Abstract
A third of the human genome encodes N-glycosylated proteins. These are co-translationally translocated into the lumen/membrane of the endoplasmic reticulum (ER) where they fold and assemble before they are transported to their final destination. Here, we show that calnexin, a major ER chaperone involved in glycoprotein folding is palmitoylated and that this modification is mediated by the ER palmitoyltransferase DHHC6. This modification leads to the preferential localization of calnexin to the perinuclear rough ER, at the expense of ER tubules. Moreover, palmitoylation mediates the association of calnexin with the ribosome–translocon complex (RTC) leading to the formation of a supercomplex that recruits the actin cytoskeleton, leading to further stabilization of the assembly. When formation of the calnexin–RTC supercomplex was affected by DHHC6 silencing, mutation of calnexin palmitoylation sites or actin depolymerization, folding of glycoproteins was impaired. Our findings thus show that calnexin is a stable component of the RTC in a manner that is exquisitely dependent on its palmitoylation status. This association is essential for the chaperone to capture its client proteins as they emerge from the translocon, acquire their N-linked glycans and initiate folding.
Keywords: calnexin, DHHC6, endoplasmic reticulum, folding, palmitoylation
Introduction
More than a third of the human genome encodes for membrane proteins or proteins secreted into the extracellular milieu. These proteins are generally synthesized with an N-terminal signal sequence that targets the translating ribosome to the endoplasmic reticulum (ER) where it docks onto the translocon (Braakman and Bulleid, 2011). The translocon is a multiprotein transmembrane complex composed of a central sec61α tetrameric pore to which associate a number of accessory proteins such as sec61β, sec62 or the TRAP (translocon-associated protein) complex (Skach, 2007). Once the ribosome is docked onto the translocon, synthesis resumes and the nascent protein is co-translationally translocated through the sec61 channel to the lumen of the ER. As the newly synthesized protein emerges from the translocon, it is handled sequentially by a variety of enzymes, in particular leading to the addition of N-linked glycans and the formation of disulphide bonds, which together increase the solubility and stability of the protein, as well as chaperones that promote folding by preventing aggregation of intermediates.
N-glycosylation is carried out by the oligosaccharyltransferase (OST), a hetero-oligomeric complex, which associates with the ribosome–translocon complex (RTC), allowing the addition of the branched oligosaccharide co- or post-translationally (Chavan and Lennarz, 2006). To prevent aggregation or too rapid folding, which might hinder N-glycosylation (McGinnes and Morrison, 1994), the OST (Li et al, 2008) and the general chaperone BiP/Grp78 (Hammond and Helenius, 1994) maintain the polypeptide in a folding competent state.
Glycoproteins are subsequently handled by the lectin-binding chaperones calnexin and calreticulin (Aebi et al, 2010). Calnexin is a type I membrane protein and calreticulin is its soluble homologue. Proteins that undergo N-glycosylation acquire en bloc a glucose3-mannose9-N-acetylglucosamine2 oligosaccharide. Calnexin/calreticulin can only bind monoglucosylated glycans. Oligosaccharide trimming is therefore required. This occurs immediately after the oligosaccharide has been transferred to the protein, first by ER glucosidase I, which removes the first glucose, and then by glucosidase II, which removes the second. The third glucose is also trimmed by glucosidase II, but its removal requires separation and repositioning of the enzyme with respect to the substrate (Deprez et al, 2005). This repositioning provides a time window during which the monoglucosylated polypeptide can interact with calnexin/calreticulin.
Glucose trimming must be rapid since calnexin was shown to bind proteins not only post-translationally but also co-translationally (Chen et al, 1995). It has been reported that upon signalling, MAP kinase and casein kinase 2-dependent phosphorylation of calnexin on Ser-563 allows its association with ribosomes (Chevet et al, 1999). We here investigated whether other mechanisms control the ability of calnexin to capture its substrates. Calnexin recently came up in four large-scale profiling studies of S-acylated proteins (Martin and Cravatt, 2009; Yang et al, 2010; Yount et al, 2010; Merrick et al, 2011). S-acylation is the addition of a fatty acid, generally C16, palmitate, but possibly also C18, stearate (Kordyukova et al, 2010), to cytosolic cysteine residues via a thioester bond (Linder and Deschenes, 2007). This modification is mediated by transmembrane palmitoyltransferases harbouring a conserved cytosolic DHHC domain, 23 of which are present in the human genome (Greaves and Chamberlain, 2011). In validation of these profiling studies and in agreement with a study published during the revision of this manuscript (Lynes et al, 2011), we show that calnexin can be palmitoylated on two juxtamembranous cysteines and that this modification is mediated by a single palmitoyltransferase, DHHC6, out of the 16 DHHC enzymes present in the mammalian ER. S-acylation controls the localization of calnexin within the ER network, favouring its targeting to the central ER. While being a single copy organelle, the ER is indeed compartmentalized into a variety of still poorly characterized domains that allow it to fulfil its multiple functions, which in addition to protein folding include quality control, synthesis of most cellular lipids and storage/control of intracellular calcium (Pendin et al, 2011). The most apparent, morphologically distinct, ER regions are the highly branched tubular network that extends to the cell periphery, the dense perinuclear ER, which is formed by sheet-like structures, and the nuclear membrane (Puhka et al, 2007; Pendin et al, 2011). The two latter regions composed the rough ER, where the RTC complexes reside.
We found that calnexin is a stable component of the RTC and this association relies on calnexin S-acylation. Formation of the calnexin–RTC supercomplex leads to the recruitment of the actin cytoskeleton, which further stabilizes the assembly. The here-identified acylation-dependent interaction of calnexin with the RTC complex allows it to efficiently capture nascent glycosylated polypeptide chains, as they emerge from the translocon, and promote their folding.
Results
Calnexin is palmitoylated in the ER on two juxtamembrane cysteines
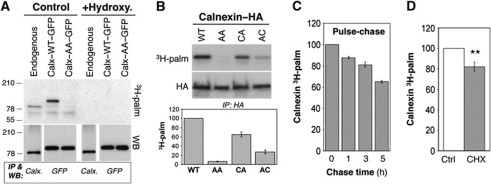
Four profiling studies (Martin and Cravatt, 2009; Yang et al, 2010; Yount et al, 2010; Merrick et al, 2011) identified, with high confidence, calnexin as an S-acylated protein in mammalian cells. By immunoprecipitation of either the endogenous or tagged calnexin from 3H-palmitate-labelled cells, we validated these profiling studies: the protein indeed incorporated radiolabelled palmitate, which could be removed by hydroxylamine hydrochloride treatment, indicating the involvement of a thioester bond (Figure 1A). This does not exclude that calnexin could be modified with either shorter or longer acyl chains as found for certain viral proteins (Kordyukova et al, 2010). 3H-palmitate incorporation occurred on both juxtamembranous cysteines (Figure 1B), although somewhat more efficiently on Cys-502, which is conserved in most species as well as in the testis-specific calnexin homologue, calmegin (Supplementary Figure S1).
Figure 1.
Calnexin is palmitoylated in the ER. (A) HeLa cells were transfected or not (endogenous) for 48 h with calnexin–WT–GFP or calnexin–AA–GFP. Cells were incubated with 3H-palmitic acid for 2 h. Cell extracts were treated or not with 1 M hydroxylamine hydrochloride pH 7.2 (1 h at room temperature) prior to immunoprecipitation using anti-calnexin or anti-GFP antibodies. Immunoprecipitates were split into two, run on SDS–PAGE and analysed by autoradiography or western blotting. (B) HeLa cells were transfected 24 h with calnexin–WT–HA or calnexin–CA–HA, calnexin–AC–HA or calnexin–AA–HA, labelled with 3H-palmitic acid and analysed as in (A) by autoradiography or western blotting (anti-HA). See Supplementary Figure S1 for positions of the cysteines. (Bottom panel) Autoradiograms were quantified using the Typhoon Imager. Errors correspond to standard deviations (n=5). (C) Non-transfected HeLa cells were labelled as in (A). The cells were then washed and incubated in the normal medium and cell lysis was performed at different time points followed by immunoprecipitation as described in (A). Samples were analysed by autoradiography and the quantification was performed using the Typhoon Imager (n=3). (D) HeLa cells were pretreated or not with cycloheximide (CHX) for 1 h followed by 3H-palmitic acid labelling in the presence or absence of the drug. Immunoprecipitation was preformed using anti-calnexin antibodies Samples were analysed by autoradiography and western blotting (anti-calnexin). Quantification was performed using the Typhoon Imager. Errors correspond to standard deviations (n=3). **P<0.01.
