Abstract
Studies have suggested that the clock regulator PER2 is a tumour suppressor. A cancer network involving PER2 raises the possibility that some tumour suppressors are directly involved in the mammalian clock. Here, we show that the tumour suppressor promyelocytic leukaemia (PML) protein is a circadian clock regulator and can physically interact with PER2. In the suprachiasmatic nucleus (SCN), PML expression and PML–PER2 interaction are under clock control. Loss of PML disrupts and dampens the expression of clock regulators Per2, Per1, Cry1, Bmal1 and Npas2. In the presence of PML and PER2, BMAL1/CLOCK-mediated transcription is enhanced. In Pml−/− SCN and mouse embryo fibroblast cells, the cellular distribution of PER2 is primarily perinuclear/cytoplasmic. PML is acetylated at K487 and its deacetylation by SIRT1 promotes PML control of PER2 nuclear localization. The circadian period of Pml−/− mice displays reduced precision and stability consistent with PML having a role in the mammalian clock mechanism.
Keywords: circadian clock, nuclear localization, PER2, PML, SIRT1
Introduction
Circadian rhythms are the daily oscillations of biological processes driven by endogenous clocks (Takahashi, 1995). The master clock is located in the suprachiasmatic nucleus (SCN; Schwartz and Gainer, 1977). However, basic clock function is also found in peripheral organs and individual nucleated cells (Welsh et al, 2004; Yoo et al, 2004). Major mammalian circadian genes identified include Casein kinases (CK1ε and CK1δ), Cryptochromes (Cry1 and Cry2), Periods (Per1, Per2 and Per3), Clock and Npas2, Rev-erbα and RoRα and Bmal1 (Ko and Takahashi, 2006). For many of these clock regulators, their levels of mRNAs and proteins oscillate during 24-h circadian time and in zeitgeber time (ZT).
The SCN and peripheral clocks are composed of interacting positive and negative-feedback loops that regulate clock gene transcription and involve proteins that shuttle between the nucleus and the cytosol (Ko and Takahashi, 2006). In the negative-feedback loop, CRYs suppress Per gene transcription by directly inhibiting the activity of BMAL1/CLOCK or BMAL1/NPAS2 heterodimers binding to the E-box of the targeted gene promoter (Kume et al, 1999; Reick et al, 2001). Genetic evidence supports CRY's negative role in the clock mechanism since mouse Period 1 (mPer1) and Period 2 (mPer2) levels are enhanced in Cry1−/−/Cry2−/− mice (Vitaterna et al, 1999). Based on co-transfection studies in NIH3T3 cells, it was proposed that CRYs are required for mPER1 and mPER2 entry into the nucleus (Kume et al, 1999). Thus, a key feature of the mammalian clock mechanism is a negative-feedback loop based on CRY's transport of PER's into the nucleus to repress BMAL1/CLOCK-mediated transcription. However, both mPER2 and mPER1 appear primarily in the nuclei of Cry1−/−/Cry2−/− SCN, mouse embryo fibroblast (MEF) and liver cells (Yagita et al, 2000, 2002). In the hypothetical positive loop, the expression of mPer2 is activated by BMAL1/CLOCK- or BMAL1/NPAS2-mediated transcription. Evidence from mPer2-deficient mice revealed that transcription of core circadian genes such as mPer1 and Cry1 was dampened in the SCN and peripheral tissues implicating mPER2 as a positive regulator (Zheng et al, 1999). Except for activation of BMAL1/NPAS2, there was no discernable enhancement of BMAL1/CLOCK transcriptional activity by mPER2 in vitro (Kaasik and Lee, 2004). Rather, transfection of an mPer2 expression construct modestly inhibited BMAL1/CLOCK-mediated transcription, leading to the view that PER2, in partner with CRYs, regulates the negative-feedback loop, a mechanism similar to dPER's role in the Drosophila clock (Hardin et al, 1990; Kume et al, 1999). These discrepancies between in-vivo and in-vitro observations on the role of PER2 in the mammalian clock remain unresolved.
Studies have implicated the circadian clock in gating of the cell division cycle and in the DNA damage response in mammals (Fu et al, 2002; Matsuo et al, 2003; Nagoshi et al, 2004; Zhao and Lee, 2010). DNA damage can phase shift the circadian rhythm of mice (Oklejewicz et al, 2008). The gene for the Neurospora circadian mutant Prd-4 encodes a mutated orthologue of the mammalian Checkpoint 2 (Chk2) gene, demonstrating that the integration of circadian function with the biological processes of cell division and DNA damage response is evolutionarily conserved (Pregueiro et al, 2006). Other studies have since implicated human PER2 (hPER2) in tumour suppression (Gery et al, 2005; Hua et al, 2006; Winter et al, 2007; Oda et al, 2009). Human tumour cells that were engineered to overexpress PER2 displayed increased apoptosis and repressed uncontrolled growth. Animal studies show that the growth rate of implanted tumour cells was negatively regulated by PER2 (Miyazaki et al, 2010). Together, these observations suggest that PER2 behaves like a tumour suppressor and likely operates in a network with other tumour suppressors. Some tumour suppressors including p53 and promyelocytic leukaemia (PML) are known to physically interact or have an interactive regulatory role (Pearson et al, 2000; de Stanchina et al, 2004). For example, the anti-proliferative activity of p53 is in part mediated through PML (de Stanchina et al, 2004). The sequestration of MDM2, the major negative regulator of p53, to the nucleolus is dependent on PML (Bernardi et al, 2004). In addition, the acetylation of p53 by CBP is modulated by PML's regulation of CBP degradation in the nucleus (St-Germain et al, 2008). The deacetylation of p53 by SIRT1 occurs in PML nuclear bodies (PML NBs; Langley et al, 2002). Recently, SIRT1 was shown to be in a complex with BMAL1/CLOCK and its deacetylation activity regulates expression of Per2 (Asher et al, 2008; Nakahata et al, 2008).
We postulate that members of the tumour suppressor network could have a role in the mammalian clock mechanism. Of particular interest to us is PML, whose roles in the DNA damage response, cell division control and chromosome instability have been well documented (Salomoni and Pandolfi, 2002; Bernardi and Pandolfi, 2007). A reciprocal chromosomal translocation t(15;17), which fuses the PML and the retinoic acid receptor alpha (RARα) genes, is the underlying cause of over 95% of acute promyelocytic leukaemia (APL) cases. Here, we show that PML is a clock regulator that acts by regulating PER2 nuclear localization. Loss of PML disrupts PER2 nuclear localization and dampens expression of mPer2 and other key circadian regulators in peripheral and SCN clocks. We show that amino acid K487 of PML is required for nuclear localization and is acetylated. Acetyl-PML deacetylation is carried out by SIRT1. Deacetylation of acetyl-PML promotes PER2 nuclear localization. Nuclear PER2 interacts with BMAL1/CLOCK resulting in enhanced heterodimer mediated transcription. Finally, we show that Pml−/− mice have an abnormal phase shift response to a light pulse. Its period displays reduced precision and stability consistent with a role for PML as a mammalian clock regulator.
