Abstract
The modification of proteins by small ubiquitin-like modifier (SUMO) is crucial for the regulation of diverse cellular processes. Protein SUMOylation is reversed by isopeptidases, collectively known as deSUMOylases. Only one family of SUMO-specific proteases has been described so far: the sentrin-specific proteases (SENP). Here, we identify and characterize a new deSUMOylase, which we have named DeSI-1 (DeSumoylating Isopeptidase 1). We describe BZEL—a new transcriptional repressor—as substrate of DeSI-1. DeSI-1 catalyses the deSUMOylation, but not the deubiquitination, of BZEL. Furthermore, the SENP substrates PML and ΔNp63 are not deSUMOylated by DeSI-1, suggesting that SENP and DeSI enzymes recognize different sets of substrates. Together, these data identify a second class of SUMO proteases.
Keywords: SUMO, SUMO-specific protease, desumoylation
Introduction
In mammalian systems, three small ubiquitin (Ub)-like modifier (SUMO) proteins, SUMO1, SUMO2 and SUMO3, have been reported [1, 2]. Moreover, sumoylation modulates many processes, such as transcription, intracellular transport, DNA repair, replication and cell signalling [2, 3]. Similar to ubiquitination, SUMO conjugation occurs through a cascade of reactions that are performed by three enzymes. The first step of sumoylation is mediated by the E1-activating enzyme heterodimer Aos1–Uba2, which requires ATP. The second step is the transfer of SUMO from Uba2 to the E2-conjugating enzyme Ubc9. Finally, SUMO is conjugated to substrate via isopeptide bonds to the ε-amino group of lysine residues on the substrate by the E3 enzyme including protein inhibitor of STAT (PIAS) family, RanBP2 and Pc2 [4, 5].
The covalent modification of proteins by SUMO is reversible, and desumoylation is mediated by SUMO-specific proteases. Although almost 100 deubiquitinases have been identified in humans [6], only one class of human desumoylase, the sentrin-specific protease (SENP) family of proteins, has been identified so far by virtue of their homology to Ulp1, the original desumoylase identified in yeast [7]. The SENP family of proteins is predicted to contain about 200-amino-acid catalytic domain, which has characteristics of a cysteine protease family [8]. The catalytic domain is typically located close to the C terminus of SENPs, whereas N-terminal domains frequently direct their subcellular localization. The catalytic domain of SENPs contains a conserved His–Asp–Cys catalytic triad, and these enzymes belong to the CE clan of cysteine proteases [9].
In mammalian systems, six SENPs have been reported [5, 10]; SENP1 localizes to nucleus and deconjugates several sumoylated proteins [11]. SENP2 localizes to a nuclear envelope-associated compartment, and when overexpressed seems to have a wide substrate specificity similar to that of SENP1 [12]. There is a spliced isoform of SENP2, called SuPr1, which could alter the distribution of nuclear PML-oncogenic domain (POD)-associated proteins, such as CBP and Daxx, and could convert Sp3 to a strong activator with diffuse nuclear localization [13]. SENP3 and SENP5 constitute a family of nucleolar SENPs with preference for SUMO2/3 [14]. SENP6 and SENP7 are nuclear proteases with deconjugating activity for poly-SUMO2/3 chains [15, 16].
As stated above, all of the SENPs localize to the nucleus or nucleus-associated structures. However, many cytoplasmic proteins, such as FAK, IκBα and NEMO [17–19], are also sumoylated, and therefore the existence of cytoplasmic desumoylases different from SENPs has been suspected. Moreover, the fact that several proteins with various natures are sumoylated predicted the existence of more desumoylase. Here we report the identification of a new class of desumoylase, which we named DeSI-1 (DeSUMOylating isopeptidase 1).
Results And Discussion
Identification of DeSI-1
We have been analysing a new BTB (Broad complex, tramtrack, bric-a-brac)-ZF (zinc-finger) protein, which we named BZEL (BTB-ZF protein expressed in effector lymphocytes). BZEL is encoded by a Zbtb46 locus (GeneID: 72147) and contains an N-terminal BTB and two Krüppel-like C2H2 zinc-finger domains at the C terminus. BZEL binds to the promoter of target genes including blimp-1 and suppresses the transcription of these genes (unpublished results).