In contrast to other lipid modifications, S-acylation is reversible (Greaves and Chamberlain, 2011). Proteins such as Ras indeed undergo dynamic (<30 min) cycles of palmitoylation–depalmitoylation, and these cycles are crucial for their function (Rocks et al, 2010). In contrast to Ras, we found that 3H-palmitate turnover on calnexin is slow (Figure 1C). As a consequence, palmitoylated calnexin must accumulate in cells and we indeed found, as described below, that at steady state the vast majority of the protein is lipid modified. Palmitoylation of calnexin mostly occurs after it has been fully synthesized, since inhibition of protein synthesis with cycloheximide 1 h prior to 3H-palmitate labelling only mildly affected palmitoylation (18±4% decrease; Figure 1D).
Calnexin palmitoylation is mediated by DHHC6
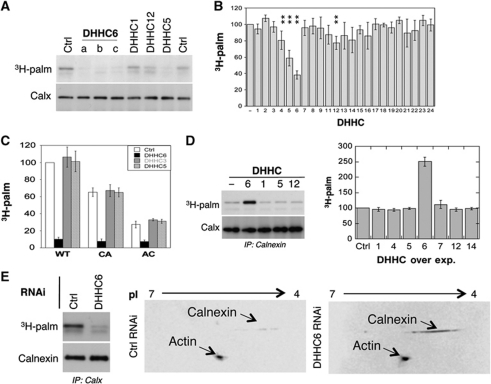
We next identified the palmitoyltransferase that modifies calnexin. PCR analysis revealed that all DHHC enzymes, with the exception of DHHC15, 19 and 22, are expressed in HeLa cells (Supplementary Figure S2A). RNAi duplexes were screened for their efficiency in silencing these enzymes (Supplementary Table S1; Supplementary Figure S2A). Silencing of DHHC enzymes had minor effects (<25% change) on 3H-palmitate incorporation into endogenous calnexin with the exception of DHHC5 and DHHC6 (Figure 2A and B). Importantly, three different siRNA duplexes against DHHC6 led to a similar decrease in calnexin palmitoylation (Figure 2A) as did an shRNA construct against the non-coding region of DHHC6 (Supplementary Figure S2B). Recomplementation of shRNA-treated cells with a DHHC6 expression plasmid restored calnexin palmitoylation (Supplementary Figure S2B), indicating that the observed effect is not due to an off target effect of the shRNA or RNAi duplexes.
Figure 2.
Calnexin is palmitoylated by DHHC6. (A) HeLa cells were transfected for 72 h with different DHHC siRNA or control siRNA (Ctrl). Cells were incubated with 3H-palmitic acid for 2 h prior to immunoprecipitation using anti-calnexin antibodies. Immunoprecipitates were split into two, run on SDS–PAGE and analysed by either autoradiography (3H-palmitate) or western blotting (anti-calnexin). (B) Autoradiograms and western blots from (A) were quantified using the Typhoon Imager. Errors correspond to standard deviations (n=4). **P<0.01. For the identity of the siRNAs see Supplementary Table S1, for their efficiency see Supplementary Figure S2A. (C) HeLa cells were transfected 72 h with calnexin–WT–HA or calnexin–CA–HA, or calnexin–AC–HA and DHHC siRNAs, labelled with 3H-palmitic acid. Immunoprecipitates using anti-HA antibodies were split into two, run on SDS–PAGE gels and analysed by either autoradiography or western blotting (see Supplementary Figure S2C). Autoradiograms and western blotting were quantified using the Typhoon Imager. Errors correspond to standard deviations (n=4). (D) HeLa cells were transfected 24 h with human DHHC cDNA, labelled with 3H-palmitic acid. Immunoprecipitates using anti-calnexin antibodies were analysed and quantified as in (B) (n=4). For analysis of cysteine mutants see Supplementary Figure S2E and F. (E) HeLa cells were transfected for 72 h with DHHC6 siRNA or control siRNA (Ctrl). (Left panel) Cells were labelled with 3H-palmitic acid as analysed as in (A). (Right panel) In all, 50 μg of cell extracts was analysed using 2D gel ZOOM IPGRunner system (Invitrogen) followed by western blotting using anti-calnexin and anti-actin antibodies.
3H-palmitate incorporation experiments were also performed upon ectopic expression of calnexin–HA. Only DHHC6 silencing led to a marked decrease in palmitoylation (Figure 2C; Supplementary Figure S2C). We currently have no clear explanation as to why DHHC5 silencing affects palmitoylation of endogenous but not ectopically expressed calnexin. DHHC6 localizes to the ER (Supplementary Figure S2D and (Gorleku et al, 2011), consistent with the calnexin localization, while DHHC5 is mostly found in the Golgi apparatus (Supplementary Figure S2D). This however does not exclude its presence in the ER. Since DHHC6 is itself palmitoylated (personal communication; Gorleku et al, 2011), our findings raise the possibility—which will be tested in future studies—that DHHC5 palmitoylates DHHC6 or somehow regulates its function.
We next performed the reverse experiments, we monitored the effect of DHHC overexpression. An increase in palmitoylation of endogenous as well as ectopically expressed calnexin was observed upon DHHC6 overexpression but for none of the other enzymes (Figure 2D; Supplementary Figure S2E and F). Analysis of the two single cysteine calnexin mutants showed that both cysteines are modified by the same DHHC6 enzyme (Supplementary Figure S2E and F). Altogether, these observations show that DHHC6 modifies calnexin on both sites. Consistent with the ubiquitous expression of calnexin, we found that the DHHC6 expression is equally broad (Supplementary Figure S3A).
Since many of the experiments described in the present work involve DHHC6 silencing, we tested whether DHHC6 siRNA triggers the unfolded protein response. A gene profiling analysis using Affimetrix chips revealed that while slight changes in expression were observed for GADD45 (−1.3-fold), BiP (1.32-fold), XBP-1 (1.4-fold), HERP (1.5-fold), these changes are minor in comparison to what is observed upon UPR activation. Usually UPR induction leads to 4–27-fold increase in the expression levels of these genes at the mRNA level (Lee et al, 2003). Also, we did not observe significant XBP-1 splicing.
As a side note, DHHC6 overexpression had no effect on the palmitoylation of four other proteins known to be S-acylated that transit through the ER: the Transferrin receptor (Alvarez et al, 1990), the Wnt co-receptor LRP6 (Abrami et al, 2008) and the two anthrax toxin receptors TEM8 and CMG2 (Abrami et al, 2006) (Supplementary Figure S3B), suggesting a certain specificity towards calnexin.
We next investigated what percentage of the calnexin population is S-acylated at steady state in HeLa cells. We previously found that glycosylphosphatidyl inositol-anchored proteins do not migrate in 2D gels unless their lipid moiety has been removed (Fivaz et al, 2000). We inferred that palmitoylation might also affect protein migration in 2D gels. Cell extracts from control and DHHC6 RNAi-treated Hela cells were analysed by 2D-PAGE followed by western blotting against calnexin, and actin as an internal control. The analysis was performed 72 h after transfection with DHHC6 siRNA. This did not affect the total level of endogenous calnexin when compared with control cells, as evidenced by 1D SDS–PAGE (Figure 2E, left panel; also seen in Figure 2A, bottom blot). As a striking validation of our strategy, DHHC6 silencing led to a 9.1±0.9 (n=4)-fold increase of the calnexin signal following 2D-PAGE, while the actin signal was unaffected (Figure 2E, right panels). The increase in calnexin staining upon DHHC6 silencing demonstrated that at least 90% of cellular calnexin is DHHC6 modified at steady state in HeLa cells. Since both sites can be modified (Figures 1B and 2C; Supplementary Figure S2C, E and F), depalmitoylation is slow and must be slow on both sites (Figure 1C), the acylated calnexin population revealed by the 2D gel analysis is most likely modified on both sites.
In Figure 2D, we show that overexpression of DHHC6 leads to a ≈2.5-fold increase in the incorporation of 3H-palmitate into calnexin. This might at first appear inconsistent with the observation that >90% of calnexin is palmitoylated. It is however important to note that only calnexin molecules with free cysteines can incorporate 3H-palmitate. Thus, the bulk of cellular calnexin (>90%) is ‘silent’ in the 3H-palmitate incorporation assay and we are monitoring the incorporation of 3H-palmitate in the remaining population. The rate at which this small population of calnexin with free cysteines becomes modified depends on the abundance of the enzyme. Thus, upon DHHC6 overexpression, incorporation rates increase due to an increased amount of the enzyme and so does the signal after our 2 h standard incorporation (Figure 2D).