Results
PML functionally and physically interacts with PER2
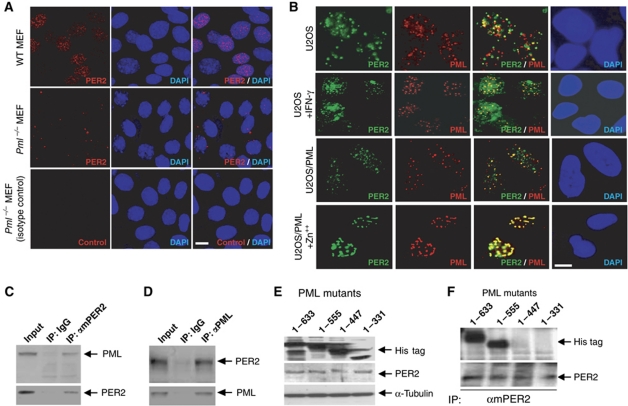
Immunofluorescence (IF) staining for PER2 and PML in MEFs revealed a significant difference in cellular localization of mPER2 between wild-type and PML-deficient (Pml−/−) cells. The majority of endogenous mPER2 appeared in the nucleus of Pml+/+ MEFs (Figure 1A; Supplementary Figure S1A). By contrast, the majority of observed endogenous mPER2 in Pml−/− MEFs was in the perinuclear region and the cytosol. This finding suggests that PML and PER2 are functionally linked. We performed double IF staining in U2OS cells to investigate whether the two proteins colocalize. We observed a limited number of the PER2 signals localized to PML NBs (Figure 1B). However, after interferon-induced expression of PML (Grotzinger et al, 1996), a significant increase in endogenous PER2 colocalization with PML NBs was observed (Figure 1B; Supplementary Figure S1B). In addition, inducible expression of PML in a stable cell line U2OS/PML by Zn++ (Wu et al, 2003) resulted in nearly 100% of endogenous PER2 localized to PML NBs. The above studies support a functional interaction between PER2 and PML.
Figure 1.
PML interacts with PER2. (A) Endogenous mPER2 in MEFs: Wild type (WT, top panel) and Pml−/− (middle and lower panel). (B) PML colocalization with PER2. Double IF staining with PER2 and PML antibodies in U2OS cells with (second row) or without (first row) treatment with 1500 U/ml γ-interferon, or stable U2OS/PML cell induced with (fourth row) or without Zn++ (third row). Western analyses were carried out with mPER2, PML or His tag as primary antibody. (C) Western analysis of immunoprecipitates using IgG or mPER2 antibody. (D) Western analysis of immunoprecipitates using IgG or PML antibody. (E) Western analysis of lysates from cells transfected with cDNA's expressing various deletion mutants of PML. (F) Western analysis of immunoprecipitates pulled down from lysates of PML mutants using an mPER2 antibody. Bars: 10 μm.
We further examined the possible physical interaction between PML and PER2 in U2OS cells using co-immunoprecipitation (Figure 1C and D). These results revealed that both the PML antibody and the mPER2 antibody co-immunoprecipitated PML and PER2 in U2OS cells. In addition, all five known isoforms of PML (Fagioli et al, 1992) were able to interact with PER2 in U2OS cells (Supplementary Figure S2). To identify the PML region that interacts with PER2, expression constructs with a His tag were generated from serial deletions of PML4 cDNA (Figure 1E). Co-immunoprecipitation with mPER2 and His-tag antibodies revealed that PML polypeptides containing amino acids 1–447 and 1–331 did not interact with PER2 in U2OS cells (Figure 1F). This study demonstrated that a domain within amino acids 448–555 of PML is required for physical interaction with PER2.
PML regulates mPER2 expression
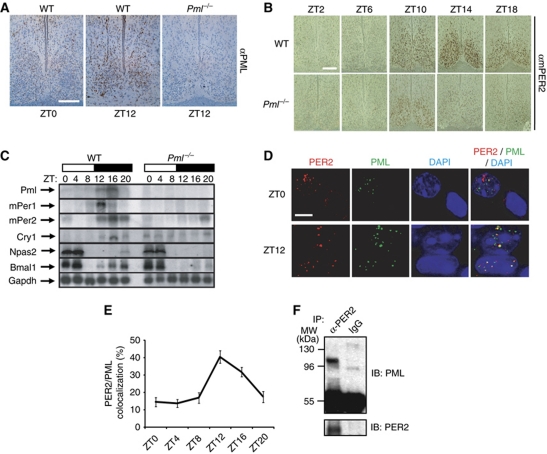
The master clock in the SCN is essential for circadian rhythm in mammals (Schwartz and Zimmerman, 1991). If the PML–PER2 interaction has biological relevance to central clock function, then PML should be expressed in the SCN. Immunohistochemical staining of SCN sections from wild-type mice with a PML antibody revealed oscillating PML levels that peak around ZT12 (Figure 2A; Supplementary Figure S3). No such immunoreactivity to PML antibody was observed in SCN sections obtained at ZT12 from Pml−/− animals. These observations indicate that PML is under clock control. Immunohistochemical staining of SCN sections with the mPER2 antibody revealed dampened mPER2 levels in Pml−/− mice when compared with wild-type animals (Figure 2B; Supplementary Figure S4). These observations suggest that PML regulates mPER2 expression.
Figure 2.
PML in the SCN and peripheral clocks upregulates mPER2 expression. (A) Immunohistochemical staining with anti-PML monoclonal antibody of SCN obtained from wild-type and Pml−/− mice. Bar: 200 μm. (B) Temporal analysis by immunohistochemical staining with mPER2 antibody of SCN sections obtained from wild-type and Pml−/− mice. Bar: 200 μm. (C) Northern analysis of liver total RNA from wild-type and Pml−/− mice obtained at various ZT points for expression of Pml, mPer1, mPer2, Cry1, Bmal1, Npas2 and Gapdh. Blots were hybridized with radiolabelled cDNA probes for the respective genes. (D) Immunohistochemical staining of PER2 (red) and PML (green) in the wild-type mice SCN at ZT0 and ZT12 observed under × 1000 magnification. Colocalization of PER2 and PML is shown as yellow. Note the nuclear or cytosolic distribution of PER2 at ZT0 in two adjacent nuclei that differentially expressed PML. Bar: 10 μm. (E) Percentage of PER2 and PML in the SCN that colocalized between ZT0 and ZT20 (n>10). (F) Western analysis with anti-PML antibodies of immunoprecipitates from anti-PER2 antibody or IgG with SCN lysates prepared from 20 mouse brains at ZT14.
The expression levels of temporally expressed core clock genes, including Npas2, Bmal1, Cry1, mPer1 and mPer2, were also examined in Pml−/− mice. Northern blot analysis of liver total RNA from Pml−/− and wild-type mice revealed moderate to severe reductions in the expressions of these genes in Pml−/− mice when compared with wild-type animals (Figure 2C; Supplementary Figure S5). Quantification analysis revealed that peak expressions of mPer1, mPer2 and Cry1 were significantly dampened in Pml−/− mice when compared with wild-type mice. Both Npas2 and Bmal1 expressions were apparently not affected significantly by the loss of PML. As in the SCN, expression of PML displayed temporal control in the liver. Together, these findings implicate PML as a positive regulator of the clock transcriptional mechanism.
We further investigated whether the PML–PER2 interaction observed in cultured cells also occurs in the SCN. Temporal interaction between PML and mPER2 in the SCN was visualized using a deconvolution microscope by staining with PML and mPER2 antibodies. Expression of both proteins was low between ZT0 and ZT8 and reached peak levels around ZT12–ZT16 (Figure 2B; Supplementary Figures S3 and S6). At ZT12, the majority of nuclei that expressed PML also expressed mPER2 in the SCN (Supplementary Figure S6). There was significantly less overlapping expression of PML and mPER2 in the SCN at ZT0 (Figure 2D). At ZT12, a significant level of mPER2 and PML colocalized in the nucleus suggesting that they interact. This interaction between mPER2 and PML appears under temporal control as the level of colocalized mPER2 and PML was observed to increase from ZT0–ZT4 to ZT12–ZT16 (Figure 2E). In addition, the mPER2 nuclear distribution pattern appears to be influenced by PML. At ZT0, a nucleus that expressed PML also displayed nuclear mPER2, while an adjacent cell nucleus that did not show PML expression displayed a perinuclear/cytosolic mPER2 distribution (Figure 2D). These observations are consistent with results from Pml−/− MEF where the mPER2 cellular distribution was primarily perinuclear/cytosolic, indicating that PML regulates PER2 nuclear localization. To further substantiate the PML–PER2 interaction in the SCN, total lysates were prepared from SCN collected from 20 mouse brains killed at ZT14. Co-immunoprecipitation assay was carried out with either anti-PER2 antibody or IgG. Western analysis with anti-PML monoclonal antibodies specific to mouse PML demonstrated that the anti-PER2 antibody but not IgG pulled down mouse PML in the SCN extract (Figure 2F). Together, these studies demonstrate that PML expression and its interaction with mPER2 in the SCN are under temporal control.