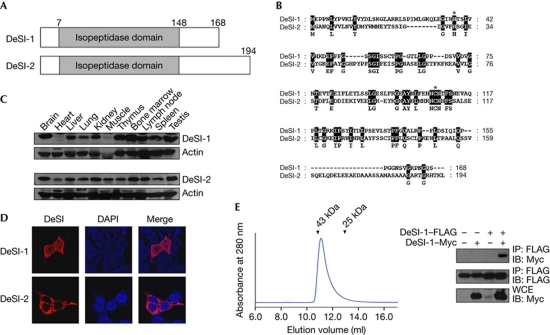
In this report, we screened for proteins that bind to BZEL by yeast two-hybrid screening. The screening of 1 × 107 independent clones of a murine T-cell lymphoma library yielded about 90 strong positives. Subsequently, we characterized a protein detected 10 times out of about 90 positive clones, which we designated DeSI-1 (Fig 1A). Murine DeSI-1 complementary DNA (previously described as PPPDE peptidase domain containing 2) [20] has an open reading frame of 504 bp, which encodes a protein of 168 amino acids (Fig 1B). A homology search revealed that DeSI-1 belongs to a group of proteins designated as PPPDE (Permuted Papain fold Peptidases of Ds-RNA viruses and Eukaryotes) [20], which were predicted to function as deubiquitinating peptidases because some of the members have a Ub-binding domain in addition to a protease domain [20]. However, no experimental data have been provided to support the deubiquitinase or desumoylase activity of PPPDE proteins. In addition to DeSI-1, one more PPPDE family of proteins, which we named DeSI-2 (previously described as PPPDE peptidase domain containing 1) [20], is present in mouse. DeSI-2 complementary DNA has an open reading frame of 582 bp, which encodes a protein of 194 amino acids that shares 23% sequence identity with DeSI-1 at the amino-acid level (Fig 1A,B). Both DeSI-1 and DeSI-2 have a peptidase domain corresponding to amino acids 7–148; however, the C-terminal segment of DeSI-2 is 47 amino acids, whereas the corresponding region in DeSI-1 is 20 amino acids (Fig 1A,B). In addition, orthologues of DeSI-1 and DeSI-2 are found in humans and rats.
Figure 1.
Identification of DeSI. (A) Schematic representation of the structure of DeSI-1 and DeSI-2. (B) Sequence alignment of DeSI-1 and DeSI-2. Amino acid identity is highlighted in black. The histidine and cysteine residues that form the catalytic site of cysteine protease are marked by asterisks. (C) Tissue expression pattern of DeSI-1 and DeSI-2. Murine tissue lysates were subjected to immunoblot analysis using rabbit anti-DeSI-1 or anti-DeSI-2 antibodies. Equal protein loading was shown by immunoblotting with anti-actin antibody. (D) Subcellular localization of DeSI-1 and DeSI-2. 293T cells were transfected with expression plasmids encoding Myc-tagged DeSI-1 or DeSI-2. Cells were visualized by staining with anti-Myc antibody along with Alexa 568-conjugated secondary antibody (red). (E) Homodimerization of DeSI-1 (left panel). The chromatogram shows that full-length DeSI-1 (Mr=20 kDa) was eluted as a ∼40-kDa protein from a Superdex 75 column. The arrowheads indicate the elution positions of the size marker proteins (right panel); 293T cells were transfected with the indicated combinations of expression plasmids encoding DeSI-1–FLAG and DeSI-1–Myc. Cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody and subsequently to immunoblot analysis with anti-Myc or anti-FLAG antibodies. DAPI, 4,6-diamidino-2-phenylindole; DeSI-1, DeSumoylating Isopeptidase 1; IB, immunoblot; WCE, whole-cell extract.
Immunoblot analysis showed that DeSI-1 and DeSI-2 are present in most of the mouse tissues tested (Fig 1C). In addition, subcellular localization analysis showed that DeSI-1 was present throughout the cytoplasm and nucleus, whereas DeSI-2 was localized mainly in the cytoplasm (Fig 1D). Interestingly, purified DeSI-1 was eluted as a dimeric protein from an analytical size-exclusion column (Fig 1E, left panel). Moreover, when coexpressed in 293T cells, FLAG- and Myc-tagged DeSI-1 proteins coimmunoprecipitated readily (Fig 1E, right panel), showing that DeSI-1 forms a dimer.