Palmitoylation is predicted to affect the conformation of calnexin
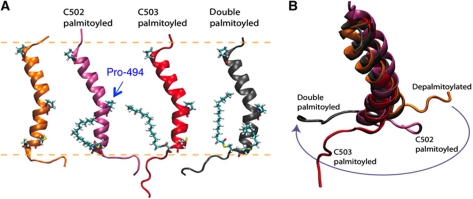
Using predictive computational methods, we next analysed whether palmitoylation is expected to modify the structure of the transmembrane domain (TMD) and/or cytosolic tail of calnexin. Using a topology prediction algorithm (Viklund et al, 2008), we first predicted that the membrane-embedded residues span from Trp-482 to Cys-502. The TMD was then modelled as an ideal α-helix based on secondary structure predictions and consistent with the UniProt annotations (P27824). We further extended the helical sequence to include residues Glu-478 through Ser-510, in order to gain information on the conformation of the cytosolic tail. The atomistic transmembrane model was inserted into a lipid membrane bilayer and refined using molecular dynamics (MD) simulations under physiological-like conditions (e.g., pH ∼7, 1 a.t.m., 298 K). Only small fluctuations of the secondary structure were observed, which support the predicted helical conformation and the match with the hydrophobic interior of the bilayer. The helix tilted by ∼30° with respect to the membrane surface normal. A distortion of 30±5° in the direction of the helix axis was observed at the level of Pro-494 (Figure 3A), as expected from the helix breaking capacity of this residue (Chang et al, 1999). This proline thus breaks the rotational symmetry of the helix with respect to its main axis. The full conservation of this residue in calnexin and calmegin (Supplementary Figure S1) suggests that the TMD kink might be of functional importance.
Figure 3.
Modelling of the effect of palmitoylation on calnexin conformation. (A) The atomistic models of the helical TMD of calnexin were equilibrated in a solvated DOPC membrane bilayer using MD simulations. Cys-502, Cys-503 and Pro-494 are shown in licorice representation. The approximate position of the membrane bilayer is shown by dashed lines. (B) Palmitoylation affects the orientation of the cytosolic tail of calnexin with respect to the axis of the transmembrane helix.
We next included palmitate moieties at positions Cys-502 (CyP502), Cys-503 (CyP503) or both (CyP502–503). Palmitoylation did not affect the proline-induced kink and only marginally affected the helix tilt. The MD simulations however predicted a palmitoyl-dependent orientation of the cytosolic tail with respect to the helix axis (Figure 3B). Interestingly, palmitoylation at position 503 is predicted to have a more pronounced effect on the conformation of the cytosolic tail, indicating that both sites are not equivalent and that one of the two might have a regulatory role, predictions that will be tested in the future. Combined with the asymmetric nature of the TMD, these simulations raise the interesting possibility that palmitoylation may affect the conformation of calnexin, which in turn could modify its affinity for membrane domains and/or its capacity to interact with proteins in the membrane or in the cytoplasm.
DHHC6 affects calnexin localization
Palmitoylation of membrane proteins has been proposed to modify their affinity for specific plasma membrane micro/nano domains (Charollais and Van Der Goot, 2009; Levental et al, 2010). We therefore investigated whether DHHC6 silencing would alter the distribution of calnexin. Under control conditions, immunofluorescence staining of calnexin is observed in all three major ER compartments (Figure 4A, for additional images see Supplementary Figure S4A), resembling the staining of the soluble lumenal ER chaperone BiP and the transmembrane RTC-associated protein TRAPα (Figure 4B, for additional images see Supplementary Figure S4A).
Figure 4.
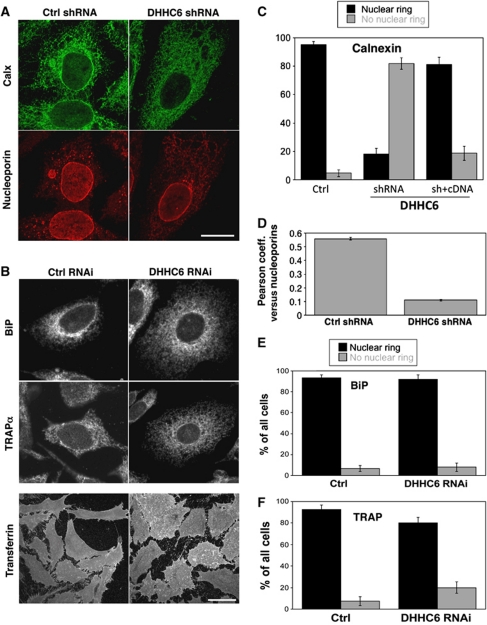
Effect of DHHC6 silencing on calnexin localization. (A) HeLa cells were transfected with either a control shRNA or shRNA against DHHC6 for 8 days, fixed in methanol and stained with anti-calnexin and anti-nucleoporin antibodies. Bar: 10 μm. (B) HeLa cells grown on cover slips were transfected for 72 h with either control siRNA or siRNA against DHHC6 and were incubated or not with fluorescent Transferrin, washed, fixed in methanol and immunostained with any one of the following antibodies: calnexin, BiP, TRAPα. Each image represents the sum of all the stacks taken in z-axis. To visualize more cells see Supplementary Figure S4A. Bar: 10 μm. (C) HeLa cells were transfected with either a control shRNA or shRNA targeting DHHC6 for 8 days. On day 6, the cells were transfected with either an empty vector or human DHHC6 cDNA bearing a myc-tag for complementation. The cells were fixed and stained with anti-calnexin, anti-nucleoporin and Hoechst dye. The cells were analysed manually for the presence or absence or restoration of the nuclear membrane staining of calnexin. In all, 50 cells were analysed per condition per experiment. Error bars represent the standard deviation (n=3). (D) The cells from (A) were imaged and the Pearson correlation coefficient for co-localization between calnexin and nucleoporins was determined by monitoring 15 cells/experiment from each condition (n=3). (E, F) Confocal stacks of cells treated as in (B) were analysed manually and the number of cells showing nuclear or no nuclear membrane staining for calnexin (C), BiP (D) or TRAPα was determined. Three independent experiments were analysed and 50 cells were counted for each experiment. Error bars represent standard deviations.
When silencing DHHC6, the first observation was a change in cell morphology: cells were somewhat less elongated (see Transferrin staining bottom of Figure 4B) and the average cell footprint was ∼30% larger (Supplementary Figure S5A). The nuclear staining and size was however unaltered (Figure 4A). When viewing through confocal stacks of a lawn of cells stained for calnexin (Supplementary Movie S1A versus Supplementary Movie S1B), it readily appeared that calnexin staining was lost from the nuclear envelope upon DHHC6 silencing. This was confirmed by quantifying the cells showing a nuclear envelope calnexin staining and those that did not, by eye for multiple cells over multiple experiments (Figure 4C). It agreement, the Pearson coefficient of co-localization of calnexin with nucleoporins strongly decreased upon DHHC6 silencing (Figure 4D; Supplementary Figure S4B). Importantly, nuclear membrane localization of calnexin was restored upon recomplementation with the DHHC6-expressing plasmid (Figure 4C; Supplementary Figure S4B). Disappearance from the nuclear membrane was specific to calnexin, since BiP and TRAPα staining was unaffected by DHHC6 silencing (Figure 4B, E and F; Supplementary Figure S4A). While disappearance from the nuclear membrane was obvious, other changes in calnexin staining, in particular in the dense and complex perinuclear ER could not be quantified in satisfactory manner. Our interpretation of the change in calnexin distribution is that palmitoylation increases the affinity of calnexin for ER sheets, which encompasses the entire rough ER and of which the nuclear membrane is the most exacerbated form.
Palmitoylation regulates the interaction of calnexin with the RTC
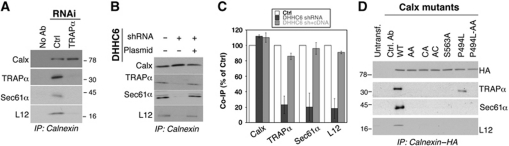
The observation that DHHC6 expression favours the localization of calnexin to the rough ER prompted us to investigate whether calnexin interacts with the RTC under steady-state conditions. The central translocon component sec61α, as well as the translocon-associated protein TRAPα and the ribosomal subunit L12 were recovered upon immunoprecipitation of endogenous calnexin (Figure 5A). The interaction of calnexin with sec61α and L12 was abolished upon RNAi silencing of TRAPα (Figure 5A), indicating first that the observed interaction of calnexin with sec61α and L12 is specific, and not due to bulk co-isolation of membrane proteins in detergent/lipid micelles. Moreover, the observation indicates that the TRAP complex mediates the interaction of calnexin with the RTC. Interestingly, TRAPα silencing did not affect the localization of calnexin to the nuclear membrane (Supplementary Figure S5B), showing that DHHC6-induced perinuclear calnexin localization is not due to its interaction with RTCs.
