Abstract
EMBO J 31 6, 1364–1378 (2012); published online January 14 2012
A large and poorly understood class of G protein-coupled receptors (GPCRs) are involved in cell adhesion and contain an autoproteolytic site known as the GPCR proteolysis site (GPS) located immediately N-terminal to the first transmembrane span. This motif of ∼50 amino acids is also found juxtaposed to the first transmembrane span of an unrelated family of proteins associated with polycystic kidney disease (PKD), but its structural and functional roles were not clear. In this issue of The EMBO Journal, Arac et al use X-ray crystallography to show that the GPS motif is merely the C-terminal end of a much larger GPCR autoproteolysis inducing (GAIN) domain. Atomic models for two of these ancient domains allow one to map the sites of mutations associated with cancer or PKD, and hint at functional roles other than autoproteolysis.
Autoproteolysis occurs in a wide variety of proteins where it typically leads to an essential structural reorganization or the release of an activated fragment. One such autoproteolytic site, called the GPS motif, is found in cell-adhesion GPCRs (Ichtchenko et al, 1999). Like other GPCRs, cell-adhesion GPCRs contain a heptahelical transmembrane domain that is expected to couple with heterotrimeric G proteins inside the cell. However, they also have unusually complex and diverse extracellular regions that contain domains homologous to those typically involved in cell adhesion (Yona et al, 2008).
The extracellular regions of cell-adhesion GPCRs all contain a conserved stalk region that ends in the GPS motif, which is cleaved in the endoplasmic reticulum just after protein synthesis. Araç et al (2012) crystallized the stalk region (∼320 amino acids) along with the preceding HormR domain (∼70 amino acids) of two different cell-adhesion GPCRs: latrophilin (also called CIRL1 or CL1) and brain angiogenesis inhibitor 3 (BAI3). The resulting structures demonstrate that the stalk and GPS motif together form a large domain with α-helical and β-sandwich subdomains (Figure 1). The site of autoproteolysis, defined by the consensus sequence HL↓(T/S), occurs in a tight turn between the last two β strands of the domain. Because the entire domain was required for autoproteolysis, the authors renamed the stalk and GPS motif the GAIN domain. Sequence analysis supports the existence of a similar domain in all cell-adhesion GPCRs and homologues of polycystic kidney disease 1 (PKD1 or polycystin-1).
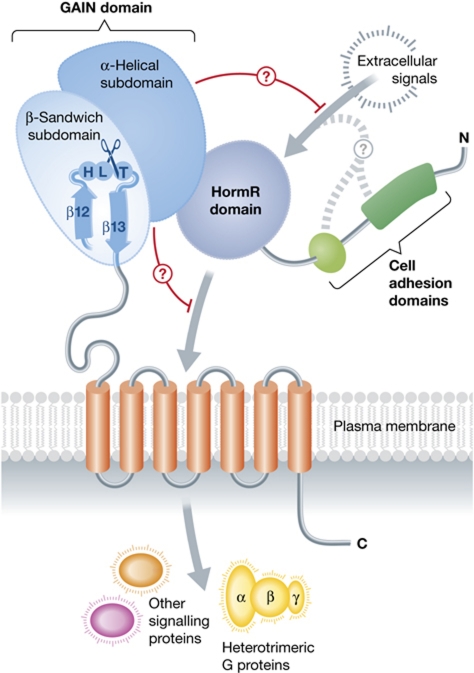
Figure 1.
The conserved structural core of the extracellular regions of cell-adhesion GPCRs, as revealed by new structures of latrophilin and BAI3. Arac et al revealed that regions formerly known as the stalk and the GPCR autoproteolyic site (GPS) fold into a single GAIN domain that is conserved in all 33 mammalian cell-adhesion GPCRs and in proteins related to PKD1. The GAIN domain catalyses its own proteolysis in a tight turn between the last two strands of the domain. The α-helical subdomain of the GAIN domains of latrophilin and BAI3 interacts with a HormR domain, although this domain is not found in all cell-adhesion GPCRs. However, this interdomain contact, and/or a direct interaction with the transmembrane helical bundle, may allow the GAIN domain to autoinhibit transmembrane signalling to heterotrimeric G proteins or other factors inside the cell.
For reasons that are not clear, the latrophilin GAIN domain used in this study was cleaved, whereas that of BAI3 was not. However, this allowed Araç et al to compare structures of wild-type GAIN domains before and after autoproteolysis. Before cleavage, the scissile bond exists in a strained conformation, and the position of adjacent catalytic residues is consistent with an Ntn hydrolase mechanism (Brannigan et al, 1995; Lin et al, 2004). After cleavage, the cleaved fragment (the β13 strand) appears firmly anchored within the GAIN domain by extensive backbone hydrogen bonds and hydrophobic interactions.