DeSI-1 acts as a BZEL desumoylase but not a deubiquitinase
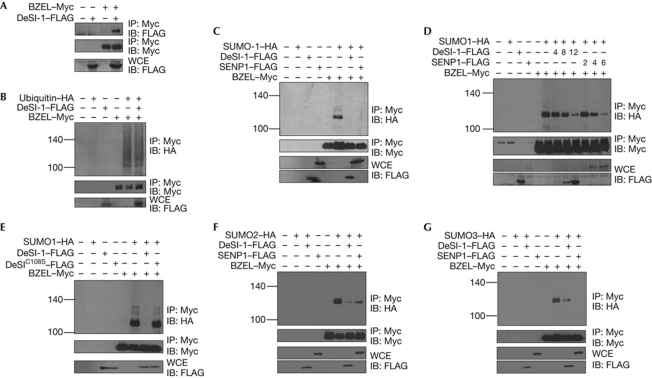
Having identified DeSI-1 as a BZEL-binding protein in the yeast two-hybrid screening, we tested whether these two proteins also interact in mammalian cells. As shown in Fig 2A, BZEL coimmunoprecipitated with DeSI-1 when these two proteins were transiently expressed in 293T cells. As BZEL can be ubiquitinated or sumoylated, we next examined whether DeSI-1 catalyses the deubiquitination or desumoylation of BZEL. To test the deubiquitinase activity of DeSI-1, the indicated combinations of BZEL, DeSI-1 and Ub were coexpressed in 293T cells, and the ubiquitination status of BZEL was analysed. As shown in Fig 2B, the levels of ubiquitinated BZEL were not affected by DeSI-1 overexpression, showing that DeSI-1 does not deubiquitinate BZEL. We then tested whether DeSI-1 acts as a desumoylase of BZEL. The indicated combinations of BZEL, SUMO1, DeSI-1 and/or SENP1 were coexpressed in 293T cells, and the sumoylation status of BZEL was analysed. BZEL was efficiently sumoylated on SUMO1 coexpression, and the level of SUMO-modified BZEL was markedly reduced on expression of DeSI-1, as well as SENP1 (Fig 2C). In addition, to examine the dose-dependent activity of DeSI-1 on BZEL, we tested the effects of increasing the amounts of DeSI-1 and found that DeSI-1 desumoylates BZEL in a dose-dependent manner (Fig 2D). As, based on sequence comparison with other cysteine proteases, the cysteine residue at position 108 of DeSI-1 is predicted to be a part of a catalytic site involved in isopeptide bond cleavage, we tested the effect of overexpression of DeSI-1C108S, in which cysteine 108 of DeSI-1 was substituted for serine. As shown in Fig 2E, the level of SUMO-modified BZEL was not affected by DeSI-1C108S, showing that the desumoylation of BZEL was mediated specifically by the isopeptidase activity of DeSI-1. Taken together, these results indicate the possibility that DeSI-1 might function as a desumoylase but not certainly as a deubiquitinase of BZEL.
Figure 2.
DeSI-1 acts as a desumoylase but not as a deubiquitinase of BZEL. (A) DeSI-1 associates with BZEL. Cells (293T) were transfected with the indicated combinations of expression plasmids encoding DeSI-1 and BZEL. Cell lysates were subjected to immunoprecipitation with the anti-Myc antibody and subsequently to immunoblot analysis with anti-FLAG (upper panel) or anti-Myc (middle panel) antibody. As a control for DeSI-1 expression, whole-cell extract (WCE) was analysed by immunoblotting with anti-FLAG antibody (lower panel). (B) DeSI-1 lacks the ability to deubiquitinate ubiquitin-modified BZEL. Cells (293T) were transfected with the indicated combinations of expression plasmids encoding DeSI-1, BZEL and/or ubiquitin. Cell lysates were subjected to immunoprecipitation with anti-Myc antibody and subsequently to immunoblot analysis with anti-HA (upper panel) or anti-Myc (middle panel) antibody. (C) DeSI-1 desumoylates SUMO1-modified BZEL. Cells (293T) were transfected with the indicated combinations of expression plasmids encoding DeSI-1, BZEL and/or SUMO1. Cell lysates were subjected to immunoprecipitation with anti-Myc antibody and subsequently to immunoblot analysis with anti-HA (upper panel) or anti-Myc (middle panel) antibody. (D) DeSI-1 desumoylates SUMO1-modified BZEL in a dose-dependent manner. The experiment was performed as described in C, except that cells were transected with increasing amounts of DeSI-1 (0, 4, 8 or 12 μg) or SENP1 (0, 2, 4, or 6 μg) expression plasmids. The expression levels of SENP1 and DeSI-1 in WCE were analysed by western blotting with anti-FLAG antibody (bottom two panels). (E) DeSI-1C108S lacks desumoylase activity against SUMO-modified BZEL. Cells (293T) were transfected with the indicated combinations of expression plasmids encoding DeSI-1, DeSI-1C108S, BZEL and/or SUMO1. Cell lysates were processed as described in B. (F) DeSI-1 desumoylates SUMO2-modified BZEL. The experiment was performed as described in C, expect that SUMO2 was coexpressed instead of SUMO1. (G) DeSI-1 desumoylates SUMO3-modified BZEL. The experiment was performed as described in C, except that SUMO3 was coexpressed instead of SUMO1. BZEL, BTB-ZF protein expressed in effector lymphocytes; DeSI-1, DeSumoylating Isopeptidase 1; HA, haemagglutinin; IB, immunoblot; IP, immunoprecipitation; SENP1, sentrin-specific proteases 1; SUMO1, small ubiquitin-like modifier 1.
