Abstract
SF3a is an evolutionarily conserved heterotrimeric complex essential for pre-mRNA splicing. It functions in spliceosome assembly within the mature U2 snRNP (small nuclear ribonucleoprotein particle), and its displacement from the spliceosome initiates the first step of the splicing reaction. We have identified a core domain of the yeast SF3a complex required for complex assembly and determined its crystal structure. The structure shows a bifurcated assembly of three subunits, Prp9, Prp11 and Prp21, with Prp9 interacting with Prp21 via a bidentate-binding mode, and Prp21 wrapping around Prp11. Structure-guided biochemical analysis also shows that Prp9 harbours a major binding site for stem-loop IIa of U2 snRNA. These findings provide mechanistic insights into the assembly of U2 snRNP.
Keywords: protein structure, RNA splicing, U2 snRNP, X-ray crystallography
Introduction
Most eukaryotic genes are interrupted by non-coding sequences known as introns. Pre-mRNA splicing removes introns and joins the coding sequences of a nascent transcript into a contiguous mature mRNA molecule (Berget et al, 1977; Chow et al, 1977). Splicing of pre-mRNA occurs in two sequential transesterification reactions and takes place in a large ribonucleoprotein complex known as the spliceosome, a dynamic assembly of five U-type small nuclear ribonucleoprotein particles (snRNPs) and additional splicing factors (Brody and Abelson, 1985; Valadkhan and Jaladat, 2010). The assembly and activation of the spliceosome follows an ordered pathway, beginning with the association of the U1 snRNP with the 5′ splice site and the interaction of the U2 snRNP with the branch point sequence. Subsequently, a preformed U4/U5.U6 tri-snRNP joins the spliceosome and initiates large dynamic rearrangements of RNA–RNA interactions, causing the dissociation of U1 and U4 snRNPs and the activation of the spliceosome (Krämer, 1996; Staley and Guthrie, 1998; Wahl et al, 2009; Will and Luhrmann, 2011). An in-depth mechanistic understanding of mRNA splicing requires structural information on spliceosome/snRNP assembly and dynamics at the atomic level; however, presently available information at this resolution is still too limited to provide a comprehensive view.
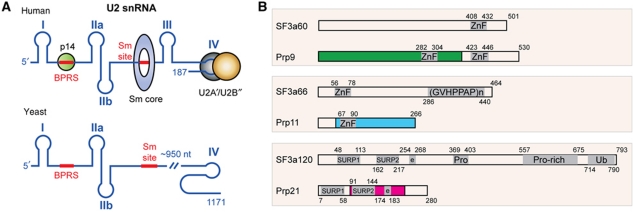
We focus on structural characterization of the heterotrimeric SF3a complex of the U2 snRNP in this study. A mature U2 snRNP is a 17S particle, which is assembled in a defined pathway revealed by in vitro characterization (Brosi et al, 1993a). First, a heptameric Sm protein complex and a U2A′–U2B″ heterodimer bind to the Sm site and stem-loop IV of the U2 snRNA, respectively, forming a 12S particle (Scherly et al, 1990; Boelens et al, 1991) (Figure 1A). Next, a multimeric SF3b complex joins the 12S particle to form a 15S particle. Finally, a functional 17S particle forms with the binding of a heterotrimeric SF3a complex (Behrens et al, 1993; Brosi et al, 1993a; Krämer et al, 1999). The composition and function of yeast and human U2 snRNPs is highly conserved, despite yeast U2 snRNA being six times longer than its human counterpart. The central ∼950 non-conserved nucleotides of yeast U2 snRNA are dispensable for splicing (Igel and Ares, 1988; Shuster and Guthrie, 1988) (Figure 1A), as human U2 snRNA can substitute for the yeast gene for splicing in vivo (Shuster and Guthrie, 1990). It should be pointed out that the structure and composition of U2 snRNP are dynamic during spliceosome assembly and splicing reactions (Jurica and Moore, 2003; Bessonov et al, 2010; Will and Luhrmann, 2011). For examples, rearrangement of U2 snRNA structure is required for pairing with U6 snRNA; and the SF3 complexes are displaced from the spliceosome prior to the first step of splicing reaction (Bessonov et al, 2010; Lardelli et al, 2010).
Figure 1.
Structures of U2 snRNA and SF3a subunits. (A) A schematic diagram of human and yeast U2 snRNA. Roman numerals denote RNA stem-loop nomenclature. Sm and the BPRS are highlighted with thick red lines. Human U2 snRNA-binding proteins with known 3D structures, including p14, Sm and the U2A′–U2B″ complexes, are indicated at appropriate positions with colour-filled circles (green: p14; grey and brown: U2A′–U2B″) or a ring (blue: Sm). (B) Domain structure of yeast and human SF3a components. Colour-filled boxes indicate regions involved in the formation of the core domain of the yeast SF3a complex. Known sequence motifs are indicated by a grey-shaded area: ZnF, zinc finger; (GVHPPAP)n, repeats of the heptamer sequence; SURP1 or 2, suppressor-of-white-apricot and prp21 motifs; Pro or Pro-rich, proline-rich regions; Ub, ubiquitin-like domain; e, charged residues. Arabic numbers indicate amino acid numbers.
The SF3a complex is composed of three subunits, namely SF3a60, SF3a66 and SF3a120 in the human complex, and their corresponding subunits Prp9, Prp11 and Prp21 in Saccharomyces cerevisiae (Chang et al, 1988; Abovich et al, 1990; Arenas and Abelson, 1993; Legrain and Chapon, 1993; Brosi et al, 1993a; Chiara et al, 1994; Krämer et al, 1994). The three SF3a subunits appear to be assembled in a 1:1:1 stoichiometry (Brosi et al, 1993b). Despite large differences in protein sizes between corresponding human and yeast subunits, biochemical analyses have identified various conserved domains (Krämer et al, 2005) (Figure 1B): SF3a60 contains a highly conserved U1C-type Cys2His2 zinc-finger domain at the C-terminal end, while Prp9 has two such domains (Legrain and Choulika, 1990; Chiara et al, 1994); both SF3a66 and Prp11 contain one U1C-type zinc-finger domain (Chang et al, 1988; Bennett and Reed, 1993), and SF3a120 and Prp21 are characterized by two suppressor-of-white-apricot and prp21/spp91 (SURP) domains, followed by a short segment of charged residues (Spikes et al, 1994; Krämer et al, 1995). It has been suggested that the zinc-finger domains are important for protein–protein interactions within U2 snRNP (Nesic and Krämer, 2001). The SURP2 domain of SF3a120/Prp21 is important for SF3a60/Prp9 binding, while no discernable function of the SURP1 domain has been found (Kuwasako et al, 2006). Deletion analyses of human SF3a proteins have implicated additional regions as important for protein–protein interactions, but a complete picture of intermolecular contacts is still lacking (Nesic and Krämer, 2001; Huang et al, 2011). Also lacking is a clear picture of protein–RNA interactions between SF3a and U2 snRNA. For example, biochemical analyses have implicated the binding of SF3a to stem-loops I and III in human 17 U2 snRNP (Dybkov et al, 2006), but in yeast cells, SF3a was found to collaborate with stem-loop IIa and its neighbouring sequences (Ruby et al, 1993; Wells and Ares, 1994; Yan and Ares, 1996) (Figure 1A).