Figure 5.
Palmitoylation regulates the interaction of calnexin with the RTC. (A) HeLa cells were transfected for 72 h with either the control siRNA or a siRNA against TRAPα. Immunoprecipitates against anti-calnexin were analysed by western blotting for calnexin, TRAPα, Sec61α and L12. (B) HeLa cells were transfected for 8 days with control shRNA or the shRNA against DHHC6. On day 6, the cells were retransfected either with an empty plasmid (lanes 1 and 2) or with a plasmid-expressing DHHC6. The cells were lysed on day 8 and immunoprecipitated using mouse anti-calnexin antibody. Immunoprecipitates were analysed SDS–PAGE followed by western blotting against calnexin, TRAPα, Sec61α and L12. For the analysis of the total cell lysates see Supplementary Figure S6A. (C) Western blots from (B) were quantified using the Image J software and the error bars represent the standard deviation (n=3). (D) HeLa cells were transfected for 48 h with either WT, single or double cysteine calnexin mutants, mutant of the phosphorylation calnexin site S563A or of the TMD proline. Immunoprecipitates against anti-HA were analysed by western blotting against: TRAPα, Sec61α, Sec62, Sec61β and L12. For the analysis of the total cell lysates see Supplementary Figure S6B.
We next silenced DHHC6 by shRNA treatment for 8 days, in order to reduce the cellular pool of S-acylated calnexin. Upon long-term DHHC6 silencing, co-immunoprecipitation of TRAPα, sec61α and L12 with calnexin decreased by ≈80% (Figure 5B and C; Supplementary Figure S6A). Importantly, interaction of calnexin with the RTC was restored by transfection with DHHC6-expressing plasmid (Figure 5B and C; Supplementary Figure S6A). RTC interaction experiments were next carried out using the single and double cysteine mutants of calnexin, the S563A mutant, where the phosphorylation site promoting ribosome association is modified (Chevet et al, 1999), and the P494L mutant, where the transmembrane proline (Figure 3A) is substituted with a hydrophobic residue to remove the kink in the TM helix. It should be noted that the two latter mutations did not prevent palmitoylation (personal communication). The calnexin–RTC interaction was abolished upon mutation of Ser-563 to alanine (Figure 5D; Supplementary Figure S6B), indicating the importance of this residue even in resting cells (Chevet et al, 1999). The RTC interaction was also severely affected by the P494L mutation, pointing out a central role of TMD conformation for the association (Figure 5D). Most strikingly, mutation of either S-acylation site abolished the calnexin–RTC interaction (Figure 5D). Thus, not only must calnexin be acylated to interact with the RTC, it has to be modified on both cysteines.
Actin is recruited and stabilizes the calnexin–RTC supercomplex
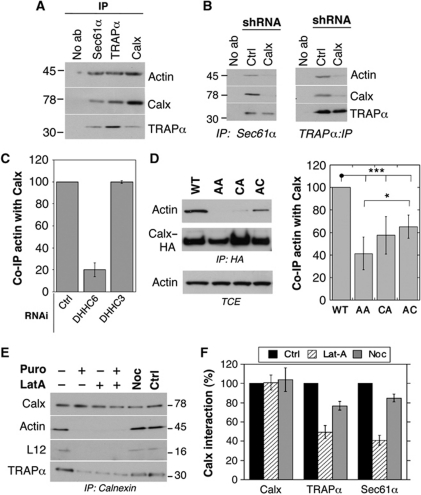
Stabilization of certain membrane protein complexes/clusters has been found to be dependent on the actin cytoskeleton (Abrami et al, 2010). We probed whether the calnexin–RTC supercomplex interacts with actin. Actin was pulled down upon immunoprecipitation of sec61α, TRAPα as well as calnexin (Figure 6A). The interaction between sec61α or TRAPα with actin was strongly diminished upon silencing of calnexin (Figure 6B), indicating first that the interaction observed in Figure 6A is specific and not due to contamination by this abundant protein, and second that calnexin plays a crucial role in the binding of actin to the RTC.
Figure 6.
Calnexin palmitoylation promotes interaction with actin. (A) HeLa cells lysates were immunoprecipitated using either anti-Sec61α anti-TRAPα, anti-calnexin antibodies or no antibody. The immunoprecipitates were analysed by western blotting against actin, calnexin and TRAPα. Blotting against Sec61α could not be performed as it migrates at the same molecular weight as the antibody heavy chain. (B) HeLa cells were transfected for 6 days with either shRNA against luciferase or shRNA against calnexin and lysed. After immunoprecipitation against Sec61α (left panel) or TRAPα (right panel), samples were analysed by western blotting against actin, calnexin and TRAPα. As in (A), Sec61α could not be probed. (C) HeLa cells were transfected with a control siRNA, siRNA against DHHC6 or DHHC3 and lysed. Immunoprecipitates against calnexin were analysed by western blotting against calnexin and actin. Quantification was performed using the Typhoon Imager. Error bars correspond to standard deviations (n=4). (D) HeLa cells were transfected or not for 48 h with cDNA-expressing calnexin–HA–WT, double or single cysteine mutants, and lysed Immunoprecipitates against HA were analysed by western blotting against HA and actin and quantified using the Typhoon Imager. Error bars correspond to standard deviations (n=4). Total cell extracts (TCE) were probed for actin. (E) HeLa cells were treated for 30 min at 37°C with either DMSO, Latrunculin A or Nocodazole or 15 min with puromycin or water, which was used as a control. The cells were lysed and immunoprecipitated with rabbit anti-calnexin antibody. The immunoprecipitates and the total cell extracts were migrated on SDS–PAGE followed by western blot to reveal calnexin, actin, TRAPα and L12 (n=3). (F) The treatment with Latrunculin A and Nocodazole was performed as in (E). Immunoprecipitates against calnexin were analysed by SDS–PAGE and followed by western blotting against calnexin, TRAPα and Sec61α. Western blots were quantified using the Image J software and the error bars correspond to standard deviations (n=3). ***P<0.001, *P<0.05.
The interaction of calnexin with actin was dependent on its palmitoylation status as indicated by the decrease in actin association upon DHHC6 silencing (Figure 6C). Calnexin–actin interaction was restored upon transfection with the DHHC6-expressing plasmid (Supplementary Figure S6C). The importance of palmitoylation for calnexin–actin interaction was confirmed by the weak association of the single and double cysteine calnexin mutants with actin (Figure 6D). The fact that DHHC6 silencing had a more pronounced effect than mutation of the palmitoylation sites might be due to indirect effects of unidentified ER DHHC6 targets.
Altogether, the above observations raised the possibility that actin can only be recruited to calnexin–RTC supercomplex when it is fully assembled. To test this possibility, we stripped off ribosomes from the ER membrane by a 15-min puromycin treatment. Strikingly, calnexin then no longer interacted with either the translocon or actin (Figure 6E), indicating that indeed actin can only bind to the fully assembled supercomplex. Moreover, the puromycin effect shows that calnexin is recruited to the translocon only when the ribosome has docked.
In reverse, we investigated whether actin polymerization was important for the stabilization of the supercomplex. Inhibiting actin polymerization by a short latrunculin A treatment was sufficient to decrease the interaction of calnexin with L12, TRAPα and sec61α (Figure 6E and F), while nocodazole, which disrupts microtubules, had no effect (Figure 6E and F).
Palmitoylation of calnexin promotes the folding of glycosylated proteins
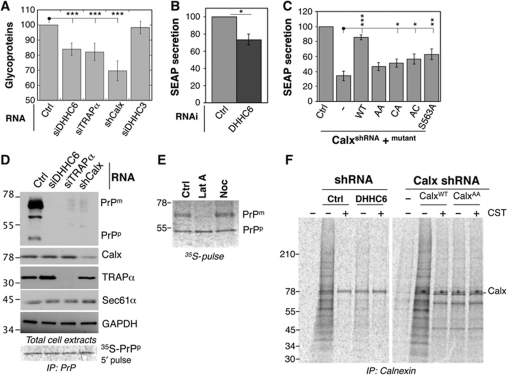
Finally we investigated whether the calnexin–RTC interaction is important for the lectin chaperone function. We first analysed the effect of silencing either calnexin, TRAPα or DHHC6 on the production—that is, an overall read out for synthesis, folding and secretion—of total glycoproteins. Silencing of any of the three proteins led to a significant decrease in total glycoproteins as monitored by a 20-min 35S Cys/Met pulse, followed by precipitation with the lectin, concavalin A (Figure 7A; Supplementary Figure S7A). The 30% drop in detected glycoproteins is consistent with the known redundancy of the folding systems for glycoproteins (Braakman and Bulleid, 2011). As a control, we monitored the production of cytosolic, that is, non-N-glycosylated, proteins and found that silencing of calnexin, TRAPα or DHHC6 had no effect (Supplementary Figure S7B).