DeSI-1 deconjugates both SUMO1 and SUMO2/3
SENP1 and SENP2 have deconjugation activity for both SUMO1 and SUMO2/3, whereas SENP3, SENP5, SENP6 and SENP7 seem to prefer SUMO2/3 [14, 21]. Having found that DeSI-1 acts as a desumoylase for SUMO1, we were interested in testing whether DeSI-1 also acts as a desumoylase for SUMO2 and SUMO3. For this, the indicated combinations of BZEL, DeSI-1, SUMO2 and SUMO3 were coexpressed in 293T cells, and the sumoylation status of BZEL was analysed. As shown in Fig 2F,G, BZEL was efficiently sumoylated on coexpression of SUMO2 or SUMO3, and the level of SUMO2- or SUMO3-modified BZEL was markedly reduced on expression of DeSI-1 and SENP1. These results show that DeSI-1 has deconjugation activity for both SUMO1 and SUMO2/3.
Recombinant DeSI-1 is a desumoylase in vitro
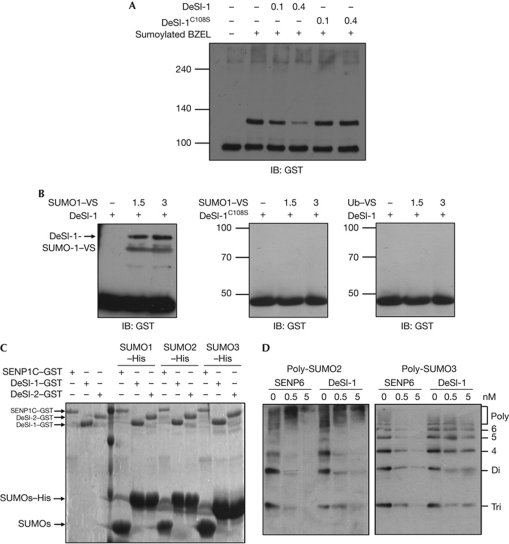
Moreover, to demonstrate that DeSI-1 itself, and not another component of the cell lysates, acts as a desumoylase, we tested the desumoylase activity of purified recombinant DeSI-1 expressed in Escherichia coli (E. coli). In vitro sumoylated BZEL was incubated with purified recombinant DeSI-1. As shown in Fig 3A, DeSI-1, but not the DeSI-1C108S mutant, effectively desumoylated SUMO1-modified BZEL in a dose-dependent manner. In addition, we compared the desumoylase activity of DeSI-1 with that of SENP1C, which corresponds to the C-terminal 200 amino acids of SENP1 known to possess desumoylating activity [22]. In vitro sumoylated BZEL was incubated with purified recombinant DeSI-1 or SENP1C. Both DeSI-1 and SENP1C effectively desumoylated SUMO1-modified BZEL (supplementary Fig S1 online).
Figure 3.
In vitro desumoylation assay using recombinant DeSI-1 and BZEL from E. coli. (A) DeSI-1 desumoylates SUMO1-modified BZEL in vitro. A concentration of 10 μM of purified GST–BZEL fusion protein from E. coli was sumoylated in vitro. Subsequently, sumoylated BZEL was incubated with 0, 0.1 or 0.4 μM of DeSI-1or DeSI-1C108S mutant purified from E. coli for 1 h at 37°C. After incubation, the reaction mixture was analysed by immunoblotting with anti-GST antibody. The arrow indicates the position of sumoylated BZEL. (B) DeSI-1 formed covalent adducts with SUMO1-VS but not with Ub–VS. Recombinant purified GST–DeSI-1, or DeSI-1C108S mutant was incubated with 0, 1.5 and 3 μg each of SUMO1–VS or Ub–VS for 1 h at 37°C. After incubation, immunoblot analysis was performed with anti-GST antibody. (C) DeSI-1 lacks the SUMO-processing activity. A measure of 30 μg of SUMO precursors (SUMOs–His) was incubated for 1 h with 2 μg each of GST–DeSI-1, GST–DeSI-2 or GST–SENP1C purified from E. coli. Subsequently, the reaction mixtures were subjected to SDS–PAGE and stained with Coomassie blue. The size of the SUMO proteins generated by cleavage after the C-terminal diglycine motif is marked by an arrow. (D) DeSI-1 cleaves polymeric SUMO2/3 chains. A measure of 0.5 μg each of poly-SUMO2 or poly-SUMO3 chains was incubated with DeSI-1 or SENP6C at three different enzyme concentrations (0, 0.5 and 5 nM). Subsequently, the reaction mixtures were analysed by immunoblotting using anti-SUMO2/3 antibody. BZEL, BTB-ZF protein expressed in effector lymphocytes; DeSI-1, DeSumoylating Isopeptidase 1; E. coli, Escherichia coli; GST, glutathione S-transferase; His, histidine; IB, immunoblot; SENP1, sentrin-specific proteases 1; SUMO1, small ubiquitin-like modifier 1; Ub, ubiquitin; VS, vinyl sulphone.