Needless to say, an atomic resolution structure of U2 snRNP or its components would greatly help in understanding the molecular basis of U2 snRNP assembly and its splicing function, as only limited structural information on U2 snRNP components is currently available (Price et al, 1998; Golas et al, 2003; Schellenberg et al, 2006; Ritchie et al, 2009). Here, we report the crystal structure of the core domain of the yeast SF3a complex, which, together with our biochemical analysis, provides the structural basis for the assembly of the SF3a complex and offers mechanistic insights into its function in U2 snRNP maturation.
Results
Structure determination and overall structure
In order to identify a core domain of the SF3a complex suitable for structural studies, we first reconstituted the S. cerevisiae SF3a complex with full-length proteins coexpressed in Escherichia coli, and carried out limited proteolytic digestion followed by MALDI-TOF analysis and N-terminal sequencing. By combining information from limited proteolysis and secondary structure prediction, we obtained a crystallizable SF3a core domain containing a large N-terminal fragment of Prp9 (Prp9ΔC, a.a. 1–389), a C-terminal fragment of Prp11 lacking the 49 N-terminal residues (Prp11ΔN, a.a. 50–266), and a middle fragment of Prp21 (Prp21M) spanning residues 87–237 (Figure 1B). The crystal belongs to the I212121 space group, with unit cell dimensions of a=100.78 Å, b=127.26 Å and c=169.17 Å, and one complex per asymmetric unit. For convenience of description, we refer here to the protein fragments of the SF3a core by their truncation variants and full-length proteins interchangeably, unless specifically noted.
The structure was first solved by multiwavelength anomalous dispersion (MAD) using three data sets collected from a crystal of selenomethionine (SeMet)-substituted proteins (Table I). Most of the Prp9ΔC and Prp21M models were built using the 3.5 Å resolution SeMet data (Supplementary Figure S1A). However, a significant percentage of the Prp11ΔN sidechains could not be assigned. To overcome this problem, 10 aliphatic residues of Prp11ΔN were individually mutated to methionine, and six single-wavelength anomalous dispersion data sets of different mutant SeMet proteins were used to aid the assignment of sidechains (Supplementary Figure S2A). The quality of the electron density map was further improved by phase extension to 3.1 Å using a data set collected from one of the Prp11ΔN mutants, L153M (Supplementary Figure S1B). The refined 3.1 Å model has an Rwork of 22.8% and Rfree of 27.8%, and the model has good stereochemistry, with 91.3% of residues lying in the most favourable regions of the Ramachandran plot and none in outlier regions. The final model includes 93% of the Prp9ΔC molecule (the 11 most C-terminal residues and residues 98–111 are disordered), 85% of Prp21M (the nine most C-terminal residues and residues 207–219 are disordered), and the C-terminal half of Prp11ΔN (a.a. 149–253) and a polyalanine model of a 36-residue fragment of Prp11ΔN (Supplementary Figure S2A). The absence of residues 50–148 of Prp11ΔN was not due to proteolysis during crystallization, as the protein sample recovered from the crystals is indistinguishable from those throughout the purification process, as judged by SDS–PAGE analysis (Supplementary Figure S2B).
Table 1. Data collection, phasing and refinement statistics.
| Data collection | Prp11 L153M mutant | SeMet derivative | ||
|---|---|---|---|---|
| Space group | I212121 | |||
| Cell dimensions (Å) | 100.78 × 127.26 × 169.17 | 99.47 × 123.78 × 167.59 | ||
| Peak | Inflection | Remote | ||
| Wavelength (Å) | 0.9793 | 0.9791 | 0.9794 | 0.9600 |
| Resolution range (Å) | 30–3.1 | 50–3.5 | 50–3.35 | 50–3.4 |
| No. reflections (total/unique) | 101 982/19 171 | 89 166/12 711 | 100 230/14 390 | 95 587/13 833 |
| Completeness (%) | 96.6 (83.0)a | 97.7 (100.0) | 97.4 (100.0) | 97.2 (100.0) |
| I/σ (I) | 19.8 (2.4) | 13.9 (3.6) | 14.0 (2.8) | 14.5 (3.7) |
| Rsym (%) | 7.0 (46.7) | 14.6 (45.6) | 14.5 (56.7) | 14.1 (44.4) |
| Refinement | ||||
| Resolution (Å) | 30–3.1 | 50–3.5 | ||
| Rwork/Rfree (%) | 22.8/27.7 | 26.3/29.3 | ||
| RMSD bond (Å) | 0.008 | 0.007 | ||
| RMSD angle (deg) | 0.625 | 0.682 | ||
| B factor (Å2) | 136.5 | 117.6 | ||
| Ramachandran values | ||||
| Most favoured (%) | 91.3 | 83.57 | ||
| Allowed (%) | 8.7 | 14.08 | ||
| Disallowed (%) | 0 | 2.35 | ||
| aValues in parentheses are for the highest-resolution shell. | ||||
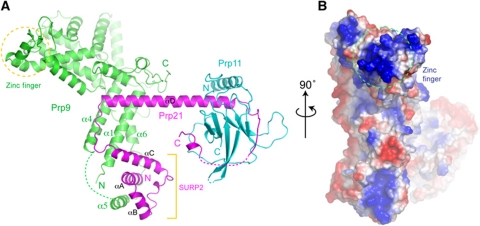
The SF3a core has a branched, Y-shaped overall structure, with a thick trunk consisting of the elongated Prp9ΔC structure and an N-terminal SURP domain of Prp21M, and a branch formed by αD, a long helix of Prp21M, to which Prp11ΔN is bound at the distal tip of αD (Figure 2A). Both Prp9ΔC and Prp21M are predominantly α-helical, with the exception of two short strands in Prp9 forming a Cys2His2-type zinc-finger domain. Prp11ΔN is mainly composed of β-strands. Notably, Prp9ΔC and Prp11ΔN do not physically interact with each other, as they are bridged through the long αD helix of Prp21M (Figure 2A). The complex has a maximal extension of ∼115 Å, measured between distal points of Prp9ΔC and Prp11ΔN, while Prp9ΔC spans ∼80 Å, and Prp21M spans ∼75 Å along the direction of αD. An examination of charge distribution on the surface of the SF3a core reveals a markedly positively charged patch near the zinc finger of Prp9ΔC, suggesting a potential role in RNA binding (Figure 2B).
Figure 2.
Overall structure of the SF3a core. (A) A ribbon representation of the core domain of the Prp9–Prp21–Prp11 complex. Prp9 is shown in green, Prp21 in magenta and Prp11 in cyan. Dashed lines denote disordered segments in the structure. A Cys2His2 U1C-type zinc finger (zinc atom shown as a sphere) of Prp9 and the SURP2 domains of Prp21 are indicated. (B) An orthogonal view of the heterotrimeric complex is shown in a surface representation with electrostatic potential distribution (blue, positively charged; red, negatively charged; white, neutral). The encircled area indicates the Prp9 region surrounding the zinc finger.