Enhancement of BMAL1/NPAS2 and BMAL1/CLOCK-mediated transcription by PML requires PER2
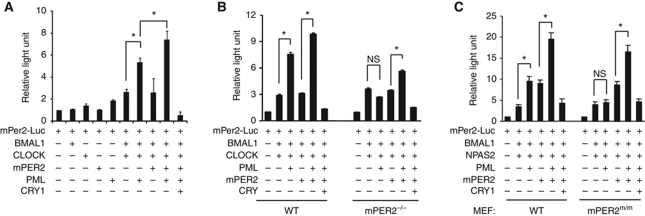
Given the above observations, we investigated whether PML could regulate mPer2 transcription. The core transcriptional complex that drives the mPer2 and mPer1 promoters consists of BMAL1/NPAS2 or BMAL1/CLOCK heterodimers in mammals (Kume et al, 1999; Reick et al, 2001). Possible PML enhancement of BMAL1/NPAS2 and BMAL1/CLOCK transcriptional activity was investigated using co-transfection of expression constructs of other clock genes into wild-type MEF or U2OS cells with an mPer2 promoter luciferase (mPer2-Luc) reporter construct (Travnickova-Bendova et al, 2002). Expression of BMAL1/NPAS2 and BMAL1/CLOCK transcriptional complexes activated mPer2 promoter activity as previously described (Kaasik and Lee, 2004). Co-expression of PML potently enhanced the transcriptional activity of both BMAL1/NPAS2 and BMAL1/CLOCK (Figure 3A–C; Supplementary Figure S7). Co-expression of mPER2 moderately enhanced BMAL1/NPAS2, consistent with previous observations (Kaasik and Lee, 2004). That PER2 can enhance BMAL1/NPAS2-mediated transcription has been independently observed (Hampp et al, 2008; Zhang et al, 2008). Co-expression of both PML and mPER2 produced further enhancement of BMAL1/NPAS2 and BMAL1/CLOCK transcriptional activities, and these activities were repressed by CRY1.
Figure 3.
PML enhances BMAL1/CLOCK and BMAL1/NPAS2 transcription requires PER2. Reporter assays were carried out using the mPer2 promoter luciferase (mPer2-luc) construct in combination with the indicated expression constructs. (A) BMAL1/CLOCK transcription of mPer2-luc in wild-type MEF cells. (B) BMAL1/CLOCK transcription in MEFs from wild-type and mPer2−/− mice. (C) BMAL1/NPAS2 transcription in MEFs from wild-type and mPer2m/m mice. Luciferase activity in samples transfected with mPer2-luc alone was arbitrarily set at 1.0. Error bars indicate s.e.m. (n=3). *P<0.05, t-test.
Using primary MEFs that were generated from wild-type, mPer2m/m and mPer2−/− mice, we investigated whether PML enhancement of heterodimer transcription requires PER2. The mPer2m/m mouse has an in-frame deletion of the PASB domain that rendered PER2 non-functional (Zheng et al, 1999). In wild-type MEFs, the results were similar to findings from U2OS cells (Figure 3A–C; Supplementary Figure S7). However, using either mPer2m/m or mPer2−/− MEFs, the enhancement of BMAL1/NPAS2 and BMAL1/CLOCK transcriptional activity by PML was abolished (Figure 3B and C). Transfection of PML or PER2 alone had no effect on mPer2-luc promoter activity using mPer2m/m or mPer2−/− MEFs (Supplementary Figure S8). The enhancement of BMAL1/NPAS2 and BMAL1/CLOCK transcriptional activity was restored by the co-expression of mPER2 and PML in mPER2-deficient cells. These reporter assay results demonstrate that the enhancement of BMAL1/NPAS2 and BMAL1/CLOCK transcriptional activity by PML is mediated through PER2.
PML is required for nuclear localization of PER2
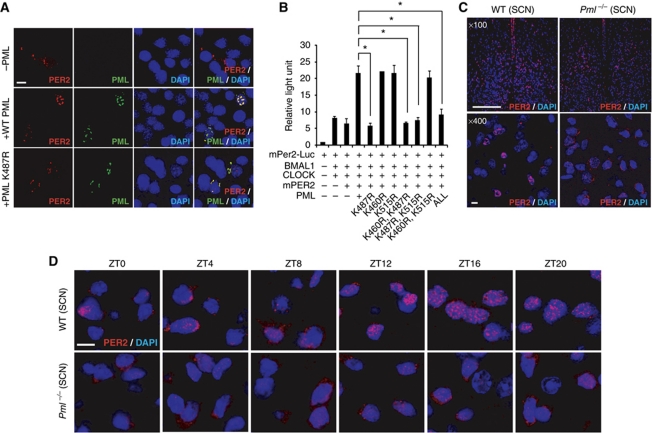
Results presented in Figure 1A demonstrate that PER2 was largely localized outside the nucleus in the absence of PML. To rule out the possibility that the PER2 distribution may be caused by its lower endogenous expression level in Pml−/− MEFs, mPER2 levels were enhanced by transfection of an mPer2 expression construct into Pml−/− MEFs. The increase in mPER2 levels did not alter its distribution in Pml−/− MEF and strongly support the notion that PML function is necessary for PER2 nuclear entry (Figure 4A, upper panel; Supplementary Figure S9). Results presented in Figure 1F indicated that the PML domain consisting of amino acids 448–555 is necessary for PER2 interaction. Within this domain, sumoylation and acetylation sites have previously been identified (Kamitani et al, 1998; Hayakawa et al, 2008). Previous studies have shown that converting lysine (K) to arginine (R) at amino-acid position 490 disrupts sumoylation of PML (Kamitani et al, 1998). However, a K490R PML mutation did not abolish PML/PER2 enhancement of BMAL1/CLOCK transcription of the mPer2-Luc promoter (Supplementary Figure S10). It was reported that amino-acid position K487 of PML is strongly acetylated while amino acids K515 and K460 might be acetylated (Hayakawa et al, 2008). Site-specific mutagenesis was carried out to substitute arginine for lysine at these amino-acid positions. Reporter assay with either wild-type MEF or U2OS cells showed that K487R, but not K515R or K460R, abolished PML/PER2 enhancement of BMAL1/CLOCK transcriptional activity (Figure 4B; Supplementary Figure S11).
Figure 4.
Nuclear localization of PER2 is regulated by PML. (A) Immunofluorescence staining with mPER2 or PML antibody. Pml−/− MEFs transfected with expression constructs: mPer2 only (top panel), mPer2 and Pml (middle panel), mPer2 and Pml K487R (lower panel). Bar: 10 μm. (B) Reporter assay using wild-type MEFs with PML carrying lysine (K) to arginine (R) substitution at amino-acid positions 460, 515 and 487 (n=2). (C) Immunohistochemical analysis with mPER2 antibody of SCN sections from wild-type or Pml−/− mice at ZT12 view at × 100 (Bar: 200 μm) and × 400 (Bar: 10 μm) magnification. (D) Immunohistochemical analysis with mPER2 antibody of the SCN sections from wild-type or Pml−/− mice from sampled every 4 h from ZT0 to ZT20 viewed at × 1000 magnification. Bar: 10 μm. *P<0.05, t-test.