Next, to further confirm the specificity of DeSI-1, we used SUMO1 and Ub tagged with vinyl sulphone derivatives (SUMO1–VS and Ub–VS), which covalently and specifically react with the nucleophilic cysteine residue within the active sites of SUMO or Ub-specific proteases [23]. As shown in Fig 3B, DeSI-1, but not the DeSI-1C108S mutant, formed an adduct with SUMO1–VS but not with Ub–VS. Therefore, these results show that DeSI-1 is a specific isopeptidase for SUMO but not for Ub.
DeSIs have isopeptidase but not SUMO-processing activity
In addition to having an isopeptidase activity that deconjugates SUMO1 from the lysine ε-amino group of the target protein, mammalian SENPs are known to function as hydrolases that cleave the carboxyl side of C-terminal Gly–Gly residues of SUMO1 precursor to produce the mature form [22]. To examine whether DeSI-1 has this SUMO-processing activity, DeSI-1, DeSI-2 and SENP1C were purified as glutathione S-transferase (GST) fusion proteins from E. coli. GST–SENP1C, but not GST–DeSI-1 and GST–DeSI-2, digested SUMOs–His to release processed SUMOs (Fig 3C). These results show that DeSI-1, unlike SENPs, has isopeptidase activity but not SUMO-processing activity.
DeSI-1 cleaves polymeric SUMO2/3 chains
Similar to Ub, SUMO2/3, but not SUMO1, can form polymeric chains [24]. SENP6 and SENP7 were previously reported to efficiently depolymerize poly-SUMO2/3 chains [15, 16]. To examine whether DeSI-1 has poly-SUMO 2/3 editing activity, DeSI-1 and SENP6C, which is the C-terminal 475 amino acids of SENP6 known to possess SUMO-editing activity [25], were purified as GST fusion proteins from E. coli and tested for SUMO2/3 chain cleaving activity. Not only SENP6C but also DeSI-1 digested poly-SUMO 2/3 chains (Fig 3D). These results show that DeSI-1 has poly-SUMO2/3-deconjugation activity on par with SENP 6C-deconjugating activity.
Substrate specificity of DeSI-1 compared with SENPs
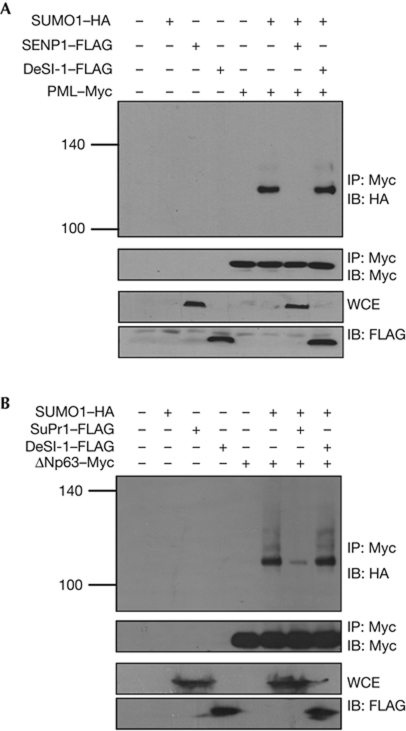
Until now, only one class of desumoylases, the SENP family, has been identified in mammals. Having identified DeSI-1 as a second class of desumoylases, we were interested in testing the substrate specificity of SENP and DeSI family proteins. For this purpose, we tested whether DeSI-1 can desumoylate PML and ΔNp63, two previously established substrates of the SENP family of proteins. PML, a tumour suppressor protein that resides in the nucleus, was established as a substrate of SENP1 [13]. PML and SUMO1 were coexpressed in 293T cells to allow sumoylation of PML, and the effects of overexpression of DeSI-1 or SENP1 were analysed. PML was immunoprecipitated from the cell lysates, and precipitates were analysed for the status of sumoylation. As shown in Fig 4A, PML was sumoylated by SUMO1 coexpression, and SUMO modification of PML was reversed by the overexpression of SENP1, but not of DeSI-1. Next, we tested whether DeSI-1 acts as a desumoylase of SUMO-modified ΔNp63. ΔNp63 is known to be modified by SUMO, and the sumoylation is reversed by a SENP family desumoylase, SuPr1 (N-terminally truncated SENP2 isoform) [26]. As shown in Fig 4B, ΔNp63 is rapidly desumoylated by SuPr1 but not DeSI-1. In addition, on incubation of in vitro sumoylated promyelocytic leukaemia (PML) and ΔNp63 with purified recombinant SENP1C, SuPr1C and DeSI-1 from E. coli, SENP1 and SuPr1, respectively, but not DeSI-1, desumoylated PML and ΔNp63 in vitro (supplementary Fig S2 online). Taken together, these observations show that SENP and DeSI-1 have different substrate specificities.