Figure 7.
Palmitoylation of calnexin promotes the folding of glycosylated proteins. (A) HeLa cells were transfected with either the control siRNA or siRNAs against DHHC6, TRAPα or DHHC3 for 72 h and shRNA against calnexin for 144 h. The cells were labelled with 35S-methionine/cysteine for 20 min. The cytosolic proteins were isolated by semi-permeabilizing the cells using digitonin. The glycoproteins were separated from the non-cytosolic fraction by using Con-A agarose beads. The glycoprotein fraction was run on SDS–PAGE and the radiolabelled products were visualized by Typhoon phosphoimager. The radiolabelled lanes were quantified using the Typhoon Imager and the histograms were plotted as the ratio of glycoproteins to the cytosolic proteins in each sample. The error bars represent the standard deviation (n=4). ***P<0.001. For the autoradiograms of glycoproteins and cytosolic proteins see Supplementary Figure S7A and B. (B) HeLa cells were co-transfected with either control siRNA or siRNA against DHHC6 and the pSEAP2 plasmid. The cell medium was replaced with fresh medium in all the plates so that the SEAP secreted in the final 24 h was quantified. The SEAP secreted was normalized to the total amount of protein present in each sample and expressed as percentage of SEAP secreted by the control. *P<0.05. (C) HeLa cells were transfected with either the control shRNA or the shRNA against calnexin. At 96 h post-transfection, cells were transfected with the pSEAP2 vector and either empty vector or the vector-expressing WT or mutant calnexins. At 120 h post-transfection, the cell medium was replaced with fresh medium in order to assess the SEAP accumulated in the last 24 h. The measured SEAP values were normalized to the total amount of the protein present in each sample and the values were plotted as a percentage of the shLuc transfection. ***P<0.001, **P<0.01, *P<0.05. (D) HeLa cells were either co-transfected with GFP–PrP and the control siRNA or the siRNA targeting DHHC6, TRAPα. Calnexin was silenced by shRNA and GFP–PrP was transfected 96 h after the shRNA transfection. Cell lysates were prepared and equal amounts of the proteins were analysed by western blotting for GFP, calnexin, TRAPα, Sec61α, GAPDH. PrPp indicates the precursor form of GFP–PrP and PrPm the mature form. (E) HeLa cells were transfected with GFP–PrPwt for 24 h, treated with either DMSO, Latranculin A or Nocodazole for 30 min at 37°C and prior to 30 min 35S-methionine–cysteine pulse labelling in the presence of the drugs. The cell lysates were immunoprecipitated with anti-GFP antibody and the immunoprecipitates were analysed by SDS–PAGE followed by Typhoon phosphoimaging. PrPp indicates the precursor and PrPm the mature form of the GFP–PrP protein. (F) To monitor the binding of calnexin to the substrates, HeLa cells were treated with either shRNA against DHHC6 (left panel) or shRNA against calnexin (right panel). The cells depleted of endogenous calnexin were recomplemented with either the WT or AA mutant. The cells were pulse labelled with 35S-methionine–cysteine for 10 min followed by a chase of 3 min either in the presence of CST or not. The lysate was immunoprecipitated using anti-calnexin antibody and migrated on a SDS–PAGE followed by fixation and drying of the cells. The radiolabelled products were revealed by Typhoon phosphoimager.
Next, we analysed the production of two specific glycoproteins: a soluble version of alkaline phosphatase (SEAP, secreted alkaline phosphatase), which provides a quantitative readout upon secretion, again used as an overall read out for synthesis, folding and secretion (Lakkaraju et al, 2008) and the prion protein (PrP), which undergoes a characteristic change in migration by SDS–PAGE upon folding and maturation (Fons et al, 2003).
DHHC6 silencing led to a 30% decrease in SEAP secretion (Figure 7B). In contrast, secretion of the non-glycosylated protein bovine preprolactin, a well-studied protein translocation substrate (Lakkaraju et al, 2008), was not affected by DHHC6 silencing (Supplementary Figure S7C).
To assess the direct involvement of calnexin and the importance of its palmitoylation status, we silenced calnexin by shRNA and recomplemented the cells with wild-type (WT) or mutant calnexins (Supplementary Figure S7D). Calnexin silencing led to a 70% drop in SEAP secretion, which could be restored to >80% of control cells, upon recomplementation with the WT protein (Figure 7C). In contrast, no recovery was observed upon recomplementation with palmitoylation-deficient calnexin (AA mutant) (Figure 7C), while single cysteine and S563A mutants led to a very weak recovery (Figure 7C). Altogether, these observations show that DHHC6-mediated palmitoylation of calnexin on both cysteines is required for optimal folding and secretion of SEAP.
We next monitored maturation of GFP-tagged PrP, which upon expression in control cells migrates by SDS–PAGE predominantly in a mature form with some precursor form (Figure 7D). Neither the mature nor the precursor PrP forms were detectable by western blotting upon silencing of calnexin, TRAPα or DHHC6. Synthesis of PrP precursor was, however, unaffected by the calnexin, TRAPα or DHHC6 silencing as determined by a 5-min 35S-pulse labelling of PrP (bottom of panel Figure 7D and quantification Supplementary Figure S7E). Thus, in the absence of these proteins, PrP is rapidly degraded following synthesis. A similar rapid post-translational degradation of PrP was observed upon Latrunculin A treatment (Figure 7E), further supporting that proper folding of PrP requires the fully assemble calnexin–RTC supercomplex.
Altogether, the above experiments indicate that S-acylation controls the ability of calnexin to acts as a chaperone. To address this point specifically, we monitored the binding of monoglucosylated substrates to calnexin. DHHC6 was silenced or not using shRNA and newly synthesized proteins were metabolically labelled. After detergent solubilizing the cells, calnexin–substrate complexes were isolated by immunoprecipitation against calnexin. As a control, cells were pretreated or not with castanospermine (CST), a specific inhibitor of α-glucosidases, which prevents the generation of monoglucosylated proteins (Hammond and Helenius, 1994). Importantly, CST had no effect on either palmitoylation or depalmitoylation of calnexin (not shown). Also, catanospermine did not alter the cellular localization of calnexin nor its interaction with the RTC (not shown).
After a pulse of metabolic labelling, a well-defined pattern of calnexin substrates was detected in control cells, which was absent in CST-treated cells (Figure 7F, left panel). DHHC6 silencing led to a drastic decrease in the amount of calnexin-bound substrates, without affecting their pattern (Figure 7F, left panel). Even when the metabolic pulse was followed by a chase period, substrate binding could not be observed in DHHC6-silenced cells, indicating that not only co- but also post-translational binding was defective. To confirm that non-acylated calnexin is unable to capture its substrates, substrate binding of the AA palmitoylation-deficient mutant calnexin was monitored in comparison to WT, in cells where endogenous calnexin was silenced by shRNA. As shown in Figure 7F (right panel), palmitoylation-deficient calnexin failed to bind its substrates. This observation, however, does not necessarily mean that the capacity of calnexin to binding monoglucosylated proteins per se is affected. It reveals that in the cellular context, calnexin must be palmitoylated and be part of the RTC complex to grab its substrates. If substrates are not captured co-translationally, they either aggregate or are handled by other folding systems in the ER, but calnexin will not bind them post-translationally.
Discussion
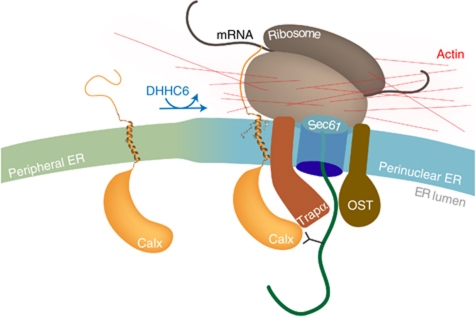
Calnexin is a transmembrane chaperone that aids in the folding of glycosylated proteins by binding to monoglucose residues on the branched N-linked oligosaccharide and thus protects the protein from aggregation. The interaction of calnexin with nascent chains has been observed >15 years ago (Chen et al, 1995), but little information was available on the specific mechanisms that may facilitate this co-translational interaction. We here report that calnexin is a stable component of the RTCs, as is the OST, leading to the formation of a supercomplex. This interaction occurs via the translocon-associated complex TRAP. Based on X-ray crystallography and single particle analysis, a model of the translocon complex has been proposed wherein TRAPα interacts with sec61α, the latter interacting via the opposing face of the molecule with sec61β (Menetret et al, 2008). Our findings thus allow a rough positioning of calnexin in this supercomplex, sandwiching TRAPα between calnexin and the translocon (Figure 8).