In Pml−/− MEFs transfected with expression constructs for wild-type Pml and mPer2, mPER2 expression in PML-positive cells colocalized with PML in the nucleus (Figure 4A, middle panel). When Pml−/− MEFs were transfected with expression constructs for Pml K487R and mPer2, immunostaining revealed that the majority of PML K487R and mPER2 colocalized in the perinuclear and cytosolic region (Figure 4A, lower panel). In contrast, PML with either the K515R or K460R mutation was localized to the nucleus (Supplementary Figure S12). These findings indicate that amino acid K487 is important for localization of PML in the nucleus. Failure of PML K487R to enter the nucleus also blocks mPER2 nuclear localization.
Next, we investigated whether the observed perinuclear and cytosolic distribution of mPER2 in Pml−/− MEFs is replicated in the master clock of the SCN in vivo. The distribution of mPER2 at ZT12 in the SCN of wild-type and Pml−/− mice was investigated by IF with an mPER2 antibody and visualized by fluorescence deconvolution microscopy (Figure 4C). The distribution of mPER2 in the SCN of wild-type mice appeared as a robust nuclear signal. In contrast, the SCN of Pml−/− mice displayed a predominantly perinuclear and cytosolic mPER2 distribution. Unlike wild-type mice, which exhibited temporal control of localization of mPER2 from the cytosol to the nucleus, the pattern of mPER2 distribution of Pml−/− mice remained predominantly perinuclear/cytosolic throughout ZT0–ZT20 in the SCN (Figure 4D; Supplementary Figure S13). Taken together, these findings in Pml−/− SCN and MEF cells (Figure 1A) demonstrate that PML regulates PER2 nuclear localization.
SIRT1 deacetylation of acetyl-PML is critical for nuclear localization of PER2
The above results demonstrate that amino-acid residue K487 of PML, the major site of PML acetylation, is essential for PML nuclear localization. SIRT1 is an NAD+-dependent deacetylase (Guarente, 2007). Previous studies have demonstrated that SIRT1 colocalizes with PML NBs (Langley et al, 2002). SIRT1 is also part of the BMAL1/CLOCK complex (Nakahata et al, 2008). Since PER2 also interacts with BMAL1/CLOCK in the nucleus, we postulated that SIRT1 could modulate the deacetylation of acetyl-PML and also regulate nuclear localization of PER2.
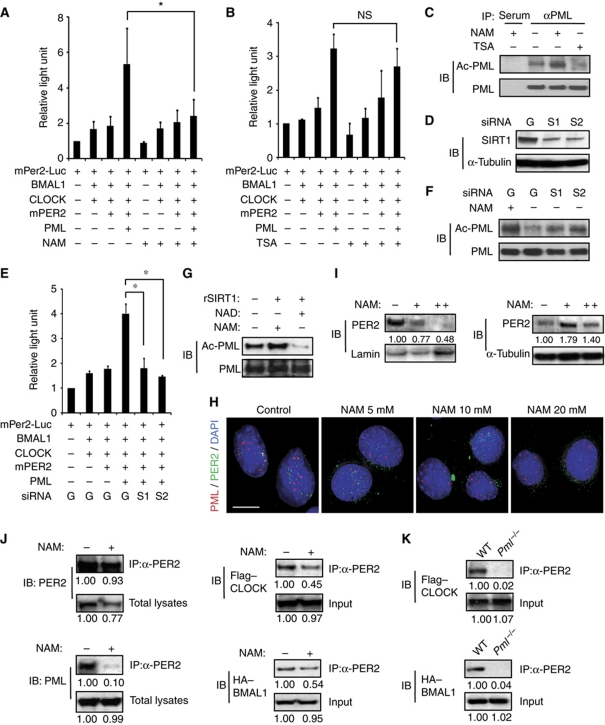
To investigate whether SIRT1 is involved in PML/PER2-mediated activation of BMAL1/CLOCK transcriptional activity, U2OS or wild-type MEF cells transfected with expression constructs for reporter assay were treated with trichostatin A (TSA) or nicotinamide (NAM) prior to harvest. SIRT1 deacetylase activity is sensitive to inhibition by NAM but not to TSA, a class I and II histone deacetylase (HDAC) inhibitor. Luciferase assay measurements showed that PER2/PML-mediated activation of BMAL1/CLOCK transcriptional activity was not affected by TSA but was inhibited by NAM (Figure 5A and B; Supplementary Figure S14).
Figure 5.
SIRT1 deacetylates acetyl-PML. Reporter assay using mPer2-Luc in U2OS cells in combination with the indicated expression constructs plus: (A) 10 mM NAM (n=3) and (B) 10 μM TSA (n=3). (C) Western analysis with anti-acetylated-lysine antibody of immunoprecipitates from PML antibody. Immunoprecipitation lysates were from U2OS/PML cells after the indicated treatment. (D) Western analysis of lysates from U2OS/PML cells treated with siRNA's to GFP [G] or SIRT1-1 [S1] and SIRT1-2 [S2]. (E) Reporter assay with the indicated expression constructs (n=3). (F) Western analysis with anti-acetylated-lysine antibody of immunoprecipitates using a PML antibody. Immunoprecipitation lysates were from U2OS/PML cells, treated as indicated. (G) Western analysis with anti-acetylated-lysine antibody of acetyl-PML after treatment with recombinant SIRT1 in the presence of NAD+ or NAM. The lower panel showed the same blot reprobed with anti-PML antibodies. (H) Wild-type MEFs were treated with the indicated amount of NAM for 24 h and stained with mPER2 and PML antibodies. Bar: 10 μm. (I) Western analysis with mPER2 antibody of fractionated NAM treated (+: 10 mM, ++: 15 mM, 14 h) MEFs. (J) Western analysis of immunoprecipitates using mPER2 antibody with lysates from wild-type MEFs transfected with FLAG–CLOCK and HA–BMAL1 expression vectors treated with or without 10 mM NAM. (K) Western analysis of anti-PER2 immunoprecipitates of wild-type or Pml−/− MEFs transfected with FLAG–CLOCK and HA–BMAL1 expression vectors. Relative densities of the bands are shown below blots. *P<0.05, t-test.
Next, the effect of NAM on the acetylation status of PML was investigated. Lysates prepared from U2OS or wild-type MEF cells stably transfected with PML and cultured with and without NAM or TSA were immunoprecipitated with PML antibody or preimmune serum. Western blot analysis showed that PML protein reactivity to anti-acetylated-lysine antibody was enhanced by NAM treatment when compared with TSA treatment or untreated controls (Figure 5C; Supplementary Figures S15 and S16). When the blot was reprobed with PML antibody, a similar level of PML protein was present in each sample except when preimmune antibody was used for immunoprecipitation (Figure 5C; Supplementary Figure S15). To establish that SIRT1 modulates PML/PER2 enhancement of BMAL1/CLOCK transcriptional activity, siRNA inhibition studies were undertaken. First, two constructs of siRNA's to SIRT1 were tested for their ability to inhibit SIRT1 expression in U2OS cells. Western analysis with SIRT1 antibody revealed that both siRNA constructs significantly inhibited SIRT1 expression (Figure 5D; Supplementary Figure S15). Then, either the two siRNAs to SIRT1 or an siRNA targeted to green fluorescence protein (GFP) as control were transfected into U2OS cells in combination with expression constructs for reporter assay. Luciferase assay measurements revealed that PML/PER2 enhancement of BMAL1/CLOCK transcription was inhibited by the siRNA to SIRT1 but not by the siRNA to GFP (Figure 5E). In addition, after the SIRT1-specific siRNA treatment, the acetylation status of PML was determined by immunoprecipitation with PML antibody followed by western analysis with anti-acetyl-lysine antibody. The results show that siRNA's to SIRT1, but not the siRNA to GFP, enhanced the acetylation state of PML (Figure 5F; Supplementary Figure S15). To further show that deacetylation of acetyl-PML is carried out by SIRT1, we undertook immunopurification of acetyl-PML. The acetyl-PML was then treated with recombinant SIRT1 protein in the presence of NAD+ or NAM. Western analysis with pan-acetyl-lysine antibodies showed that acetyl-PML treated with SIRT1/NAM and an untreated control were each strongly reactive (Figure 5G; Supplementary Figure S15). In contrast, after SIRT1/NAD+ treatment, the level of acetyl-PML reactive to pan-acetyl-lysine antibodies was reduced. Reprobing of the blot with PML antibodies showed comparable levels of PML in each lane (Figure 5G; Supplementary Figure S15). Jointly, these findings demonstrate that acetyl-PML deacetylation is carried out by SIRT1 in the presence of NAD+.