Figure 4.
PML and ΔNp63, substrates of SENP, are not desumoylated by DeSI-1. (A) 293T cells were transfected with the indicated combinations of expression plasmids encoding PML, SUMO1, DeSI-1 and/or SENP1. Cell lysates were subjected to immunoprecipitation with the anti-Myc antibody, followed by immunoblot analysis with anti-HA or anti-Myc antibodies. To control for the expression of DeSI-1 and SENP1, WCE was analysed by immunoblot (IB) analysis using the anti-FLAG antibody. (B) The experiment was performed as described in A, except that SuPr1 was expressed instead of SENP1, and the sumoylation of ΔNp63, instead of PML, was analysed. DeSI-1, DeSumoylating Isopeptidase 1; HA, haemagglutinin; IP, immunoprecipitation; SENP1, sentrin-specific proteases 1; SUMO1, small ubiquitin-like modifier 1; WCE, whole-cell extract.
DeSI-1 modulates BZEL transcriptional repressor activity
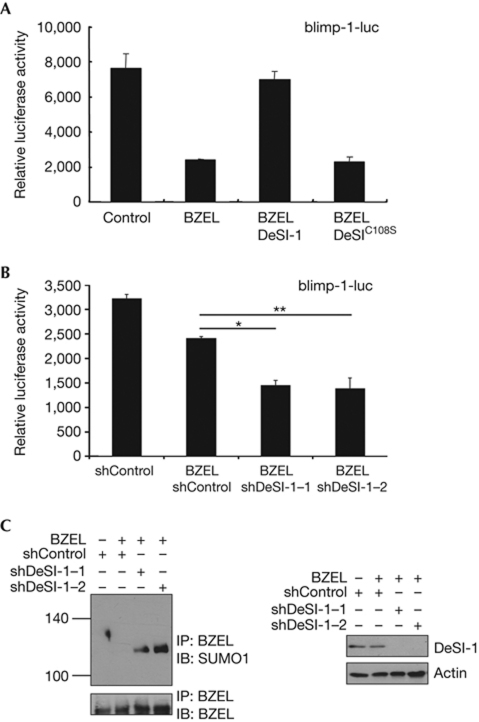
Having established that DeSI-1 acts as a desumoylase of BZEL, we were interested in testing the effect of DeSI-1 on the function of BZEL. As BZEL acts as a transcriptional repressor of the blimp-1 gene (unpublished results), we tested whether desumoylation mediated by DeSI-1 influences the repressor activity of BZEL. When transiently expressed in Raji B cells, DeSI-1, but not the DeSI-1C108S mutant, reversed the BZEL-mediated repression of blimp-1 promoter-driven reporter activity (Fig 5A). Furthermore, on examination of the effects of the depletion of endogenous DeSI-1 by introducing DeSI-1-specific short hairpin RNAs (shRNAs; Fig 5B), the repressor activity of BZEL was enhanced. These results indicate that DeSI-1-mediated desumoylation might regulate the transcription of the endogenous blimp-1 gene. As shown in Fig 5C, the expression of DeSI-1 shRNAs affected only endogenous DeSI-1 without altering the protein levels of BZEL or actin in A20 cells. Furthermore, the level of SUMO-modified BZEL was significantly enhanced. Altogether, these results support the notion that DeSI-1 acts as a desumoylase of endogenous BZEL to regulate downstream target gene expression.
Figure 5.