Structure of Prp9
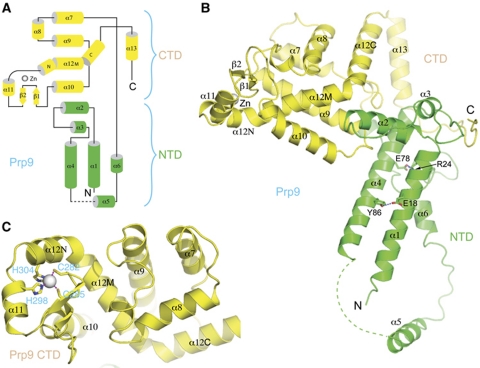
Prp9ΔC consists of 13 α-helices and a pair of short β-strands, arranged in an elongated structure that can be roughly divided into two domains (Figure 3A). The N-terminal domain (NTD), where Prp21M binds, is composed of helices α1–α6 (Figure 3B). A pair of amphipathic helices, α1 and α4, pack against each other in an antiparallel manner and are held together primarily by hydrophobic interactions, with notable exceptions of an Arg24–Glu78 charge pair, and a hydrogen bond between Glu18 and Tyr86. The stabilization by hydrogen bonds is functionally important, as the prp9 mutant, which is a temperature sensitive mutant lead to the discovery of the PRP9 gene, encodes an E78K mutation (Legrain and Choulika, 1990). Helix α2 is positioned perpendicular to α1 and α4, and together with the N-terminal portion of α4, forms the major contacting area for the C-terminal domain (CTD) of Prp9ΔN, principally via polar interactions. A segment of 14 disordered residues C-terminal to α4 tethers α5, which makes no contact with the rest of the structure apart from covalent joining via its C-terminal loop. α5 is involved in interactions with the SURP2 domain of Prp21M, indicating that the conformation of α5 is stabilized by this interaction.
Figure 3.
Structure of Prp9ΔC (a.a. 1–389). (A) A topological diagram of the Prp9ΔC fold. Helices are shown as cylinders and strands with arrows. A grey sphere represents the zinc atom. Secondary structural elements shown in green belong to the NTD, and those in yellow form the CTD. (B) A ribbon diagram showing the relative positioning of NTD and CTD. (C) A direct view of the zinc finger in CTD. The two cysteines and two histidines coordinating the zinc ion are shown in a stick representation, and the zinc ion is shown as a sphere. Magenta dashed lines indicate tetrahedral coordination of the zinc ion.
The CTD of Prp9ΔC is centred on an unusual helix, α12, which is sharply bent at both ends (Figure 3C). The middle portion of this helix, α12M, is placed between helices α9 and α10, and projects out of the approximately planar arrangement of helices α7, α9 and α10. The N-terminal portion, α12N, harbours a histidine (His304), which, together with another histidine (His298) in the preceding helix, α11, and two cysteines located in the loop connecting β1 and β2 (Cys282 and Cys285), coordinates the binding of a zinc ion, forming a Cys2His2-type zinc finger that resembles the RNA-binding structural motif of U1C (Muto et al, 2004). The C-terminal portion of α12, α12C, is almost perpendicular to α12M and runs antiparallelly next to α8 (Figure 3C). The C-terminal helix, α13, makes a V-shaped turn towards NTD and engages in extensive polar interactions with the region connecting NTD and CTD (Figure 3B). The very C-terminal tail of CTD snakes around to make numerous polar and charged interactions with the NTD loop connecting α1 and α2, and the long loop connecting NTD and CTD. Additional interdomain contacts involve regions surrounding α9-turn-α10 and the junction between α12M and α12C.
Structures of Prp21 and Prp11
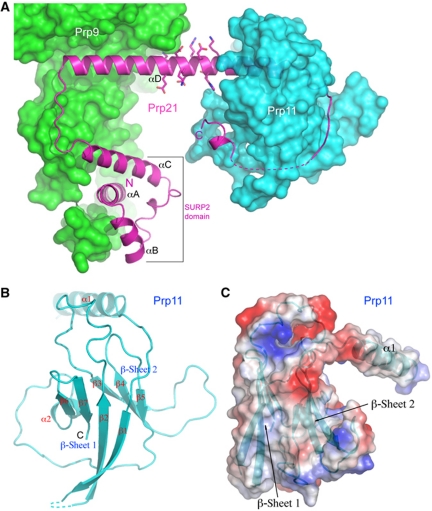
Prp21M contains a SURP domain, which is a compact domain composed of three helices (αA to αC) and a 310 turn (Figure 4A). Prp21 has two predicted SURP domains, the one in the structure corresponds to the second SURP domain (SURP2), as the extreme N-terminal SURP domain has no known function and is not included in the crystallization construct. SURP2 has a structure similar to that of human SF3a120 determined by NMR (Kuwasako et al, 2006). In brief, αA and αB are positioned in a V-shape, and αC crosses over αA on the same side of the αA–αB plane with a 310 turn. The SURP2 domain is followed by a long helix, αD. Interestingly, SURP2 is not in contact with αD, suggesting that the spatial positioning of SURP2 with respect to αD is likely to be flexible in the absence of Prp9. Consistent with αD's role in binding Prp9 and Prp11 at opposite ends, which are decorated with hydrophobic residues important for protein–protein interactions, the middle region of αD has a contiguous stretch of conserved charged amino acids, Glu174 to Lys183, which are completely solvent exposed (Figure 4A). The C-terminal extension of αD is composed of a loop that wraps around Prp11, and a disconnected short C-terminal helix interacting with one of the β-sheets of Prp11ΔN.
Figure 4.
Structure of Prp21M and Prp11ΔN. (A) Prp21M structure shown as a ribbon diagram (magenta) is superimposed on the surface representations of Prp9ΔC (green) and Prp11ΔN (cyan). A contiguous stretch of charged amino acids on αD is shown as sticks. (B) Structure of Prp11ΔN shown in a ribbon representation. (C) An orthogonal view of Prp11ΔN, shown in an electrostatic potential surface representation, displays a ‘baseball glove’-shaped binding pocket (thumb, α1; palm, β-sheet 2) for Prp21M. A ribbon diagram of Prp11ΔN is superimposed onto the semi-transparent surface.