Next, we investigated whether PML deacetylation by SIRT1 regulates PER2 localization in the nucleus. Given that K487R PML was unable to enter the nucleus (Figure 4A), our mutation studies could not address whether changes in PML acetylation status via the loss of K487 regulates PER2 nuclear localization. Therefore, using wild-type MEF cells, we investigated whether treatment with NAM, which inhibits PML deacetylation by SIRT1, would affect PER2 nuclear localization. Wild-type MEFs were treated with increasing concentrations of NAM and nuclear localization of endogenous mPER2 and PML was monitored by IF using PML and mPER2 antibodies (Figure 5H). In the absence of NAM, both mPER2 and PML appear primarily in the nucleus. Increasing concentrations of NAM, which increases K487PML acetylation (Figure 5C; Supplementary Figures S15 and S16), did not affect PML nuclear localization (Figure 5H; Supplementary Figure S17). By contrast, increasing NAM concentrations correlate with an increased perinuclear/cytosol distribution of mPER2 (Figure 5H; Supplementary Figure S17). In addition, nuclear and cytosolic fractionation studies followed by western analysis demonstrate that NAM treatment decreased mPER2 levels in the nuclear fraction (Figure 5I, left panel). In contrast, the cytosolic fraction showed increased mPER2 levels (Figure 5I, right panel). Western blotting of an MEF cell lysate from untreated cells and cells treated with 10 mM NAM demonstrated that co-immunoprecipitation of PML and PER2 was significantly disrupted by NAM treatment (Figure 5J). Consistently, mPER2 interaction with the BMAL1/CLOCK complex was significantly decreased as shown by co-immunoprecipitation using epitope-tagged proteins (Figure 5J). Supportive of these observations is that in Pml−/− MEF cells the interaction of PER2 with CLOCK and BMAL1 was also significantly reduced or not detectable (Figure 5K). Taken together, these studies show that deacetylation of PML by SIRT1 regulates PML control of mPER2 nuclear localization and its subsequent interaction with the BMAL1/CLOCK heterodimer.
The period length of Pml−/− mice displays reduced precision and stability
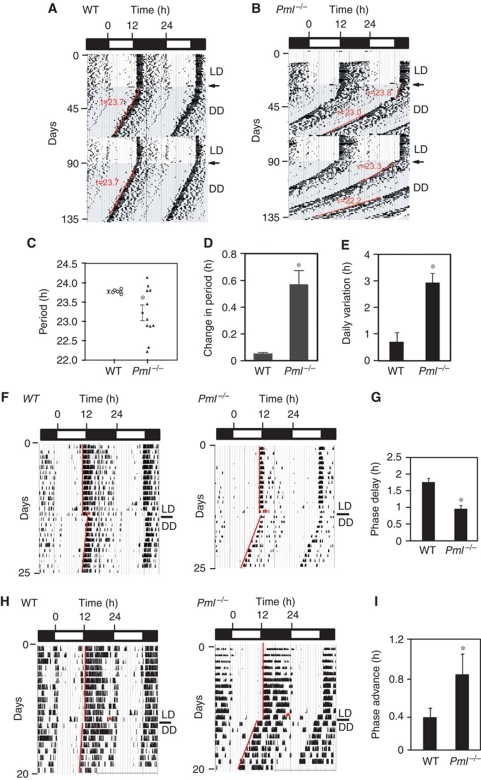
Finally, we investigated whether PML regulates circadian rhythm behaviour. We assessed the behavioural phenotype of Pml−/− mice by monitoring their wheel-running activity. Wild-type and inbred Pml−/− (129sv) mice were entrained initially to a 12 h:12 h light-dark (LD) cycle for about 3 weeks and then released into a 12 h:12 h dark-dark (DD) or free-running environment. Wild-type mice exhibited an average period length of 23.6±0.1 h (mean±s.e.m., n=6) (Figure 6A and C). For Pml−/− mice, the average period length was shorter at 23.2±0.2 h (mean±s.e.m., n=12) but exhibited significant variability (Figure 6B and C). In addition, under prolonged free-running conditions, an individual Pml−/− mouse would gradually or suddenly change its endogenous period length (Figure 6B).
Figure 6.
Circadian phenotype of Pml−/− mice. The wheel-running actograms are double plotted showing a 12 h light/12 h dark cycle (LD) with the shaded background indicating a free-running period of 12 h dark/12 h dark (DD) cycles. Transition from LD to DD is indicated by an arrow. Actograms are representative of (A) a wild-type mouse and (B) A Pml−/− mouse. Note the instability in period (τ). (C) Summary of periods obtained from wild-type and Pml−/− mice. Error bar indicates s.e.m. *P<0.05. (D) Average difference in observed period length after re-entrainment of individual wild-type (n=6) and Pml−/− (n=12) mice. Error bar indicates s.e.m. *P<0.05. (E) Average daily variant from period length of individual wild-type (n=6) and Pml−/− (n=12) mice. Error bar indicates s.e.m. *P<0.001. (F, G) Phase delay response to ZT14 light pulse (15 min) of wild-type and Pml−/− mice. (H, I) Phase advanced response to ZT22 light pulse (15 min) of wild-type and Pml−/− mice.
To assess this phenotype further, the same Pml−/− and wild-type mice were re-entrained to an LD cycle followed by a second release into a free-running condition. For wild-type mice, the periods measured from the second release into DD were all within ±0.1 h of their first measurements, a clear indication that their endogenous clocks were stable and precisely controlled. However, the majority of the Pml−/− mice had periods that differed significantly from the first measurement (Figure 6B and D). Both period lengthening and shortening were observed and there was no particular bias to the direction of these changes. Comparing the longest and shortest period length of an individual mouse measured by linear regression sampled over at least 10 days revealed an average difference of 0.5±0.1 h (mean±s.e.m., n=12) for Pml−/− mice compared with 0.06±0.03 h (mean±s.e.m., n=6) for wild-type mice. Next, the precision of activity onset was compared between Pml−/− and wild-type mice. The comparison revealed a 3 × greater daily variation in activity onset from mean free-running period for Pml−/− mice (Figure 6E). Finally, we examined the loss of PML on the phase shift response of mice using a protocol in which a light pulse was given at either ZT14 or ZT22 on the last day of entrainment prior to their release into a free-running environment (Albrecht et al, 2001; Spoelstra et al, 2004). Using this protocol, the level of phase delay in response to a ZT14 light pulse averaged −1.9±0.1 h (mean±s.e.m., n=11) in wild-type, whereas Pml−/− mice displayed a significantly reduced phase delay response that averaged −1.0±0.1 h (mean±s.e.m., n=6) (Figure 6F and G). When the light pulse was given at ZT22, wild-type mice displayed a phase advance response that averaged +0.4 h±0.1 h (mean±s.e.m., n=11). In contrast, the Pml−/− mice displayed enhanced phase advance that averaged +0.9 h±0.3 h (mean±s.e.m., n=8) (Figure 6H and I). These observations indicate that the loss of PML affects the phase shift response, period length and stability of the endogenous circadian clock. Together, the wheel-running behaviours of Pml−/− mice indicate a compromised endogenous clock, which is consistent with PML having a regulatory role in the mammalian clock mechanism.
