]
- Russ AP et al. (2000) Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404: 95–99 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ (2008) Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson CR, Griffin KJ, Farr GH III, Terashima A, Himeda C, Kikuchi Y, Kimelman D (2005) Eomesodermin is a localized maternal determinant required for endoderm induction in zebrafish. Dev Cell 9: 523–533 [DOI] [PubMed] [Google Scholar]
- David R et al. (2008) MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol 10: 338–345 [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C (2008) Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 3: 69–84 [DOI] [PubMed] [Google Scholar]
- Lindsley RC et al. (2008) Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 3: 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C (2011) Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol 192: 751–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Blanpain C (2010) Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res 107: 1414–1427 [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186 [DOI] [PubMed] [Google Scholar]
- Gaspard N et al. (2008) An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455: 351–357 [DOI] [PubMed] [Google Scholar]
- Levine AJ, Brivanlou AH (2007) Proposal of a model of mammalian neural induction. Dev Biol 308: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspard N, Vanderhaeghen P (2010) Mechanisms of neural specification from embryonic stem cells. Curr Opin Neurobiol 20: 37–43 [DOI] [PubMed] [Google Scholar]
- Iacovino M, Bosnakovski D, Fey H, Rux D, Bajwa G, Mahen E, Mitanoska A, Xu Z, Kyba M (2011) Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29: 1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Daley GQ (2002) HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109: 29–37 [DOI] [PubMed] [Google Scholar]
- Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH (2006) Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 127: 1137–1150 [DOI] [PubMed] [Google Scholar]
- Moretti A et al. (2006) Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127: 1151–1165 [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM (2006) Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 11: 723–732 [DOI] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G (2004) Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432: 625–630 [DOI] [PubMed] [Google Scholar]
- Izumi N, Era T, Akimaru H, Yasunaga M, Nishikawa S (2007) Dissecting the molecular hierarchy for mesendoderm differentiation through a combination of embryonic stem cell culture and RNA interference. Stem Cells 25: 1664–1674 [DOI] [PubMed] [Google Scholar]
- Green MD, Chen A, Nostro MC, d’Souza SL, Schaniel C, Lemischka IR, Gouon-Evans V, Keller G, Snoeck HW (2011) Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat Biotechnol 29: 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon FL, Fairclough L, Price BM, Casey ES, Smith JC (2001) Determinants of T box protein specificity. Development 128: 3749–3758 [DOI] [PubMed] [Google Scholar]
- Oginuma M, Hirata T, Saga Y (2008) Identification of presomitic mesoderm (PSM)-specific Mesp1 enhancer and generation of a PSM-specific Mesp1/Mesp2-null mouse using BAC-based rescue technology. Mech Dev 125: 432–440 [DOI] [PubMed] [Google Scholar]
- Haraguchi S, Kitajima S, Takagi A, Takeda H, Inoue T, Saga Y (2001) Transcriptional regulation of Mesp1 and Mesp2 genes: differential usage of enhancers during development. Mech Dev 108: 59–69 [DOI] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ (2011) The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 13: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SE, Taniguchi K, Yen W, Melhuish TA, Shen J, Walsh CA, Sutherland AE, Wotton D (2010) Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development 137: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S (2000) Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med 10: 345–352 [DOI] [PubMed] [Google Scholar]
- Kitajima S, Takagi A, Inoue T, Saga Y (2000) MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 127: 3215–3226 [DOI] [PubMed] [Google Scholar]
- David R, Jarsch V, Schwarz F, Nathan P, Gegg M, Lickert H, Franz WM (2011) Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc Res 92: 115–122 [DOI] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Davidson B, Levine M (2009) Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev Biol 328: 552–560 [DOI] [PubMed] [Google Scholar]
- Gaspard N, Bouschet T, Herpoel A, Naeije G, van den Ameele J, Vanderhaeghen P (2009) Generation of cortical neurons from mouse embryonic stem cells. Nat Protoc 4: 1454–1463 [DOI] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R (2006) Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem 281: 5106–5119 [DOI] [PubMed] [Google Scholar]
- Rustighi A et al. (2009) The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol 11: 133–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.