Prp11ΔN contains a β-sandwich core, formed by back-to-back stacking of two β-sheets (Figure 4B). The first sheet (β-sheet 1) consists of four antiparallel strands arranged in a β1–β2–β7–β6 configuration, and the second sheet (β-sheet 2) is composed of three contiguous strands, β3–β4–β5. The two sheets are connected via two long loops at opposite ends of the β-sheets, one between β2 and β3, and another with a short helix (α2) between β5 and β6. The β-sandwich core of Prp11ΔN is stabilized by extensive hydrophobic interactions between the two sheets, and the folding topology resembles that of the α-crystallin domain of hsp26, which is a small heat shock protein (sHsps) belonging to a superfamily of molecular chaperones that suppress protein aggregation (Kim et al, 1998). The connection between β1 and β2 in the first sheet was not modelled due to weak electron densities. In fact, for the same reason, only a polyalanine model was built for the N-terminal 36-amino-acid segment encompassing α1 and β1. Overall, the electron density for Prp11ΔN is weaker than that for Prp9ΔC and Prp21M, but the β-sandwich core was unambiguously assigned with the assistance of selenium anomalous signals from six leucine-to-methionine mutants (L153M, A174M, V183M, V200M, I211M and V252M). Another four mutants (L70M, V81M, L95M, V143M) did not give rise to detectable anomalous difference peaks in electron density maps calculated from the diffraction data of the SeMet-substituted mutants, indicating that the N-terminal region is not well ordered, explaining our inability to assign sidechains to the 36-residue N-terminal segment. Helix α1, together with the exposed surface of β-sheet 2, forms a pronounced binding cleft resembling the shape of a baseball glove, with α1 being the thumb and the β-sheet surface the palm (Figure 4C).
Prp9–Prp21 interaction
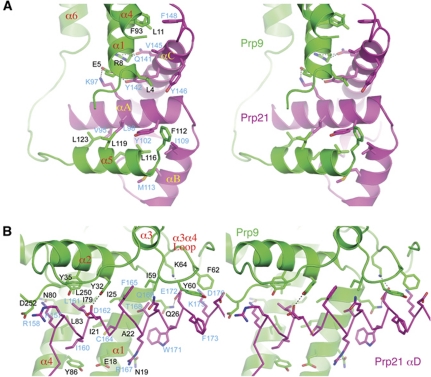
The interaction between Prp9ΔC and Prp21M buries a pairwise surface area of 3679 Å2, indicating a stable association. Two major Prp9-binding sites on Prp21M include the SURP2 domain and the N-terminal half of αD (Figures 2 and 5). The Prp9–SURP2 interaction shares ∼60% of the contact area (2216 Å2), in which helix α5 of Prp9 binds the SURP2 domain by antiparallel packing with αA of Prp21, and this binding is further strengthened by contacts with the angled αB of SURP2 (Figure 5A). The interaction between α5 and SURP2 is mostly hydrophobic and involves Phe112, Leu116, Leu119 and Leu123 of Prp9, and conserved residues on αA (Val95, Leu98 and Tyr102) and αB (Ile109 and Met113) of Prp21M (Figure 5A). The mode of α5–SURP2 interaction is similar to that found in the NMR structure of the SURP2 domain of human SF3a120 in complex with an isolated helix of SF3a60 (Kuwasako et al, 2006). However, in the context of the SF3a complex in our structure, helix α1 of Prp9 makes extensive interactions with SURP2 via its αA and αC helices (Figure 5A). A notable interaction between α1 and αA occurs though a pair of oppositely charged residues, Glu5 of Prp9 and Lys97 of Prp21. The contact between α1 and αC is more extensive than that between α1 and αA and involves a mixture of hydrophobic and hydrogen bond interactions; hydrophobic interactions between Leu4, Leu11 and the aliphatic part of Arg8 of Prp9 and Tyr142, Val145, Tyr146 and Phe148 of Prp21M, and hydrogen bonds between Glu5 and Tyr142, and Arg8 and Gln141 of Prp9 and Prp21, respectively (Figure 5A). In addition, Phe93 on α4 of Prp9 also contributes to the hydrophobic interaction with Phe148 of Prp21 located at the end of αA. The newly discovered intermolecular interactions involving α1 and α4 of Prp9 provide a complete picture of how the SURP2 domain of Prp21 binds Prp9.
Figure 5.
Prp9–Prp21 interactions. (A) Interactions mediated by the SURP2 domain of Prp21. The Prp9 and Prp21 residues involved are shown as stick models (carbon: green for Prp9, magenta for Prp21; nitrogen: blue; oxygen: red; sulphur: gold) superimposed with the ribbon diagrams of Prp9 (green) and Prp21 (magenta). Black dashed lines denote hydrogen bonds. (B) Interactions mediated through the N-terminal portion of Prp21's αD, which is shown as a Cα trace. Residues labelled in black and blue are those of Prp9 and Prp21, respectively.
The second major area of interaction involves the N-terminal portion of αD from Prp21M and a wedge in Prp9ΔC formed principally by helices α1, α2, α4, and a hairpin loop connecting α3 and α4 (Figure 5B). A collection of hydrophobic residues from α1, α2 and α4 cluster together to form a cleft that houses the binding of αD via the Ile160, Leu161, Cys164, Phe165, Thr168 and Trp171 residues of Prp21M. The mainly hydrophobic contact is decorated with hydrogen bond interactions, including those between Lys157 and Asn80, Asp162 and Tyr32, Arg167 and Asn19, Glu172 and Gln26 of Prp21 and Prp9, respectively (Figure 5B). The Prp21-binding site in Prp9 is extended by the α3–α4 hairpin loop region, in which two aromatic residues, Tyr60 and Phe62, each make hydrophobic interactions with Prp21 by inserting between Glu172 and Phe173, and Lys175 and Asp176 in αD, respectively. In addition, a charge pair between Lys64 of Prp9 and Glu172 of Prp21 further highlights the importance of the α3–α4 hairpin loop of Prp9ΔC in binding Prp21M.
The bidentate Prp9–Prp21 interaction sites are connected by a narrow corridor that involves an interaction between helix α4 of Prp9 and the linker connecting αC and αD of Prp21, which runs antiparallelly along α4 (Figure 2A). The linker is rigidly held by a network of intermolecular hydrogen bond interactions between its mainchain carbonyl and amide groups and the sidechain groups of Gln90, Asn94 and Asn97 of Prp9.
Prp21–Prp11 interaction
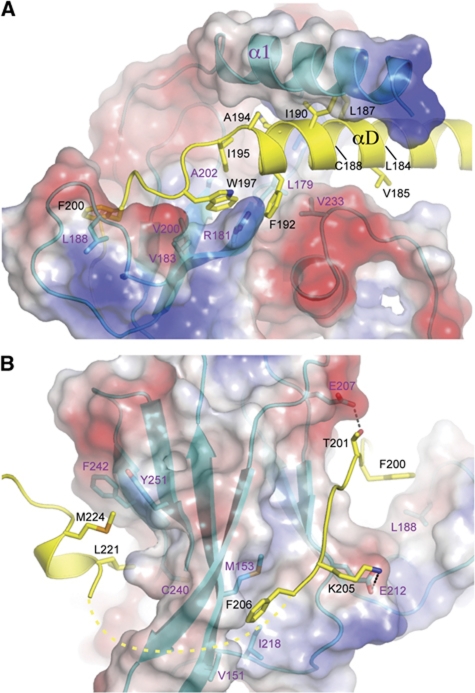
The interaction between Prp11ΔN and Prp21M occludes a pairwise surface area of 2603 A2, also indicating a stable association between the two proteins. The baseball glove-shaped binding cleft of Prp11, formed by helix α1 and the surface of β-sheet 2, grabs helix αD of Prp21 at its C-terminal portion and its immediate C-terminal extension (Figure 6A). The interactions are mainly hydrophobic in nature, as most of the Prp21 residues facing Prp11, which include Leu184, Val185, Leu187, Cys188, Ile190, Phe192 and Ala194 on αD, and Ile195 and Trp197 immediately following αD, are hydrophobic. The precise mode of interaction involving amino acids on α1 of Prp11 cannot be discerned due to weak electron densities for the sidechain groups. However, the interactions involving residues located on the palm area of Prp11 are clearly defined. Notably, Phe192 on αD of Prp21M is bound in a hydrophobic pocket formed by Leu179, Val 233 and the aliphatic part of Arg181; and an adjacent hydrophobic pocket formed by Leu179, the aliphatic part of Arg181, Val200 and Ala202, is bound by Trp197 on the C-terminal loop of αD (Figure 6A).