Discussion
Our current study was prompted by observations suggesting that some tumour suppressors and circadian regulators could have an interactive regulatory role. Expression of p53 and key cell-cycle regulators has been shown to be under circadian control (Bjarnason et al, 1999). Our own studies have implicated mPER2 in tumour suppression and DNA damage control (Fu et al, 2002). Several studies have now demonstrated that increased PER2 expression in a variety of human and mouse cancer cell lines inhibits uncontrolled tumour growth, suggesting a tumour suppressor function (Gery et al, 2005; Hua et al, 2006; Oda et al, 2009; Miyazaki et al, 2010). A recent study showed that loss of mPER2 function further enhances the spontaneous tumour rate in p53−/− mice (Lee et al, 2010). Our present study focusing on the tumour suppressor PML was prompted by our initial observations in cultured cells that PER2 and PML were colocalized in the nucleus and that the cellular distribution of PER2 was altered in PML-deficient cells. Our locomotor behaviour analysis demonstrated that PML-deficient mice exhibit a compromised endogenous clock that lacks precision and stability, consistent with the notion that PML plays a regulatory role in the mammalian clock. Under free-running conditions, a loss of rhythm can be observed in PER2-deficient mice but not in Pml−/− mice. However, both genotypes have enhanced phase advance and reduced phase delay responses to ZT22 and ZT14 light pulse, respectively (Albrecht et al, 2001).
Expression of PER2 and other key clock regulators is dampened in Pml−/− mice, indicating that PML is a positive regulator in the mammalian clock mechanism. Co-transfections and reporter assays with the mPer2 promoter showed that PML enhancement of BMAL1/CLOCK-mediated transcription required PER2. Co-immunoprecipitation experiments revealed that PML interacts with PER2 through a domain between amino acids 448 and 555, a region known to contain the nuclear localization signal and post-translational modification sites involving SUMOylation (K490) and acetylation (K487) (Kastner et al, 1992; Kamitani et al, 1998; Hayakawa et al, 2008). Our studies revealed that a K487R but not a K490R mutation affected PML/PER2 enhancement of BMAL1/CLOCK-mediated transcription. Previous studies have demonstrated that sumoylation at K490 was required for PML NB formation but a K490R mutation did not block PML nuclear entry (Brand et al, 2010). Consistent with previous findings, we observed that K487 is required for PML nuclear entry and this residue is acetylated (Hayakawa et al, 2008; Jul-Larsen et al, 2009). In Pml−/− MEF or those expressing PML K487R, mPER2 remained primarily outside the nucleus. Interestingly, cytosolic PML K487R and mPER2 remained colocalized demonstrating that the loss of PML K487 did not abolish PML/PER2 interaction. This observation is consistent with deletion mapping studies implicating the carboxyl region of PML as necessary for interaction with PER2. That PML is essential for nuclear localization of PER2 is further supported by the observation that the SCN of Pml−/− mice displayed a perinuclear/cytoplasmic distribution of mPER2 throughout a diurnal cycle. Recently, PML has also been shown to be involved in the nuclear localization of two other nuclear proteins, CBP and Rb (Jul-Larsen et al, 2009; Regad et al, 2009). It was proposed that PML promotes the nucleocytoplasmic transport of nuclear proteins through its interaction with nuclear membrane proteins including nucleoporins and lamin (Jul-Larsen et al, 2009, 2010). The acetylation of PML is enhanced by CBP, which has histone acetyltransferase activity (Hayakawa et al, 2008). Interestingly, CBP is known to interact with the BMAL1/CLOCK complex (Takahata et al, 2000). In transgenic Drosophila, CBP knockdown increases period length and its overexpression results in arrhythmic circadian behaviour (Lim et al, 2007). These observations suggest that CBP has a role in the clock mechanism. Therefore, PML may play a broader role in the nucleocytoplasmic transport of core clock or clock-controlled proteins.
Inhibition of SIRT1 by siRNA resulted in an increase in acetyl-PML. In addition, we demonstrated that acetyl-PML is deacetylated by recombinant SIRT1 in the presence of NAD+, a process inhibited by NAM. NAM, an SIRT1 inhibitor, which increases PML acetylation did not block PML entry into the nucleus. However, nuclear entry of PER2 was inhibited by NAM treatment. Consequently, PER2 interaction with CLOCK and BMAL1 in the nucleus was reduced. Therefore, we propose that acetyl-PML entry into the nucleus is independent of its interaction with PER2, and deacetylation of lysine 487 mediates PML regulation of PER2 nuclear entry. SIRT1 is also involved in the deacetylation of acetyl-BMAL1 (Nakahata et al, 2008). The acetylation of BMAL1 facilitates the recruitment of CRY1 to the BMAL1/CLOCK complex leading to inhibition of transcription (Hirayama et al, 2007). Observation showing that Sirt1−/− cells have highly dampened mPer2 expression is supportive of this model (Nakahata et al, 2008). Thus, the deacetylation of both acetyl-BMAL1 and acetyl-PML by SIRT1 can act in concert to enhance BMAL1/CLOCK-mediated transcription of its target genes since this modification blocks CRY1 (negative regulator) recruitment to BMAL1 and enhances PML transport of PER2, a positive regulator, into the nucleus. In turn, the deacetylase activity of SIRT1 is regulated by the circadian control of NAD+ salvage, creating a closed loop feedback regulation (Nakahata et al, 2009; Ramsey et al, 2009). Our studies demonstrate that SIRT1 deacetylation of PML is part of the positive loop that regulates PER2 nuclear localization leading to enhanced BMAL1/CLOCK-mediated transcription.
PER2 has been proposed to act as a negative regulator of BMAL1/CLOCK or BMAL1/NPAS2 transcription (Kume et al, 1999). However, such a role contradicts observations that a dampened expression of several key clock genes in the SCN and peripheral tissues was observed in PER2-deficient mice (Zheng et al, 1999, 2001; Kaasik and Lee, 2004). In the studies of Kume et al (1999), only a modest inhibition of CLOCK/BMAL1-mediated transcription of the Per1 promoter by PER2 was observed. These authors further showed that PER2 failed to inhibit BMAL1/MOP4-mediated transcription of the AVP promoter. In contrast, CRY inhibition of BMAL1/MOP4 and BMAL1/CLOCK-mediated transcription was equally potent with both Per1 and AVP promoters. Given that MOP4 (NPAS2) is the redundant homologue of CLOCK, the proposal that PERs act together with CRY to inhibit core clock heterodimer transcription activity is inconsistent with these and other observations. Substantial evidence supporting the notion that PER2 activates rather than represses BMAL1/NPAS2 transcription has been reported by our group as well as by other investigators (Kaasik and Lee, 2004; Hampp et al, 2008; Zhang et al, 2008). Furthermore, recent reconstitution studies using recombinant proteins demonstrated that PER2 actually blocks CRY interaction with BMAL1/CLOCK when the heterodimer complex was bound to its targeted E-box DNA (Ye et al, 2011). These studies further showed that even when CRY was bound to the BMAL1/CLOCK:E-box it was effectively removed by the addition of PER2. Thus, these studies demonstrate that PER2 acts opposite to CRY in the clock mechanism.