DeSI-1 modulates the transcriptional repressor activity of BZEL. (A) Raji B cells were transfected with reporter genes driven by the blimp-1 promoter along with a combination of expression plasmids encoding BZEL (7 μg), DeSI-1 (7 μg) and DeSI-1C108S (7 μg). As a control for the transfection efficiency, the cells were also cotransfected with pCMV-β-galactosidase control vector. Subsequently, cell lysates were assayed for luciferase activity, and the luciferase activity was normalized against β-galactosidase activity. Values are the averages of three independent experiments performed in duplicate. Error bars represent standard deviations. (B) A20 B cells were electroporated with reporter genes driven by the blimp-1 promoter along with a combination of expression plasmids encoding BZEL (2 μg), shDeSI-1–1 (10 μg), shDeSI-1–2 (10 μg) or control shRNA (10 μg). Subsequently, luciferase activity was determined as described in A. Values are the averages of three independent experiments performed in duplicate, and error bars representing standard deviations are shown. Data were also analysed by Student's t-test; *P<0.005 and **P<0.02. (C) Cell lysates from the experiment shown in B were subjected to immunoprecipitation (IP) with anti-BZEL antibody and subsequently to immunoblot (IB) analysis with anti-SUMO1 (upper panel) or anti-BZEL (lower panel) antibodies (left). As a control for DeSI-1 expression, whole-cell extracts (WCE) were analysed by immunoblotting with anti-DeSI-1 or anti-actin antibody (right). BZEL, BTB-ZF protein expressed in effector lymphocytes; DeSI-1, DeSumoylating Isopeptidase 1; shRNA, short hairpin RNA; SUMO1, small ubiquitin-like modifier 1.
In addition, the total cellular pattern of SUMO1- and SUMO2/3-modified proteins were not altered significantly on introduction of DeSI-1 shRNAs (supplementary Fig S3A and B online), indicating that DeSI-1 substrates are more specific compared with those of SENPs, and therefore the modification of these DeSI-1 substrates were not detected in the total cellular sumoylation pattern.
DeSI-1 differs from the SENP family of desumoylases in several aspects. The PPPDE superfamily, including DeSIs, comprises cysteine proteases with a papain-like fold, which is characteristic of clan CA proteases. In contrast, SENPs are members of clan CE proteases [9]. Importantly, it is likely that the substrates of DeSIs and SENPs are different, as we have shown in this paper that PML and ΔNp63, known substrates of SENPs, are not desumoylated by DeSI-1 (Fig 4). Moreover, the subcellular localization of DeSIs (Fig 1D) and SENPs [27] is different. DeSI-1 is located in both the cytosol and nucleus, and DeSI-2 is located in the cytoplasm only. In contrast, the members of SENP are predominantly localized to the nucleus or nucleus-related structure; SENP1 and SENP2 are localized to the nucleus, although these two molecules have both a nuclear localization signal and a nuclear export signal. SENP3 and SENP5 are both localized to the nucleolus. SENP6 and SENP7 are also localized to the nucleoplasm [15]. Consistent with the localization of SENPs in the nucleus, most SUMO-modified proteins studied until now are nuclear proteins. However, many cytoplasmic proteins, such as FAK, IκBα and NEMO [17–19], are also modified by SUMO. It will be interesting to test whether the cytoplasmic SUMO-modified proteins are desumoylated by DeSI-1 or DeSI-2.
Coimmunoprecipitation between DeSI-1 and its substrate BZEL (Fig 2A) indicates that DeSI-1 forms a stable complex with its substrate. It will be interesting to define the substrate-binding domain of DeSI-1, and also to test whether stable binding is required for the recognition of substrate. Interestingly, no report has suggested the stable binding between SENP family proteins and their substrates.
Recently, Wss1, a member of Wss1p-like metalloproteases (WLM) family of metalloproteases, was suggested to have a SUMO-dependent isopeptidase activity in yeast, although direct biochemical data supporting the desumoylating activity were not provided [28]. The desumoylase activity of DeSI-1 found in this report opens up the possibility that several classes of desumoylases exist in mammalian cells.
In summary, our data indicate the presence of a new class of desumoylases. Further studies are required to define the signalling pathways that function upstream of DeSIs and the other substrates of DeSIs, as well as the biological processes that are regulated by these new desumoylases.
Methods
Detailed descriptions for yeast two-hybrid screening, plasmids, isolation of murine tissue proteins, construction of shRNA-expressing plasmids, immunofluorescent microscopy, immunoprecipitation and immunoblot analysis, antibodies and expression and purification of recombinant proteins from E. coli are provided in the supplementary information online.
In vitro desumoylation assays. A measure of 1 μg of purified GST–BZEL from E. coli was subjected to SUMO modification using an in vitro sumoylation kit (Active motif). For in vitro desumoylation assay, purified GST–BZEL was sumoylated as described above, followed by incubation at 37°C with purified DeSI-1, SENP1C or DeSI-1C108S mutant in 10 mm Tris–HCl (pH 8.0), 150 mm NaCl and 1 mm dithiothreitol and 2 mM ATP. The reaction was terminated by the addition of 3 × SDS–PAGE sampling buffer, followed by boiling. The samples were resolved by SDS–PAGE followed by immunoblotting with anti-GST antibody (Cell Signaling).