Figure 6.
Prp21–Prp11 interactions. (A) The C-terminal portion of Prp21's αD is bound between Prp11's α1 and β-sheet 2. The side chains of Prp21 in the Prp11 interaction region are shown as stick models superimposed onto a ribbon representation (yellow). Prp11ΔN is shown in a semi-transparent electrostatic potential surface representation superimposed onto a ribbon diagram (cyan). Selected Prp11 residues involved in the interaction with Prp21 are shown as sticks. Prp21 residues are labelled in black and those of Prp11 are labelled in magenta. (B) Prp21–Prp11 interactions viewed from the opposite direction to that of (A).
Prp21 has a long loop C-terminal to αD that traverses β-sheet 2 and curls back towards β-sheet 1 (Figure 6B). Besides the aforementioned hydrophobic interactions involving Ile195 and Trp197, two aromatic residues, Phe200 and Phe206, are engaged in apparent hydrophobic interactions with Prp11: Phe200 is partially exposed and interacts with Leu188 of Prp11, and Phe206 is situated in a hydrophobic pocket formed by Prp11 residues Val151, Leu153 (Met153 in the L153M mutant structure) and Ile218. The Prp11–Prp21 binding is further strengthened by intermolecular hydrogen bonds involving mainchain atoms, as well as those between the sidechains of Thr201 and Glu207, and Lys205 and Glu212 of Prp21 and Prp11, respectively (Figure 6B). A segment of the Prp21M loop (a.a. 207–219) cannot be reliably modelled, leaving a short orphan helix not connected to the rest of Prp21M. This helix is anchored on the β-sheet 1 surface primarily through hydrophobic interactions between Met224 and Leu221 of Prp21 and a hydrophobic surface patch formed by Cys240, Phe242 and Tyr251 of Prp11 (Figure 6B).
RNA-binding properties of SF3a
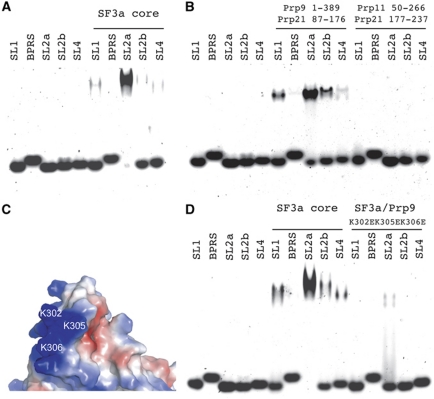
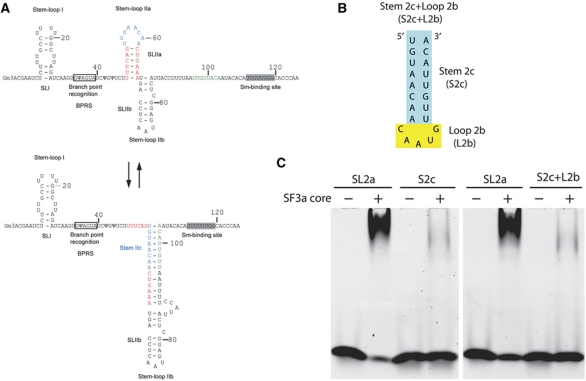
With the structure in hand, we explored the U2 snRNA-binding properties of yeast SF3a. We used chemically synthesized yeast U2 snRNA fragments, including fragments encompassing stem-loops I, IIa, IIb, IV, and the branch point recognition sequence (BPRS), and probed the binding of the SF3a complex by electrophoretic mobility shift assay (EMSA). A stem-loop III fragment was not tested because it is dispensable for splicing in yeast (Igel and Ares, 1988; Shuster and Guthrie, 1988; Ares and Igel, 1990). The EMSA result shows that the yeast SF3a complex preferentially binds stem-loop IIa (SL2a) (Figure 7A). This binding pattern of the SF3a core domain is indistinguishable from the complex assembled from the full-length proteins (Supplementary Figure S3). SL2a was shown to rearrange into an alternative conformation, termed stem IIc (Figure 8A), before the first step of splicing reaction, and change back to the SL2a structure prior to the second step of splicing reaction (Hilliker et al, 2007; Perriman and Ares, 2007). EMSA with the stem IIc (S2c) duplex or a stem loop joining S2c with loop IIb (L2b) shows that SF3a does not bind S2c (Figure 8B and C).
Figure 7.
Interactions between the SF3a core domain and U2 snRNA. (A) Binding of the SF3a core to synthetic yeast U2 snRNA fragments detected using EMSA. SL1, SL2a, SL2b and SL4 indicate stem-loops I, IIa, IIb and IV, respectively, and BPRS denotes the BPRS of U2 snRNA. (B) EMSA shows that a mini complex of Prp9ΔC and its interacting portion of Prp21M (a.a. 87–176) harbour the full RNA-binding capacity of the core domain of SF3a, since the C-terminal portion of Prp21M (a.a. 177–237) in complex with Prp11ΔN displays no detectable binding to the U2 snRNA fragments. (C) The three positively charged residues of Prp9, K302, K305 and K306, located on the surface next to the zinc finger are potentially important for RNA binding. (D) RNA binding is significantly reduced by the K302E/K305E/K306E triple mutant of Prp9ΔC.
Figure 8.
SF3a does not bind a rearranged form of SL2a. (A) A schematic diagram showing the region of yeast U2 snRNA having alternative conformations. The stem and loop regions of SL2a are indicated with red and blue letters, respectively. The downstream sequences involved in the formation of stem IIc are shown in green letters. (B) A schematic diagram of the RNA elements, S2c and S2c+L2b, used for binding studies. (C) EMSA results of the binding of the SF3a core complex to S2c and S2c+L2b elements of U2 snRNA, in comparison to that of SL2a.
Next, we explored which components of the SF3a complex are responsible for SL2a binding. Since the structure shows that Prp9ΔC and Prp11ΔN are not in direct contact and that they are bridged by the long helix αD of Prp21M, we separated the SF3a core complex into two halves by truncating Prp21M in the middle of αD (between a.a. 176 and 177). One half, termed Prp9–21N, contains Prp9ΔC and the N-terminal half of Prp21M (a.a. 87–176), while the other half, termed Prp11–21C, contains Prp11ΔN and the C-terminal region of Prp21M (a.a. 177–237). EMSA results show that the Prp9–21N complex is responsible for the observed RNA-binding properties of the SF3a core, as no binding was detected for the Prp11–21C complex (Figure 7B). To reveal whether full-length Prp11 has additional U2 snRNA-binding ability, we reconstituted the complex with the C-terminal half of Prp21M, and found that it too has no RNA-binding activity (data not shown). Thus, we conclude that the U2 snRNA-binding region of SF3a is located in the Prp9–21N region.