In summary, we demonstrate that PML is a circadian clock regulator. Our studies further demonstrate the integration of tumour suppression and clock function, raising the possibility that other tumour suppressors could have direct roles in circadian control.
Materials and methods
Animals
Wild-type (129sv), Pml−/− (129sv), mPer2−/− and mPer2m/m mice were housed in a standard animal maintenance facility under a 12 h light:12 h dark cycle. The PML−/− animals were obtained from the Mouse Models of Human Cancers Consortium repository, National Cancer Institute (Wang et al, 1998). All animals studies were carried under Institutional approved animal protocol: HSC—AWC-06-077 and ACUF 05-07-05731.
Cell lines, cell culture and plasmids
Immortalized Pml−/− and wild-type MEFs were established as described previously (Xu et al, 2005). Primary MEFs were generated from 13.5 days gestation embryos as previously described (Fu et al, 2002). U2OS, stably transfected U2OS/PML cells were cultured in DMEM as previously reported (Wu et al, 2003). The full-length cDNAs of PML1, PML2, PML3, PML4 and PML5 were constructed as previously reported (Wu et al, 2001). PML deletion mutants were constructed as described (Wu et al, 2001). Plasmid constructs for Clock, Bmal1, Npas2, Cry1 and mPer2 were as previously described (Kaasik and Lee, 2004).
Antibodies
Mouse anti-tubulin monoclonal antibody was purchased from Sigma-Aldrich. The rabbit PML polyclonal antibody was raised against the GST–PML fusion protein (Wu et al, 2003). The mPer2 antibody was as described previously (Zheng et al, 2001). To demonstrate the specificity of our mPer2 antibody, Supplementary Figure S18 shows that it recognized the 170-kDa PER2 protein in wild-type but not in Per2−/− MEFs. In addition, the antibody exhibited negative IF signal in Per2−/− MEF cells. The mouse PML monoclonal antibody was from Upstate Biotechnology. The anti-acetyl lysine antibody was from Cell Signaling Technology. The hSIRT1 antibody was from Santa Cruz Biotechnology.
siRNA knockdown
The siRNAs for SIRT1 (#133051-HSS177403: SIRT1-1, -HSS177404: SIRT1-2) were from Invitrogen. The siRNA for EGFP was from Ambion. For SIRT1 knockdown, cells were seeded in a D100 dish at a density of 3 × 106/dish and transfected with 50 μM of siRNAs using LipofectAmine 2000 (Invitrogen).
IF staining
Cells were cultured in six-well plates on sterile cover slips. Cells were placed on ice for 15 min, then treated with cytoskeleton buffer (10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100) for 5 min. After washing three times with phosphate-buffered saline (PBS), cells were either treated or untreated with cytoskeleton stripping buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% Tween-20, 0.25% sodium deoxycholate) for another 3 min. Cells were then fixed in cold 4% neutral paraformaldehyde in PBS for 30 min on ice, washed in PBS, and then permeabilized in a 1% Triton X-100/0.5% NP-40/PBS solution. Incubation with the primary antibody was carried out for 2 h at room temperature. Incubation with the secondary antibody was carried out for 1 h at room temperature followed by staining of DNA with 4,6-diamidino-2-phenylindole (DAPI) for 5–10 min. The mPER2 or PML antibody was detected with rabbit Alexa Fluor 555 or mouse Alexa Fluor 488 antibody (Invitrogen), respectively. Slides were then mounted with Vectashield antifade medium (Vector Laboratories) after three washes with washing buffer and examined with a fluorescence deconvolution microscope (Applied Precision, Issaquah, WA, USA).
Immunohistochemistry
Mice were killed at the indicated times. Whole brains were fixed in a 10% formaldehyde solution overnight, embedded in paraffin, and cut into 5 μm sections. Antigen retrieval was performed in citric acid salt buffer and boiling the buffer in a microwave for 15 min. The tissue slides were then treated with 3% hydrogen peroxide for 30 min, washed and blocked in 1% BSA and 5% goat serum. The PML monoclonal antibody and mPER2 polyclonal antibody were incubated for 48 h at 4°C, washed with 0.1% Tween-20 in PBS. The primary antibody was detected with biotinylated secondary antibody followed by a peroxidase-conjugated avidin, and treatment with DAB substrate and haematoxylin counterstain (Vector Laboratories). For IF, the mPER2 antibody was detected with rabbit Alexa Fluor 555 antibody (Invitrogen).
Northern blot analyses
For all northern blot analyses, 20 μg of total RNA was resolved by gel electrophoresis and transferred onto a nylon membrane. Blots were hybridized with 32P-labelled cDNA probes for mPer1, mPer2, Cry1, Bmal1, Npas2 and Gapdh as previously described (Kaasik and Lee, 2004).
Locomotor activity
Wheel-running activity was monitored as described (Zheng et al, 1999). Male wild-type and PML−/− mice aged about 16 weeks were used. Briefly, mouse were initially entrained to 12:12 h LD cycle for at least 2 weeks, followed by their released into 12:12 h DD free-running condition. For phase shift studies, the protocol was as previously described (Albrecht et al, 2001; Spoelstra et al, 2004). Zeitgeber Time zero (ZT0) is light on and ZT12 is light off.
Reporter assays
Reporter assays were carried out as previously described (Kaasik and Lee, 2004). U2OS and primary MEFs (1.3 × 105) were seeded into six-well dishes and transfected with the 1.7 k mPer2-Luc reporter construct (Travnickova-Bendova et al, 2002). Co-transfections were performed with the respective expression constructs for Clock, Bmal1, Npas2, Cry1, PML and mPer2. Cells were harvested 24 h post transfection, and luciferase assay was performed with the assay kit obtained from Promega using a TD-20/201 luminometer (Turner Designs). Co-transfection of Renilla-luc is used as internal control.
Immunoprecipitation and immunoblotting
The cells were lysed in an ice-cold lysis buffer containing 50 mM Tris–HCl, pH 7.4, 0.25 mM deoxycholate, 150 mM NaCl, 2 mM EGTA, 0.1 mM Na3VO4, 10 mM NaF, 1 mM PMSF, 10 mM NAM, 10 μM TSA and complete Protease Inhibitor (Roche). Immunoprecipitation of PML was carried out by using rabbit PML antibody and collected on protein G-agarose beads (PIERCE), washed and eluted. The immunoprecipitates and cell lysates were separated by SDS–PAGE and used for immunoblotting as described previously (Miki et al, 2007).
In-vitro SIRT1 deacetylation assay
U2OS/PML cells were transfected with pRSV-CBP and pRSV-CREB expression vectors and incubated with 10 mM NAM and 100 μM ZnSO4. Cells were lysed with lysis buffer without NAM and TSA at 24 h post transfection. Immunoprecipitation of PML was performed using anti-PML antibody. The final wash was performed with SIRT1 deacetylation buffer (50 mM Tris–HCl, pH 9.0, 5% glycerol, 50 mM NaCl, 4 mM MgCl2, 0.5 mM DTT, 0.1 mM PMSF) and the immunoprecipitated PML protein was used as a template. IP-purified PML was incubated with recombinant SIRT1 (ActiveMotif) and 1 mM NAD or 40 mM NAM for 2 h at 30°C. The reaction was stopped by adding equal volume of 2 × sample buffer. Samples were then subjected to western analysis and immunoblotting.
Supplementary Material
Acknowledgments
We thank Dr J Lever for comments on the manuscript and Dr Jianping Jin, Jolene Li and Wenxin Zou for technical support. This work was supported by grants from the National Institute of Health to CCL and KSC. CCL is also support in part by a Pioneer Award from NIH.