Figure 8.
Molecular consequences of calnexin palmitoylation. When its palmitoylation sites are free, calnexin preferentially localizes to the peripheral tubular ER. Upon S-acylation by DHHC6, calnexin partitions preferentially into the rough ER and associates with TRAPα, altogether ensuring that calnexin efficiently interacts with the RTC. Once the calnexin–RTC has assembled into a supercomplex, actin is recruited and stabilizes the assembly. In this configuration, calnexin is positioned to efficiently grab nascent glycoproteins as they emerge from the translocon pore, acquire their N-linked glycan from which the two external glucoses have been trimmed.
Formation and stabilization of calnexin–RTC supercomplex requires several factors. As previously described (Chevet et al, 2010), Ser-563 is critical possibly due to its phosphorylation. The kinked conformation of the TMD also influences the interaction, as indicated by the disruptive effect of mutating the highly conserved TM Pro-494. Importantly, calnexin must be palmitoylated.
Despite the presence of 12 (Ohno et al, 2006) to 16 (our unpublished observations in Hela cells) palmitoyltransferase in the mammalian ER, the S-acylation of calnexin is performed by a single enzyme, DHHC6. The involvement of a single DHHC enzyme was rather unexpected since these enzymes are thought to have broad overlapping specificities at least for soluble substrates (Greaves and Chamberlain, 2011). Also, in contrast to soluble substrates such as Ras, calnexin does not undergo dynamic cycles of palmitoylation–depalmitoylation. As a consequence, at steady state, the S-acylated form, and most likely the dual acylated form, accumulates and almost the entire pool of cellular calnexin is modified in resting cells as shown by our 2D gel analysis. The observed slow rate of palmitate turnover might be due to the close proximity, and possibly even embedding, of the cysteine residues in the membrane, which would render them inaccessible to cytosolic thioesterases, of which only one has so far been identified, the soluble enzyme APT1 (Dekker et al, 2010). Recently, it has been observed for β1-adrenergic receptor that the susceptibility of cysteines to thioesterases indeed correlates with distance from the TMD (Zuckerman et al, 2011).
S-acylation of calnexin promotes its interaction with the RTC in two ways: First, palmitoylation affects the ability of calnexin to interact with the TRAP complex, possibly by influencing the conformation of the TMD and/or the cytosolic tail as suggested by the MD simulations. Second, palmitoylation favours the association of calnexin with the nuclear membrane, and probably with sheet-like ER structures in general. Calnexin spans the membrane only once and it is therefore unlikely that it has any membrane curvature sensing capacity. Since protein and lipid compositions are likely different in ER tubules versus ER sheets (Shibata et al, 2008, 2010), palmitoylation may favour the partitioning of calnexin to sheet-like structures, in a manner similar to raft partitioning of transmembrane proteins at the plasma membrane (Levental et al, 2010). Calnexin has also been shown to localize to mitochondrial-associated membranes (MAM), sites of interaction between the ER and mitochondria (Myhill et al, 2008; de Brito and Scorrano, 2010). During the revision of this manuscript, localization of calnexin as well as of TMX, a transmembrane ER-thioredoxin, to MAMs was reported to depend on palmitoylation of these two type I membrane proteins (Lynes et al, 2011). Future studies should elucidate whether and how S-acylation-dependent localization of calnexin to the various ER domains is controlled.
Interestingly, once fully assembled, the calnexin–RTC supercomplex recruits the actin cytoskeleton, which in turn is required to stabilize the assembly. While in yeast and plants where cortical ER is known to move along actin cables (Du et al, 2004), few reports describe interactions between actin and the ER in animal cells (Du et al, 2004). Actin is well known to interact with the nuclear envelope via Nesprins, large transmembrane proteins involved in nuclear positioning and nuclear envelope architecture (Lombardi et al, 2011). However, interactions with the rest of the ER have not been reported with the exception of a study showing that actin controls the diffusion of newly synthesized glycoproteins in the ER lumen and thereby reduces aggregation during folding (Nagaya et al, 2008). Our observations raise the possibility that this is due to the handling of these glycoproteins by the calnexin–RTC supercomplexes.
Altogether, the present work leads to a model where DHHC6-mediated S-acylation favours the partitioning of calnexin into the rough ER and, in addition, allows it to interact with RTC components leading to the formation of a supercomplex, which subsequently recruits the actin cytoskeleton, the polymerization of which in turn is required to maintain the assembly (Figure 8). When the nascent polypeptide chain emerges from the translocon pore, it arrives into a folding prone microenvironment: the OST and BiP initially keep it in an unfolded state to allow optimal N-glycosylation and also await the full synthesis of folding units. By being an integral part of the ribosome–translocon supercomplex, calnexin is ideally positioned to capture its client protein following appropriate glucose trimming, protecting the protein from aggregation as it undergoes oxidative folding, further benefiting from the calnexin-associated oxidoreductase ERp57.
Materials and methods
Cell culture, plasmids, transfections and real-time PCR
HeLa cells (ATCC) were grown in complete modified Eagle's medium (MEM) (Sigma) at 37°C supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, penicillin and streptomycin. RPE1 cells were grown in complete Dulbeccos MEM (DMEM) (Gibco) supplemented with 10% FBS, 2 mM L-glutamine, penicillin and streptomycin. Mammalian expression plasmids harbouring human C-terminally tagged Calnexin–HA and Calnexin–GFP were generated by cloning the calnexin cDNA into pCDNA3 vector bearing the HA tag or GFP. Constructs to express single- or double-point mutation of calnexin were generated using the Quickchange (Stratagene) strategy following the manufacturer's instructions. Isoform 1 of human tumour endothelial marker 8 (TEM8) cDNA tagged with a HA epitope was cloned in the pIREShyg2 vector. Plasmid encoding human myc–LRP6 was provided by Dr G Davidson. GFP–PrP was provided by Dr Chiara Zurzolo. PPL-3f was provided by Dr Katharina Strub. Human DHHC cDNA were cloned in pCE puro-His–myc or flag-expressing plasmids. Plasmids were transfected into cells for 24 or 48 h (2 μg cDNA/9.6 cm2 plate) using Fugene (Roche Diagnostics Corporation). For control transfections, we used an empty pCDNA3 plasmid. shRNA against calnexin were generated from the p.Super.Retro.Puro vector containing the hairpin insert against the following target sequence present at the 3′ UTR of calnexin gene: 5′-gagcttgatctgtgatttc-3′ (Supplementary Table S1). As a control, we used shRNA against the target sequence in firefly luciferase gene: 5′-CGTACGCGGAATACTTCGA-3′. shRNA against DHHC6 was generated by cloning the hairpin insert against the target region in the 3′ UTR of DHHC6: 5′CCTAGTGCCATGATTTAAA3′ (Supplementary Table S1).
For the real-time PCR, RNA was extracted from a six-well dish using the RNeasy kit (Qiagen). In all, 1 mg of the total RNA extracted was used for the reverse transcription using random hexamers and superscript II (Invitrogen). A 1:40 dilution of the cDNA was used to perform the real-time PCR using SyBr green reagent (Roche). mRNA levels were normalized using three housekeeping genes: TATA-binding protein, β-microglobulin and β-glucoronidase. Total RNA of different human tissues was obtained from Amsbio.
Antibodies and reagents
Polyclonal antibodies against calnexin were generated against the C-terminal peptide: CDAEEDGGTVSQEEEDRKPK in rabbit; rat antibodies against purified CMG2 were produced in our laboratory; anti-HA and anti-GFP were from Roche; Anti-flag M2 antibody from Sigma; anti-HA–agarose-conjugated beads, used for the immunoprecipitations from Roche (Applied Science, IN); anti-calnexin monoclonal and anti-myc antibodies from Santa Cruz; anti-actin antibodies from Millipore; anti-Transferrin receptor antibodies from Zymed; anti-Nuclear Pore Complex Proteins (Mab414), anti-Sec61α, anti-BiP antibodies from Abcam. Anti-L12 antibodies were provided Dr K Strub. Anti-TRAPα antibody was provided by Dr R Hegde. Protein G-agarose-conjugated beads were from GE Healthcare, HRP secondary antibodies were from Pierce, and Alexa-conjugated secondary antibodies from Molecular Probes. Latrunculin A (used at a concentration of 0.4 mg/ml for 30 min or 1 h) and Nocodazole (used at a concentration of 10 μM for 30 min or 1 h) were from Sigma. CST was purchased from Calbiochem and used at a concentration of 1 mM. Puromycin was purchased from Calbiochem and used at a concentration of 200 μM for 15 min to strip the ER of ribosomes in HeLa cells.