Prp9 has an U1C-type Cys2His2 zinc finger, and we reasoned that this region might be important for RNA binding. We first changed the two cysteines (Cys282 and Cys285) independently into alanines, but the recombinant mutant complex was completely insoluble, most likely due to the disruption of the structure introduced by the mutations. Next, we changed three positively charged residues surrounding the zinc finger, Lys302, Lys305 and Ly306, to negatively charged glutamate residues (Figure 7C). The K302E/K305E/K306E/triple mutant SF3a core complex significantly impaired the RNA-binding ability of the SF3a core complex (Figure 7D). In contrast, little effect on RNA binding was observed by mutating a pair of positively charged residues, R114 and K115, located at the NTD of Prp9, distal from the zinc-finger motif (Supplementary Figure S4A and B). Therefore, we conclude that the positively charged region surrounding the zinc-finger motif of Prp9 is important for U2 snRNA binding.
Discussion
Pre-mRNA splicing is a major form of post-transcriptional regulation of eukaryotic gene expression, and the molecular machine catalysing the splicing reactions is extremely complex. While determination of the atomic resolution structure of the spliceosome is met with serious difficulties, subspliceosomal particles, represented by snRNPs, are the current frontiers of structural studies of mRNA splicing (Pomeranz Krummel et al, 2009; Leung et al, 2011). With U2 snRNP, the two major building blocks are the SF3a and SF3b complexes, and determination of their structures would greatly shed the light on the assembly of U2 snRNP and clear the way for the determination of high-resolution structures of U2 snRNP by cryo-EM or X-ray crystallography. We focused on structural and biochemical characterizations of the yeast SF3a complex here, and the structural information obtained here should be useful for understanding the assembly of the U2 snRNP.
The structure of the yeast SF3a complex provides direct evidence that two subunits, Prp9 and Prp11, do not contact each other, but are bridged together via Prp21. This result is consistent with previous biochemical and genetic analyses of yeast and human SF3a complexes (Rain et al, 1996; Nesic and Krämer, 2001; Huang et al, 2011). Furthermore, the structure reveals a bidentate mode of interactions between Prp9 and Prp21 that involves both the SURP2 domain and the Prp21 N-terminal half of αD. This discovery reconciles the apparent discrepancies between an NMR study and biochemical analyses of interactions between human homologues of Prp21 and Prp9, SF3a120 and SFa60 (Kuwasako et al, 2006; Huang et al, 2011). The NMR structure of an isolated SURP2 domain of SF3a120 (a.a. 134–217) in complex with a short fragment of SF3a60 (a.a. 71–107) closely resembles the interaction between the SURP2 domain of Prp21 and α5 of Prp9 in our structure (Kuwasako et al, 2006). However, in the context of the heterotrimeric complex, helix α1 of Prp9 also makes important contacts with the SURP2 domain of Prp21. More importantly, the structure of the SF3a core uncovered a novel binding site in Prp9 that recognizes αD of Prp21. This discovery rationalizes the observation from biochemical analyses that the SURP2 domain is not sufficient for Prp9 binding, and provides a complete picture of the complexity of Prp9–Prp21 interactions. The interaction between Prp21 and Prp11 is much less well understood. A recent study mapped a region important for SFa66 binding to a 27-amino-acid segment C-terminal to the SURP2 domain of SF3a120, but this region lacked any identifiable sequence motifs helpful for understanding the interaction between the two SF3a subunits (Huang et al, 2011). Our structure reveals that the core domain of Prp11 has an unexpected β-sandwich fold of the α-crystallin domain shared by sHsps (Kim et al, 1998). One of the β-sheet surfaces, together with an N-terminal helix, forms the major binding site of Prp21. Taken together, our structural information unambiguously reveals the architecture of the evolutionarily conserved core domain of the SF3a heterotrimeric complex, and provides a wealth of information about protein–protein interactions governing the assembly of the complex.
The structure of the SF3a core domain lacks the SURP1 domain at the N-terminus of Prp21, an U1C-type zinc-finger domain at the N-terminus of Prp11, and a similar domain at the C-terminus of Prp9. These domains are conserved between the yeast and human SF3a proteins. The SURP1 domain of Prp21 has no known functions and is dispensable for interaction with Prp9 and splicing in vivo. The U1C-type zinc fingers in the human counterparts of Prp9 and Prp11 are required for association with the 15S U2 snRNP, and it has been suggested that these zinc fingers may be involved in interacting with Sm proteins and/or components of the SF3b complex, rather than interacting with U2 snRNA (Nesic and Krämer, 2001), leaving the interaction between SF3a and U2 snRNA poorly understood. Previous biochemical studies have indicated that human SF3a makes direct contacts with stem-loops I and III of U2 snRNA, whereas stem-loop IIa and its adjacent sequences collaborate with SF3a in yeast pre-mRNA splicing (Dybkov et al, 2006). Given that human U2 snRNA can complement splicing in yeast, it is unlikely that human and yeast U2 snRNPs are significantly different. We show here that the yeast SF3a complex preferentially binds to the SL2a element of U2 snRNA. This finding fits nicely with results from previous genetic studies, indicating that SL2a collaborates with the SF3a complex in splicing in yeast (Ruby et al, 1993; Wells and Ares, 1994; Yan and Ares, 1996). Further analyses show that the SL2a-binding ability residues within Prp9, although the presence of the C-terminal half of Prp21 and Prp11 may be beneficial for the binding specificity of the trimeric complex (Figure 7A and B; Supplementary Figure S4C). Lastly, SL2a was shown to be rearranged into an alternative structure, stem IIc, before the first step of splicing reaction and toggle back to the SL2a configuration prior to the second step of splicing reaction (Hilliker et al, 2007; Perriman and Ares, 2007). Our analysis reveals that SF3a does not bind S2c, which may account for the displacement of SF3a from the spliceosome before the first step of splicing reaction (Bessonov et al, 2010; Lardelli et al, 2010).