But what is the functional role of autoproteolysis? Early work suggested that it may be required for efficient membrane transport because mutation of residues in the GPS motif were known to impair proper membrane trafficking (Krasnoperov et al, 2002). However, other studies, including that of Araç et al, have shown that this is not always the case (Qian et al, 2002). More likely, mutations in the core of the GAIN domain lead to protein folding defects, which would in turn impair trafficking (Lin et al, 2004). Autoproteolysis may therefore represent a mechanism by which the GAIN domain is locked into its functional and presumably more stable state after proper folding occurs. For this reason, it would be interesting to determine the relative thermostability of a GAIN domain before and after cleavage. Still, there remains evidence that GAIN domain autocleavage has functional consequences. Mice that express non-cleavable PKD1 exhibit abnormal kidney development (Yu et al, 2007). Comparison of the latrophilin and BAI3 GAIN domain structures also reveals a subtle conformational change in the β13 strand after cleavage that could, in principle, impact the ability of the domain to interact with other signalling domains. Finally, it has been reported that the cleaved extracellular region of latrophilin is dissociable from its transmembrane domain, and that ligand-induced reassociation may play a role in signalling (Silva and Ushkaryov, 2010).
The GAIN domains of latrophilin and BAI homologues are mutated in human cancer, whereas the PKD1 GAIN domain is frequently mutated in autosomal dominant PKD (ADPKD). Most of the cancer-associated mutations map to the surface of the GAIN domain and did not affect autoproteolysis, implying that they disrupt the intermolecular contacts of the domain. In support of this idea, the authors showed that the latrophilin GAIN domain is the binding site for the black widow toxin α-latrotoxin. In contrast, most of the ADPKD mutations in the PKD1 GAIN domain interfered with autoproteolysis and/or protein folding, hinting at a different role for the domain in PKD1-related proteins. Furthermore, the GAIN domains of latrophilin and BAI3 interact with the preceding HormR domain in a manner that would block the interaction of homologous HormR domains with hormones. The GAIN domain may therefore serve to autoinhibit signalling activity, either by blocking the interactions of other extracellular domains or, in the case of cell-adhesion GPCRs, by modulating the activity of the adjacent seven transmembrane bundle (Figure 1). Indeed, truncation of the N-terminus of GPR56, including the bulk of the GAIN domain, leads to constitutive activation (Paavola et al, 2011). Thus, the structural stage is now set for studies aimed at understanding the molecular basis for signal transduction through cell-adhesion GPCRs and PKD1 proteins, and how defects in these proteins lead to disease.
Acknowledgments
This study was supported by NIH Grants HL086865 and HL071818.
Footnotes
The author declares that he has no conflict of interest.
References
- Araç D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Südhof TC, Brunger AT (2012) A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J 31: 1364–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378: 416–419 [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Bittner MA, Krasnoperov V, Little AR, Chepurny O, Holz RW, Petrenko AG (1999) A novel ubiquitously expressed α-latrotoxin receptor is a member of the CIRL family of G-protein-coupled receptors. J Biol Chem 274: 5491–5498 [DOI] [PubMed] [Google Scholar]
- Krasnoperov V, Lu Y, Buryanovsky L, Neubert TA, Ichtchenko K, Petrenko AG (2002) Post-translational proteolytic processing of the calcium-independent receptor of α-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J Biol Chem 277: 46518–46526 [DOI] [PubMed] [Google Scholar]
- Lin HH, Chang GW, Davies JQ, Stacey M, Harris J, Gordon S (2004) Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem 279: 31823–31832 [DOI] [PubMed] [Google Scholar]
- Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA (2011) The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem 286: 28914–28921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian F, Boletta A, Bhunia AK, Xu H, Liu L, Ahrabi AK, Watnick TJ, Zhou F, Germino GG (2002) Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc Natl Acad Sci USA 99: 16981–16986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, Ushkaryov YA (2010) The latrophilins, ‘split-personality’ receptors. Adv Exp Med Biol 706: 59–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Lin HH, Siu WO, Gordon S, Stacey M (2008) Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem Sci 33: 491–500 [DOI] [PubMed] [Google Scholar]
- Yu S, Hackmann K, Gao J, He X, Piontek K, Garcia-Gonzalez MA, Menezes LF, Xu H, Germino GG, Zuo J, Qian F (2007) Essential role of cleavage of polycystin-1 at G protein-coupled receptor proteolytic site for kidney tubular structure. Proc Natl Acad Sci USA 104: 18688–18693 [DOI] [PMC free article] [PubMed] [Google Scholar]