Immunoprecipitation and RNAi experiments
siRNA against human DHHC were purchased from Qiagen (see Supplementary Table S1). As control siRNA we used the following target sequence of the viral glycoprotein VSV-G: 5′attgaacaaacgaaacaagga3′. For gene silencing, Hela cells were transfected for 72 h with 100 pmol/9.2 cm2 dish of siRNA using interferrin (Polyplus) transfection reagent.
For all immunoprecipitations unless specified, cells were lysed 30 min at 4°C in IP buffer (0.5%NP-40, 500 mM Tris–HCl pH 7.4, 20 mM EDTA, 10 mM NaF, 2 mM benzamidine, and a cocktail of protease inhibitors; Roche) followed by centrifugation for 3 min at 2000 g. The supernatants were precleared with protein G-agarose-conjugated beads and incubated 16 h at 4°C with antibodies and beads. The beads were washed for three times with the IP buffer and resuspended in the sample buffer (2 ×) after the final wash. The samples were heated at 95°C for 5 min and migrated on SDS–PAGE. Western blotting was performed using the iBlot (Invitrogen) according to the manufacturer's instructions. Quantification of the blots was done using either the Typhoon Imager or Image J software.
Radiolabelling experiments
To monitor palmitoylation, HeLa cells transfected with either the WT or mutant calnexin constructs were incubated for 2 h at 37°C in IM (Glasgow minimal essential medium buffered with 10 mM Hepes, pH 7.4) with 200 μCi/ml 3H palmitic acid (9,10-3H(N)) (American Radiolabeled Chemicals, Inc.). The cells were washed and the cell lysate was extracted followed by immunoprecipitation with tag specific antibodies in the case of exogenously expressed calnexin and calnexin antibody in case of endogenous protein. After the washes, beads were incubated for 5 min at 90°C in non-reducing sample buffer (2 ×) prior to SDS–PAGE. After the SDS–PAGE, the gel was incubated with a fixative solution (25% isopropanol, 65% H2O, 10% acetic acid), followed by a 30-min incubation with signal enhancer Amplify NAMP100 (Amersham). The dried gels were exposed to a Hyperfilm MP (Amersham). Chemical removal of S-palmitoylation was performed by treating cell extracts for 1 h at room temperature with 1 M hydroxylamine hydrochloride (Sigma) pH 7.2. Protein synthesis was blocked by 1 h treatment with 10 μg/ml cycloheximide (Sigma) at 37°C.
To monitor the ability of calnexin to bind substrates, HeLa cells were first transfected with the shRNA against calnexin for 7–9 days. After 96 h, cells were retransfected with either the WT or the AA mutant calnexin for 48 h.
Cells were pretreated or not for 1 h with 1 mM CST, a specific inhibitor of α-glucosidases and pulse labelled with 100 μCi/ml 35S-methionine/cysteine for 10 min followed by a 3-min chase in the presence or absence of drug. Cells were harvested in an isotonic HEPES buffer (pH 6.8) containing 2%CHAPS and protease inhibitor cocktail. Post-nuclear supernatants, obtained by centrifuging the sample at 10 000 g for 10 min, were submitted to immunoprecipitation overnight with anti-calnexin antibody followed by incubation with protein G agarose beads for 2 h at 4°C.
To monitor the synthesis and maturation of GFP–PrP protein, HeLa cells transfected with GFP–PrP under different experimental conditions were labelled with 100 μCi/ml 35S-methionine/cysteine for 30 min. The cells were then lysed in lysis buffer composed of 0.1 M Tris–HCl pH 8 and 1% SDS. The cell lysate was heated at 95°C for 10 min with occasional stirring until the lysate is no more viscous. The cleared lysate was diluted in the RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris–HCl (pH 8), 1 mM EDTA, and 1 × protease inhibitor cocktail). Immunoprecipitation was performed using an anti-GFP antibody.
For all the experiments, samples were analysed by 4–12% gradient SDS–PAGE, followed by fixing and drying of the gels. The radiolabelled products were revealed using Typhoon phosphoimager and the quantified using the Typhoon Imager (Image QuantTool, GE Healthcare).
Glycoprotein analysis
Fractionation of newly synthesized glycoproteins was performed in 35 mm dishes. Cells were metabolically labelled with 50 μCi/ml of 35S-Methionine/cysteine mix for 20 min, followed by a wash with ice-cold PBS. Cytosolic proteins were extracted by treating the cells with 150 μg/ml of digitonin in KHM buffer (110 mM KAc, 20 mM Hepes, pH 7.2, 2 mM MgAc2) for 5 min. After the recovery of cytosolic extract, the cells were washed once again in the KHM buffer and resuspended in 500 μl of the IP buffer to extract the non-cytosolic proteins. The glycoproteins were separated from the non-cytosolic protein fraction by incubation with Con-A beads (GE Healthcare) for 1 h. Equal amounts of total protein were loaded on the Con-A beads from all the samples Con-A beads selectively bind to the glycoproteins. The beads were washed for three times with the IP buffer and the glycoproteins were eluted by 300 μl of IP buffer containing 0.25 M α-methyl-D-mannopyranoside. Both the cytosolic and glycoprotein fractions were migrated on 4–20% SDS–PAGE gradient gels. The gels were fixed, dried and further analysed for autoradiography.
Calnexin complementation and SEAP assay
For the complementation assay, HeLa cells were transfected with shRNA against calnexin and the transfected cells were selected by treating with puromycin (3 μg/ml) for 24 h. At 72 h post-transfection, the cells were split into six-well plates and the day after were transfected with the control plasmid, the WT or the mutant calnexin cDNAs and when required with the pSEAP2 reporter plasmid.
SEAP assay was performed using Great EscAPe SEAP Chemiluminescence Kit 2.0 (Clonetech). The cell medium was changed 24 h before the SEAP assay was done in order to monitor the SEAP secreted for 24 h when the RNAi or the over expression is most effective. Assay was performed in a 96-well plate, using 15 μl of the growth medium according to the manufacturer's instructions. The Chemiluminescence signal was collected by Spectra Max multiwell plate reader and the data were analysed by Soft Max Pro 5 software.
DHHC6 recomplementation
For the complementation assay, HeLa cells were transfected with shRNA against DHHC6 followed by selection of transfected cells by treating with puromycin (3 μg/ml) for 24 h. The cells were further transfected on day 6 with the cDNA-expressing human DHHC6 without its 3′ UTR for complementation. The functional assays were performed on day 8 to analyse the restoration of phenotypes observed by the loss of DHHC6.
Immunofluorescence microscopy
Immunofluorescence in HeLa was done as described previously with the exception of the fixative used. To label the plasma membrane of the cells, the cells were treated with 5 μg/ml of FITC-labelled Transferrin (Molecular Probes) for 1 h at 4°C. This was followed by washes at 4°C and fixation. Cells were fixed and permeabilized with methanol at −20°C for 4 min. Cells were further labelled with the appropriate primary antibody, followed by labelling with Alexa-conjugated goat anti-rabbit or anti-mouse IgG (405, 488, 568 nm). The nuclei were stained by Hoechst dye. Images were acquired using a × 63 lens on LSM-710 Laser scanning microscope (Carl Zeiss Microimaging, Inc.). The Zen software was used for the processing of the images and the 3D stacks. The movie animations and the summation of the z-stacks were done using the Image J software. To calculate the Pearson correlation coefficient, four different regions on the nuclear membrane were analysed for 15 cells. Each condition was subjected to a total of 60 regions and the Pearson coefficient was calculated using Coloc 2 plugin in FIJI software.
MD simulations
We used MD simulations to characterize the calnexin TM domain at the atomistic level in the membrane environment. As a first step, the ideal helical TM model was inserted and equilibrated in a 60 × 60 Å2 palmitoyl oleoly phosphatidyl choline membrane patch (Humphrey et al, 1996) to characterize its structure and dynamics in a phospholipid bilayer. A Palmitoyl group was then covalently linked to C502 and C503 sulphur atoms to form the single and double palmitoylated calnexin models.