It is extremely interesting that the SL2a-binding zinc finger in Prp9 is not conserved in the human protein. Previous genetic studies showed that mutating one of the histidine residues of this zinc finger in Prp9 interfered with its function, while changing each of the cysteines to a serine or replacing the other histidine with a leucine was benign (Legrain and Choulika, 1990). Our structure and in vitro mutagenesis data indicate that the zinc finger in Prp9 is playing a structural role, holding key residues in place for interacting with RNA. One possible explanation of the genetic results is that, at least in an in vivo environment, the RNA-binding surface of Prp9 is resilient to amino-acid substitutions of the zinc-finger motif. Related to the structural stability of the zinc-finger region is the absence of a corresponding zinc-finger motif in human SF3a60, which is expected to have a structure similar to Prp9 based on the sequence similarity between the yeast and human proteins (Supplementary Figure S5). The scenario of sharing a highly similar structure between proteins with and without zinc-binding motif is not unprecedented: the RING finger and U-box motifs of ubiquitin E3 ligases share a highly similar structure but the former is a zinc finger and the latter is free of zinc (Aravind and Koonin, 2000; Ohi et al, 2003). Even though the zinc finger is absent in human SF3a60, the corresponding region also shows the presence of positively charged residues (Supplementary Figure S5). It is conceivable that the overall shape and charge distribution on the protein surface of this region is similar between Prp9 and SF3a60, hence the conserved U2 snRNA-binding property of yeast and human SF3a complexes. Future structural studies of SF3a–U2 snRNA complexes or an intact U2 snRNP should provide a definitive answer to the mode of interaction between SF3a and U2 snRNA.
Materials and methods
Identification of the SF3a core
Recombinant, full-length S. cerevisiae SF3a was produced in the BL21(DE3)-RIL strain of E. coli by coexpression. Prp9 was expressed as a GST-fusion protein using the pGEX-KG vector, and a poly(His)-tagged Prp11 and an untagged Prp21 were expressed using a single pRSF-Duet vector (Novagen). Protein production was induced by the addition of 0.5 mM IPTG at 18°C for 12 h. The trimeric complex was purified by sequential column chromatography using HisTrap HP, glutathione sepharose, HiTrap Phenyl HP and Sephadex-200 columns. The GST-tag of Prp9 was removed by thrombin digestion on the glutathione sepharose column. The purified SF3a was analysed by limited proteolysis with Glu-C at room temperature for 1–2 h. Large proteolytic fragments were analysed by MALDI-TOF mass spectrometry and N-terminal sequencing by Edman degradation.
SF3a core preparation
Based on the results from proteolysis, a Prp9 fragment lacking the C-terminal 141 residues (Prp9ΔC, a.a. 1–389), a Prp11 fragment encompassing amino acids 50–266 (Prp11ΔN) and a Prp21 fragment spanning residues 87–237 (Prp21M) were coexpressed and purified using the same strategy described above for the full-length proteins, except that the HiTrap Phenyl column was replaced with a HiTrap S column. SeMet-substituted proteins were prepared by first growing the cells in M9 medium to mid-log phase, followed by addition of 30 mg/l SeMet along with 100 mg/l Thr, Lys and Phe, and 50 mg/l Leu, Ile and Val to the growing culture, and induction of protein expression by IPTG at 18°C for ∼12 h. The SeMet-substituted SF3a core was purified following the same procedure used for the native SF3a core, and the protein complex was eluted from the Sephadex-200 column in a buffer containing 10 mM HEPES pH 8.0, 0.2 M NaCl and 1 mM DTT and then concentrated to ∼15 mg/ml for crystallization.
Crystallization and data collection
Initial crystals of the SF3a core grew in several conditions containing ethanol, isopropanol or PEGs of varying molecular weights (from PEG 2000MME to PEG 8000) as precipitants. The best diffracting crystals were obtained by hanging drop vapour diffusion at 17°C in a condition containing 8–12% ethanol, 100 mM MES pH 6.0. The rod-shaped crystals were cryo-protected for data collection at 100 K by serial transfer of crystals into solutions of mother liquor supplemented with 10, 15, 20, 25 and 30% glycerol. Three SeMet MAD data sets at wavelengths of λpeak=0.9791 Å, λinflection=0.9794 Å and λremote=0.9600 Å were collected at beamline X29 of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory, and data were processed using the HKL2000 program suite (Otwinowski and Minor, 1997). SeMet data sets from single methionine mutants of Prp11ΔN were collected at the Se peak wavelength. Statistics of data collection are summarized in Table I.
Structure determination and model building
In all, 11 out of the 13 possible Se sites were determined using the AutoSol program of the PHENIX suite, and the 3.5 Å MAD phased electron density map was readily traceable (Adams et al, 2010). The polypeptide chains of Prp9ΔC and Prp21M were built into the solvent-flattened electron density map, and building of the Prp21M model was assisted by knowledge of the NMR structure of the SURP2 domain of SF3a120 (Kuwasako et al, 2006). The electron density of Prp11ΔN is relatively weak, making the assignment of side chains difficult. To overcome this problem, several residues (Leu70, Val81, Leu95, Val143, Leu153, Ala174, Val183, Val200, Ieu211 and Val252) of Prp11ΔN were individually mutated to Met, and SeMet-substituted mutant SF3a core complex crystals were prepared. SeMet anomalous difference peaks for L153M, A174M, V183M, V200M, I211M and V252M were used as landmarks for the sequence assignment of Prp11ΔN. Model building and rebuilding were carried out with the program COOT (Emsley et al, 2010), and refinement was performed using the PHENIX Refine program. Phasing and refinement statistics are summarized in Table I. The PDB accession code of the atomic coordinates and diffraction data of the SF3a core is 4DGW.
In vitro RNA-binding assay
Amino-acid substitution and truncation variants of the SF3a subunits were generated by PCR and the protein samples were prepared in a manner similar to the wild-type complex. U2 snRNA oligos were commercially synthesized (Thermo Fisher Scientific), and resuspended with RNase-free sterile water to a final concentration of 50 μM. The sequences of the five oligos used for binding are SL1: UCUCUUUGCCUUUUGGCUUAGAUC; BPRS: AAGUGΨAGUAUC, SL2a: UUUUCAGUGUAACAACUGAAA; SL2b: AUGACCUCAAUGAGGCUCAU and SL4: CCCUCGCACUUGUGG. In addition, a Stem 2c (S2c) duplex formed by the oligo pair 5′-UGUAACAA-3′ and 5′-UUGUUACA-3′, and a hybrid stem loop with the S2c sequence linked by a 5nt SL2b loop sequence (S2c+L2b), 5′- UGUAACAACAAUGUUGUUACA-3′, were used for testing the binding of SF3a to an alternative structure of the SL2a region. The oligos were annealed by heating them to 80°C for 10 min, followed by slowly cooling them down to room temperature over a period of 30 min. Binding reactions were conducted by incubating 0.1 nmol of the purified proteins with 0.05 nmol of individual RNA oligos in 20 μl reaction mixtures, containing 10 mM Tris 7.0, 250 mM NaCl (300 mM NaCl when the mixture contained the full-length SF3a complex), 2 mM MgCl2 and 1 mM DTT. After 2 h incubation at room temperature, samples were analysed by electrophoresis at 100 V for 60 min on a 4% non-denaturing polyacrylamide gel pre-equilibrated with 1 × Tris-borate-EDTA buffer. Gels were stained by SYBR-Gold and detected by UV transillumination.
Supplementary Material
Acknowledgments
We thank Dr Rong-Huay Juang of the National Taiwan University for protein N-terminal sequencing, Dr Adrian Krainer of Cold Spring Harbor Laboratory for advice and help with RNA-binding experiments, beamline scientists at NSLS for technical support during data collection, Drs Ying Huang and Mingzhu Wang of the Chinese Academy of Sciences (CAS) for help with structure determination and Joy Fleming for critical reading of the manuscript. This work was supported in part by a grant from the National Natural Science Foundation of China (Grant No. 30988001). RMX was also supported by a Novo Nordisk-CAS Great Wall Professorship.