Labelling reactions and detection. The assay was performed using the SUMO1–VS or Ub–VS (Boston Biochem). A 0.1 nM concentration of recombinant DeSI-1 or SENP1C and 0, 0.5 or 1 μg each of SUMO1–VSVS or Ub–VS were allowed to react for 1–2 h at 37°C in 50 mm Tris–HCl (pH 8.0), 50 mm NaCl, 5 mM MgCl2 and 2 mm dithiothreitol and 2 mM ATP. Reactions were terminated by 3 × SDS–PAGE sampling buffer, followed by boiling. The samples were resolved by SDS–PAGE followed by immunoblotting with anti-GST antibody (Cell Signaling).
SUMO2/3 chain-editing assays. Equal amounts of poly-SUMO2 or poly-SUMO3 (0.5 μg/μl Boston Biochem) were incubated with purified DeSI-1or SENP6C at 30°C in 25 mm Tris–HCl (pH 8.0), 150 mm NaCl, 0.1% tween 20 and 2 mm dithiothreitol. Reactions were stopped after 1 h by addition of 3 × SDS–PAGE sampling buffer followed by boiling. The samples were resolved by SDS–PAGE followed by immunoblotting with anti-SUMO2/3 antibody (Abcam).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported by grant from the Korea Health 21 R&D Project (grant no. A090224). We appreciate the members of the laboratory and E. Suh for helpful discussions and H. Lee for expression plasmids of SENP1, SuPr1, ΔNp63 and PML.
Author contributions: H.M.S., E.N., W.S.K. and J.-H.K. performed experiments; E.J.S. performed the experiments and wrote the manuscript; and B.-H.O. and Y.Y. designed the experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Mukhopadhyay D, Dasso M (2007) Modification in reverse: the SUMO proteases. Trends Biochem Sci 32: 286–295 [DOI] [PubMed] [Google Scholar]
- Gill G (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18: 2046–2059 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Cheng J, Bawa T, Lee P, Gong L, Yeh ET (2006) Role of desumoylation in the development of prostate cancer. Neoplasia 8: 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Sun SC (2008) Deubiquitylation and regulation of the immune response. Nat Rev Immunol 8: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398: 246–251 [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Rawlings ND (2001) Evolutionary lines of cysteine peptidases. Biol Chem 382: 727–733 [DOI] [PubMed] [Google Scholar]
- Drag M, Salvesen GS (2008) DeSUMOylating enzymes—SENPs. IUBMB Life 60: 734–742 [DOI] [PubMed] [Google Scholar]
- Lee MH et al. (2006) SUMO-specific protease SUSP4 positively regulates p53 by promoting Mdm2 self-ubiquitination. Nat Cell Biol 8: 1424–1431 [DOI] [PubMed] [Google Scholar]
- Gong L, Millas S, Maul GG, Yeh ET (2000) Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem 275: 3355–3359 [DOI] [PubMed] [Google Scholar]
- Nishida T, Kaneko F, Kitagawa M, Yasuda H (2001) Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J Biol Chem 276: 39060–39066 [DOI] [PubMed] [Google Scholar]
- Best JL, Ganiatsas S, Agarwal S, Changou A, Salomoni P, Shirihai O, Meluh PB, Pandolfi PP, Zon LI (2002) SUMO-1 protease-1 regulates gene transcription through PML. Mol Cell 10: 843–855 [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET (2006) Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem 281: 15869–15877 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M (2006) SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol 174: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LN, Geoffroy MC, Jaffray EG, Hay RT (2009) Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J 421: 223–230 [DOI] [PubMed] [Google Scholar]
- Kadare G, Toutant M, Formstecher E, Corvol JC, Carnaud M, Boutterin MC, Girault JA (2003) PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J Biol Chem 278: 47434–47440 [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT (1998) SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 2: 233–239 [DOI] [PubMed] [Google Scholar]
- Mabb AM, Wuerzberger-Davis SM, Miyamoto S (2006) PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol 8: 986–993 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L (2004) Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle 3: 1440–1450 [DOI] [PubMed] [Google Scholar]
- Yeh ET (2009) SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem 284: 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Chau SF, Lam KH, Chan HY, Ng TB, Au SW (2006) Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease. Biochem J 398: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J et al. (2004) Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol Cell Biol 24: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276: 35368–35374 [DOI] [PubMed] [Google Scholar]
- Lima CD, Reverter D (2008) Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem 283: 32045–32055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Lee JH, Kim TY, Lee H (2007) Regulation of DeltaNp63alpha by tumor necrosis factor-alpha in epithelial homeostasis. FEBS J 274: 6511–6522 [DOI] [PubMed] [Google Scholar]
- Melchior F, Schergaut M, Pichler A (2003) SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci 28: 612–618 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Chen CF, Brill SJ (2010) Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase. 30: 3737–3748 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.