All simulations were performed using the NAMD (Phillips et al, 2005) engine, in combination with the CHARMM27 force field brooks, including CMAP corrections. TIP3P water (Jorgensen et al, 1983) parameterization was used to describe the water molecules. The spatial overlapping of lipid molecules and protein were removed and the resulting protein-membrane system was solvated in variable-size water box, at a salt concentration of 150 mM NaCl. The periodic electrostatic interactions were computed using the particle-mesh Ewald summation with a grid spacing smaller than 1 Å. All systems were first minimized by 2000 conjugate gradient steps, and subsequently gradually heated from 0 to 300 K in 800 ps with a constraint on the protein backbone scaffold. Finally, the systems were equilibrated for 10 ns at 300 K. Free MD of all equilibrated system were run for not less than 50 ns with a 2-fs integration time step using the RATTLE algorithm applied to all bonds, and trajectories collected for analysis. Constant temperature (300 K) was imposed by using Langevin dynamics (Brunger and Brooks, 1984), with damping coefficient of 1.0 ps. Constant pressure of 1 a.t.m. was maintained with a Langevin piston dynamics (Feller et al, 1995) 200 fs decay period and 50 fs time constant.
Statistical analysis
All experiments were performed at least three times, independently. Two-tailed t-tests were performed to evaluate the significance of the data.
Supplementary Material
Acknowledgments
We thank L Symul for the graphics work in Figure 8, G Davidson for the myc–LRP6 plasmid, C Zurzolo for the PrP plasmid, K Strub for the PPL-3f plasmid and anti-L12 antibodies, R Hedge for anti-TRAPα antibodies. This work was supported by the Swiss National Science Foundation and the EuroMembrane in the context of the EUROCORES program from the European Science Foundation.
Author contributions: AKKL and LA conceived and performed the experiments, analysed the data and wrote the manuscript. TL and MDP performed the molecular dynamics experiments and critically read the manuscript. SB and BK performed the experiments. AK provided the DHHC-expressing plasmids. FGVDG conceived the experiments, analysed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abrami L, Bischofberger M, Kunz B, Groux R, van der Goot FG (2010) Endocytosis of the anthrax toxin is mediated by clathrin, actin and unconventional adaptors. PLoS Pathog 6: e1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrami L, Kunz B, Iacovache I, van der Goot FG (2008) Palmitoylation and ubiquitination regulate exit of the Wnt signaling protein LRP6 from the endoplasmic reticulum. Proc Natl Acad Sci USA 105: 5384–5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrami L, Leppla SH, van der Goot FG (2006) Receptor palmitoylation and ubiquitination regulate anthrax toxin endocytosis. J Cell Biol 172: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M, Bernasconi R, Clerc S, Molinari M (2010) N-glycan structures: recognition and processing in the ER. Trends Biochem Sci 35: 74–82 [DOI] [PubMed] [Google Scholar]
- Alvarez E, Girones N, Davis RJ (1990) A point mutation in the cytoplasmic domain of the transferrin receptor inhibits endocytosis. Biochem J 267: 31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ (2011) Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem 80: 71–99 [DOI] [PubMed] [Google Scholar]
- Brunger A, Brooks CL (1984) Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem Phys Lett 105: 495–500 [Google Scholar]
- Chang DK, Cheng SF, Trivedi VD, Lin KL (1999) Proline affects oligomerization of a coiled coil by inducing a kink in a long helix. J Struct Biol 128: 270–279 [DOI] [PubMed] [Google Scholar]
- Charollais J, Van Der Goot FG (2009) Palmitoylation of membrane proteins (review). Mol Membr Biol 26: 55–66 [DOI] [PubMed] [Google Scholar]
- Chavan M, Lennarz W (2006) The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem Sci 31: 17–20 [DOI] [PubMed] [Google Scholar]
- Chen W, Helenius J, Braakman I, Helenius A (1995) Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci USA 92: 6229–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevet E, Smirle J, Cameron PH, Thomas DY, Bergeron JJ (2010) Calnexin phosphorylation: linking cytoplasmic signalling to endoplasmic reticulum lumenal functions. Semin Cell Dev Biol 21: 486–490 [DOI] [PubMed] [Google Scholar]
- Chevet E, Wong HN, Gerber D, Cochet C, Fazel A, Cameron PH, Gushue JN, Thomas DY, Bergeron JJ (1999) Phosphorylation by CK2 and MAPK enhances calnexin association with ribosomes. EMBO J 18: 3655–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L (2010) An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J 29: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, Rusch M, Kramer JW, Rauh D, Coates GW, Brunsveld L, Bastiaens PI, Waldmann H (2010) Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol 6: 449–456 [DOI] [PubMed] [Google Scholar]
- Deprez P, Gautschi M, Helenius A (2005) More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell 19: 183–195 [DOI] [PubMed] [Google Scholar]
- Du Y, Ferro-Novick S, Novick P (2004) Dynamics and inheritance of the endoplasmic reticulum. J Cell Sci 117: 2871–2878 [DOI] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103: 4613 [Google Scholar]
- Fivaz M, Vilbois F, Pasquali C, van der Goot FG (2000) Analysis of GPI-anchored proteins by two-dimensional gel electrophoresis. Electrophoresis 21: 3351–3356 [DOI] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS (2003) Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol 160: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorleku OA, Barns AM, Prescott GR, Greaves J, Chamberlain LH (2011) Endoplasmic reticulum localization of DHHC palmitoyl transferases mediated by lysine-based sorting signals. J Biol Chem 286: 39573–39584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Chamberlain LH (2011) DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci 36: 245–253 [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A (1994) Folding of VSV G protein: sequential interaction with BiP and calnexin. Science 266: 456–458 [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926 [Google Scholar]
- Kordyukova LV, Serebryakova MV, Baratova LA, Veit M (2010) Site-specific attachment of palmitate or stearate to cytoplasmic versus transmembrane cysteines is a common feature of viral spike proteins. Virology 398: 49–56 [DOI] [PubMed] [Google Scholar]
- Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K (2008) SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133: 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I, Lingwood D, Grzybek M, Coskun U, Simons K (2010) Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci USA 107: 22050–22054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chavan M, Schindelin H, Lennarz WJ (2008) Structure of the oligosaccharyl transferase complex at 12 A resolution. Structure 16: 432–440 [DOI] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ (2007) Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol 8: 74–84 [DOI] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J (2011) The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem 286: 26743–26753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynes EM, Bui M, Yap MC, Benson MD, Schneider B, Ellgaard L, Berthiaume LG, Simmen T (2011) Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J 31: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR, Cravatt BF (2009) Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods 6: 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnes LW, Morrison TG (1994) The role of the individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology 200: 470–483 [DOI] [PubMed] [Google Scholar]
- Menetret JF, Hegde RS, Aguiar M, Gygi SP, Park E, Rapoport TA, Akey CW (2008) Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure 16: 1126–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick BA, Dhungana S, Williams JG, Aloor JJ, Peddada S, Tomer KB, Fessler MB (2011) Proteomic profiling of S-acylated macrophage proteins identifies a role for palmitoylation in mitochondrial targeting of phospholipid scramblase 3. Mol Cell Proteomics 10: M110.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhill N, Lynes EM, Nanji JA, Blagoveshchenskaya AD, Fei H, Carmine Simmen K, Cooper TJ, Thomas G, Simmen T (2008) The subcellular distribution of calnexin is mediated by PACS-2. Mol Biol Cell 19: 2777–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaya H, Tamura T, Higa-Nishiyama A, Ohashi K, Takeuchi M, Hashimoto H, Hatsuzawa K, Kinjo M, Okada T, Wada I (2008) Regulated motion of glycoproteins revealed by direct visualization of a single cargo in the endoplasmic reticulum. J Cell Biol 180: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y, Kihara A, Sano T, Igarashi Y (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta 1761: 474–483 [DOI] [PubMed] [Google Scholar]
- Pendin D, McNew JA, Daga A (2011) Balancing ER dynamics: shaping, bending, severing, and mending membranes. Curr Opin Cell Biol 4: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E (2007) Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol 179: 895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, Waldmann H, Bastiaens PI (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141: 458–471 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA (2010) Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK (2008) The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283: 18892–18904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR (2007) The expanding role of the ER translocon in membrane protein folding. J Cell Biol 179: 1333–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund H, Bernsel A, Skwark M, Elofsson A (2008) SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology. Bioinformatics 24: 2928–2929 [DOI] [PubMed] [Google Scholar]
- Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics 9: 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, Lopez CB, Hang HC (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol 6: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman DM, Hicks SW, Charron G, Hang HC, Machamer CE (2011) Differential regulation of two palmitoylation sites in the cytoplasmic tail of the {beta}1-adrenergic receptor. J Biol Chem 286: 19014–19023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.