Author contributions: RMX conceived the project, and both authors participated in the experimental design, data analysis and manuscript preparation. PCL carried out all the experiments, and RMX participated in crystallographic data collection and structure determination.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abovich N, Legrain P, Rosbash M (1990) The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol Cell Biol 10: 6417–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (2000) The U box is a modified RING finger—a common domain in ubiquitination. Curr Biol 10: R132–R134 [DOI] [PubMed] [Google Scholar]
- Arenas JE, Abelson JN (1993) The Saccharomyces cerevisiae PRP21 gene product is an integral component of the prespliceosome. Proc Natl Acad Sci USA 90: 6771–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M Jr, Igel AH (1990) Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev 4: 2132–2145 [DOI] [PubMed] [Google Scholar]
- Behrens SE, Tyc K, Kastner B, Reichelt J, Lührmann R (1993) Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol 13: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Reed R (1993) Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science 262: 105–108 [DOI] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA (1977) Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA 74: 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Luhrmann R (2010) Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16: 2384–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens W, Scherly D, Beijer RP, Jansen EJR, Dathan NA, Mattaj lW, Venrooij WJv (1991) A weak interaction between the U2A′ protein and U2 snRNA helps to stabilize their complex with the U2B″ protein. Nucleic Acids Res 19: 455–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody E, Abelson J (1985) The ‘spliceosome’: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228: 963–967 [DOI] [PubMed] [Google Scholar]
- Brosi R, Groning K, Behrens SE, Lührmann R, Krämer A (1993a) Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science 262: 102–105 [DOI] [PubMed] [Google Scholar]
- Brosi R, Hauri HP, Krämer A (1993b) Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem 268: 17640–17646 [PubMed] [Google Scholar]
- Chang TH, Clark MW, Lustig AJ, Cusick ME, Abelson J (1988) RNA11 protein is associated with the yeast spliceosome and is localized in the periphery of the cell nucleus. Mol Cell Biol 8: 2379–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Champion-Arnaud P, Buvoli M, Nadal-Ginard B, Reed R (1994) Specific protein-protein interactions between the essential mammalian spliceosome-associated proteins SAP 61 and SAP 114. Proc Natl Acad Sci USA 91: 6403–6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ (1977) An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8 [DOI] [PubMed] [Google Scholar]
- Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Lührmann R (2006) U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol 26: 2803–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Lührmann R, Stark H (2003) Molecular architecture of the multiprotein splicing factor SF3b. Science 300: 980–984 [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP (2007) U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Ferfoglia F, Raleff F, Krämer A (2011) Interaction domains and nuclear targeting signals in subunits of the U2 small nuclear ribonucleoprotein particle-associated splicing factor SF3a. J Biol Chem 286: 13106–13114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igel AH, Ares M (1988) Internal sequences that distinguish yeast from metazoan U2 snRNA are unnecessary for pre-mRNA splicing. Nature 334: 450–453 [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12: 5–14 [DOI] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim SH (1998) Crystal structure of a small heat-shock protein. Nature 394: 595–599 [DOI] [PubMed] [Google Scholar]
- Krämer A (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annl Rev Biochem 65: 367–409 [DOI] [PubMed] [Google Scholar]
- Krämer A, Ferfoglia F, Huang CJ, Mulhaupt F, Nesic D, Tanackovic G (2005) Structure-function analysis of the U2 snRNP-associated splicing factor SF3a. Biochem Soc Trans 33: 439–442 [DOI] [PubMed] [Google Scholar]
- Krämer A, Gruter P, Groning K, Kastner B (1999) Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol 145: 1355–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Legrain P, Mulhauser F, Gröning K, Brosi R, Bilbe G (1994) Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res 22: 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Mulhauser F, Wersig C, Gröning K, Bilbe G (1995) Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of Saccharomyces cerevisiae. RNA 1: 260–272 [PMC free article] [PubMed] [Google Scholar]
- Kuwasako K, He F, Inoue M, Tanaka A, Sugano S, Güntert P, Muto Y, Yokoyama S (2006) Solution structures of the SURP domains and the subunit-assembly mechanism within the splicing factor SF3a complex in 17S U2 snRNP. Structure 14: 1677–1689 [DOI] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR III, Stevens SW (2010) Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Chapon C (1993) Interaction between PRP11 and SPP91 yeast splicing factors and characterization of a PRP9-PRP11-SPP91 complex. Science 262: 108–110 [DOI] [PubMed] [Google Scholar]
- Legrain P, Choulika A (1990) The molecular characterization of PRP6 and PRP9 yeast genes reveals a new cysteine/histidine motif common to several splicing factors. EMBO J 9: 2775–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Nagai K, Li J (2011) Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473: 536–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto Y, Pomeranz Krummel D, Oubridge C, Hernandez H, Robinson CV, Neuhaus D, Nagai K (2004) The structure and biochemical properties of the human spliceosomal protein U1C. J Mol Biol 341: 185–198 [DOI] [PubMed] [Google Scholar]
- Nesic D, Krämer A (2001) Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP, and prespliceosome formation. Mol Cell Biol 21: 6406–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL (2003) Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol 10: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Perriman RJ, Ares M Jr (2007) Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K (2009) Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K (1998) Crystal structure of the spliceosomal U2B″-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature 394: 645–650 [DOI] [PubMed] [Google Scholar]
- Rain JC, Tartakoff AM, Kramer A, Legrain P (1996) Essential domains of the PRP21 splicing factor are implicated in the binding to PRP9 and PRP11 proteins and are conserved through evolution. RNA 2: 535–550 [PMC free article] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, MacMillan AM (2009) Spliceosome structure: piece by piece. Biochim Biophys Acta 1789: 624–633 [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J (1993) Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev 7: 1909–1925 [DOI] [PubMed] [Google Scholar]
- Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Lührmann R, Glover JNM, MacMillan AM (2006) Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci USA 103: 1266–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW (1990) Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B″ and their cognate RNAs. Nature 345: 502–506 [DOI] [PubMed] [Google Scholar]
- Shuster EO, Guthrie C (1988) Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell 55: 41–48 [DOI] [PubMed] [Google Scholar]
- Shuster EO, Guthrie C (1990) Human U2 snRNA can function in pre-mRNA splicing in yeast. Nature 345: 270–273 [DOI] [PubMed] [Google Scholar]
- Spikes DA, Kramer J, Bingham PM, Doren KV (1994) SWAP pre-mRNA splicing regulators are a novel, ancient protein family sharing a highly conserved sequence motif with the prp21 family of constitutive splicing proteins. Nucleic Acids Res 22: 4510–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Jaladat Y (2010) The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics 10: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wells SE, Ares M Jr (1994) Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol 14: 6337–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R (2011) Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: pii: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Ares M Jr (1996) Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits [published erratum appears in Mol Cell Biol 1996 Jul;16(7):3980]. Mol Cell Biol 16: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.