Author contributions: Figures 1A, 2D, 2E, 3A, 4, 5, 6F-6I and Supplementary Figures S1, S3–S6 and S8–S18 were generated by TM. Figure 1B–F and Supplementary Figures S2 and S7 were generated by ZX, Figures 2C and 6A–E were generated by MCG, Figure 2A and B were generated by ML. ZZ and AV were responsible for generating site-specific PML mutants. MAR contributed to the wheel-running analysis. CCL and KSC initiated and directed the study. This manuscript was written by CCL and KSC.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC (2001) mPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms 16: 100–104 [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP (2004) PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol 6: 665–672 [DOI] [PubMed] [Google Scholar]
- Bjarnason GA, Jordan RC, Sothern RB (1999) Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am J Pathol 154: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, Lenser T, Hemmerich P (2010) Assembly dynamics of PML nuclear bodies in living cells. PMC Biophys Mar 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW (2004) PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13: 523–535 [DOI] [PubMed] [Google Scholar]
- Fagioli M, Alcalay M, Pandolfi PP, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci PG (1992) Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene 7: 1083–1091 [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee CC (2002) The circadian gene Period2 plays an important role in tumour suppression and DNA damage response in vivo. Cell 111: 41–50 [DOI] [PubMed] [Google Scholar]
- Gery S, Gombart AF, Yi WS, Koeffler C, Hofmann WK, Koeffler HP (2005) Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood 106: 2827–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger T, Jensen K, Will H (1996) The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J Biol Chem 271: 25253–25260 [DOI] [PubMed] [Google Scholar]
- Guarente L (2007) Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol 72: 483–488 [DOI] [PubMed] [Google Scholar]
- Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, Perreau-Lenz S, Brunk I, Spanagel R, Ahnert-Hilger G, Meijer JH, Albrecht U (2008) Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol 18: 678–683 [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540 [DOI] [PubMed] [Google Scholar]
- Hayakawa F, Abe A, Kitabayashi I, Pandolfi PP, Naoe T (2008) Acetylation of PML is involved in histone deacetylase inhibitor-mediated apoptosis. J Biol Chem 283: 24420–24425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P (2007) CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450: 1086–1090 [DOI] [PubMed] [Google Scholar]
- Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, Wang X, Wang Z, Cornelissen-Guillaume G, Halberg F (2006) Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci 97: 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jul-Larsen A, Grudic A, Bjerkvig R, Bøe SO (2009) Cell-cycle regulation and dynamics of cytoplasmic compartments containing the promyelocytic leukemia protein and nucleoporins. J Cell Sci 122: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Jul-Larsen A, Grudic A, Bjerkvig R, Bøe SO (2010) Subcellular distribution of nuclear import-defective isoforms of the promyelocytic leukemia protein. BMC Mol Biol 11: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik K, Lee CC (2004) Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430: 467–471 [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Wada H, Fukuda-Kamitani T, Yeh ET (1998) Identification of three major sentrinization sites in PML. J Biol Chem 273: 26675–26682 [DOI] [PubMed] [Google Scholar]
- Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub MP, Durand B, Lanotte M, Berger R, Chambon P (1992) Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J 11: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15: 271–277 [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T (2002) Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J 21: 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Donehower LA, Herron AJ, Moore DD, Fu L (2010) Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 5: e10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Lee J, Choi C, Kim J, Doh E, Choe J (2007) Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol Cell Biol 27: 4876–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H (2003) Control mechanism of the circadian clock for timing of cell division in vivo. Science 302: 255–259 [DOI] [PubMed] [Google Scholar]
- Miki T, Takegami Y, Okawa K, Muraguchi T, Noda M, Takahashi C (2007) The reversion-inducing cysteine-rich protein with Kazal motifs (RECK) interacts with membrane type 1 matrix metalloproteinase and CD13/aminopeptidase N and modulates their endocytic pathways. J Biol Chem 282: 12341–12352 [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Wakabayashi M, Hara Y, Ishida N (2010) Tumor growth suppression in vivo by overexpression of the circadian component, PER2. Genes Cells 15: 351–358 [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U (2004) Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705 [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda A, Katayose Y, Yabuuchi S, Yamamoto K, Mizuma M, Shirasou S, Onogawa T, Ohtsuka H, Yoshida H, Hayashi H (2009) Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res 29: 1201–1209 [PubMed] [Google Scholar]
- Oklejewicz M, Destici E, Tamanini F, Hut RA, Janssens R, van der Horst GT (2008) Phase resetting of the mammalian circadian clock by DNA damage. Curr Biol 18: 286–291 [DOI] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406: 207–210 [DOI] [PubMed] [Google Scholar]
- Pregueiro AM, Liu Q, Baker CL, Dunlap JC, Loros JJ (2006) The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science 313: 644–649 [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324: 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T, Bellodi C, Nicotera P, Salomoni P (2009) The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat Neurosci 12: 132–140 [DOI] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL (2001) NPAS2: an analog of clock operative in the mammalian forebrain. Science 293: 506–509 [DOI] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP (2002) The role of PML in tumor suppression. Cell 108: 165–170 [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Gainer H (1977) Suprachiasmatic nucleus: use of 14C-labeled deoxyglucose uptake as a functional marker. Science 197: 1089–1091 [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Zimmerman P (1991) Lesions of the suprachiasmatic nucleus disrupt circadian locomotor rhythms in the mouse. Physiol Behav 49: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Spoelstra K, Albrecht U, van der Horst GT, Brauer V, Daan S (2004) Phase responses to light pulses in mice lacking functional per or cry genes. J Biol Rhythms 19: 518–529 [DOI] [PubMed] [Google Scholar]
- St-Germain JR, Chen J, Li Q (2008) Involvement of PML nuclear bodies in CBP degradation through the ubiquitin-proteasome pathway. Epigenetics 3: 342–349 [DOI] [PubMed] [Google Scholar]
- Takahashi JS (1995) Molecular neurobiology and genetics of circadian rhythms in mammals. Ann Rev Neurosci 18: 531–553 [DOI] [PubMed] [Google Scholar]
- Takahata S, Ozaki T, Mimura J, Kikuchi Y, Sogawa K, Fujii-Kuriyama Y (2000) Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes Cells 5: 739–747 [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P (2002) Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci USA 11: 7728–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA 96: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP (1998) Role of PML in cell growth and the retinoic acid pathway. Science 279: 1547–1551 [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14: 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL (2007) Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia 9: 797–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS, Vallian S, Seto E, Yang WM, Edmondson D, Roth S, Chang KS (2001) The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol Cell Biol 21: 2259–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS, Xu ZX, Hittelman WN, Salomoni P, Pandolfi PP, Chang KS (2003) Promyelocytic leukemia protein sensitizes tumor necrosis factor alpha-induced apoptosis by inhibiting the NF-kappaB survival pathway. J Biol Chem 278: 12294–12304 [DOI] [PubMed] [Google Scholar]
- Xu ZX, Zou WX, Lin P, Chang KS (2005) A role for PML3 in centrosome duplication and genome stability. Mol Cell 17: 721–732 [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H (2002) Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J 21: 1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Yamaguchi S, Tamanini F, van Der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H (2000) Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev 14: 1353–1363 [PMC free article] [PubMed] [Google Scholar]
- Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A (2011) Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem 286: 25891–25902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS (2004) PERIOD2: LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 101: 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, Lee CC, Albrecht U, Tamanini F, Meijer JH, Oostra BA, Nelson DL (2008) Fragile X-related proteins regulate mammalian circadian behavioural rhythms. Am J Hum Genet 83: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Lee CC (2010) Circadian clock, cell cycle and cancer -circadian rhythm and cell growth regulation. The Circadian Clock. Edited by Urs Albrecht (Springer Science + Business Media, New York). Prot Reviews 12: 139–155 [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC (2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105: 683–694 [DOI] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400: 169–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.