Abstract
E2F transcription factors are implicated in diverse cellular functions. The founding member, E2F-1, is endowed with contradictory activities, being able to promote cell-cycle progression and induce apoptosis. However, the mechanisms that underlie the opposing outcomes of E2F-1 activation remain largely unknown. We show here that E2F-1 is directly methylated by PRMT5 (protein arginine methyltransferase 5), and that arginine methylation is responsible for regulating its biochemical and functional properties, which impacts on E2F-1-dependent growth control. Thus, depleting PRMT5 causes increased E2F-1 protein levels, which coincides with decreased growth rate and associated apoptosis. Arginine methylation influences E2F-1 protein stability, and the enhanced transcription of a variety of downstream target genes reflects increased E2F-1 DNA-binding activity. Importantly, E2F-1 is methylated in tumour cells, and a reduced level of methylation is evident under DNA damage conditions that allow E2F-1 stabilization and give rise to apoptosis. Significantly, in a subgroup of colorectal cancer, high levels of PRMT5 frequently coincide with low levels of E2F-1 and reflect a poor clinical outcome. Our results establish that arginine methylation regulates the biological activity of E2F-1 activity, and raise the possibility that arginine methylation contributes to tumourigenesis by influencing the E2F pathway.
Keywords: arginine methylation, cancer, E2F-1, growth control
Introduction
E2F is a complex family of transcription factors implicated in different cell fates, including proliferation and apoptosis (Stevens and La Thangue, 2003; Frolov and Dyson, 2004; Polager and Ginsberg, 2008; van den Heuvel and Dyson, 2008). The first family member identified, E2F-1, physically interacts with the retinoblastoma tumour suppressor protein pRb, which negatively regulates E2F-1 activity (Bandara and La Thangue, 1991; Zamanian and La Thangue, 1992; Weinberg, 1995; Stevens and La Thangue, 2003; Frolov and Dyson, 2004). While it has been established that E2F-1 promotes proliferation and therefore is potentially oncogenic, it has also become clear that E2F-1 is endowed with apoptotic activity (Polager and Ginsberg, 2008; van den Heuvel and Dyson, 2008). Thus, many E2F target genes are connected with apoptosis (Ren et al, 2002), and in Rb−/− mice the enhanced level of tissue-specific apoptosis reflects deregulated E2F-1 activity (Tsai et al, 1998; Iaquinta and Lees, 2007). Further, E2F-1−/− mice suffer from an increased incidence of tumours, which is consistent with a role for E2F-1 as a tumour suppressor, possibility reflecting its role in apoptosis (Field et al, 1996; Yamasaki et al, 1996). In contrast, the frequency of tumours seen in Rb+/− mice is reduced upon inactivating E2F-1, arguing that E2F-1 also contributes to tumour progression (Yamasaki et al, 1998). The molecular mechanisms that dictate the opposing outcomes of E2F-1 activity, and basis of the context dependency of these events, remain largely unknown.
Not only is E2F-1 regulated during cell-cycle progression through its cyclical interactions with pRb and phosphorylation by Cdk kinases (Stevens and La Thangue, 2003; van den Heuvel and Dyson, 2008), but also under conditions of DNA damage (Pediconi et al, 2003; Stevens and La Thangue, 2003; Stevens et al, 2003). In DNA-damaged cells, E2F-1 is induced in a fashion that follows similar kinetics to p53 (Blattner et al, 1999; Hofferer et al, 1999; Pediconi et al, 2003; Stevens and La Thangue, 2003). DNA damage activates phosphokinases, such as ATM/ATR and Chk1/Chk2, which in turn phosphorylate effector proteins that mediate the outcome of the DNA damage response (Jackson and Bartek, 2009). Both families of DNA damage responsive kinases phosphorylate E2F-1 and augment apoptosis (Stevens and La Thangue, 2003; Stevens et al, 2003). Because tumour cells remain sensitive to the apoptotic effects of E2F-1 (Rodicker et al, 2001; Polager and Ginsberg, 2008), it is possible that mechanisms exist that counter balance and suppress any apoptosis that could result from the inadvertent E2F-1 activity.
Arginine methylation is becoming increasingly recognized as an important type of modification in protein control (Bedford and Richard, 2005; Bedford and Clarke, 2009; Kowenz-Leutz et al, 2010), and a variety of processes are know to be influenced by arginine methylation, including RNA processing, chromatin regulation and transcriptional control (Meister et al, 2001; Pal et al, 2004; Kowenz-Leutz et al, 2010). The methylation of arginine residues is catalysed by two groups of protein arginine methyltransferases (PRMT), the type I enzymes that catalyse formation of asymmetric modifications, and type II enzymes that catalyse symmetric modifications (Bedford and Clarke, 2009). In previous studies, p53 was shown to undergo arginine methylation, and a role established for PRMT5 in the methylation process; PRMT5 directed methylation of p53 occurred in cells upon DNA damage, and coincided with activation of the p53 response (Jansson et al, 2008). Further, PRMT5 activity is enhanced by cyclin D/Cdk4 kinase, which thereby contributes to oncogenesis (Aggarwal et al, 2010).
In exploring the wider role of arginine methylation in proliferation control, we reasoned that there might be an influence on other key growth regulating pathways, and considered the E2F pathway as one such possibility. Here, we demonstrate that E2F-1 is a direct target for arginine methylation mediated by PRMT5, which occurs on a central motif in the protein, which shares similarity to p53. Arginine methylation regulates E2F-1 DNA-binding and transcriptional activity. Depleting PRMT5 causes an E2F-1-dependent decrease in cell growth, and renders cells sensitive to E2F-1-dependent apoptosis, which coincides with increased levels of E2F-1 protein and expression of downstream E2F target genes. Significantly, E2F-1 is methylated in tumour cells, and demethylated under DNA damage conditions, which cause E2F-1 stabilization and associated apoptosis. Human biopsies taken from malignant disease frequently exhibit high expression of PRMT5, and in colorectal cancer (CRC) a subgroup of tumours exist where high levels of PRMT5 and low levels of E2F-1 correlate with poor prognosis. Our results establish a new mechanism that influences the biological properties and physiological outcome of E2F-1 activity, and highlight the interplay between arginine methylation and E2F-1 that might promote tumourigenesis.
Results
E2F-1 is methylated by PRMT5
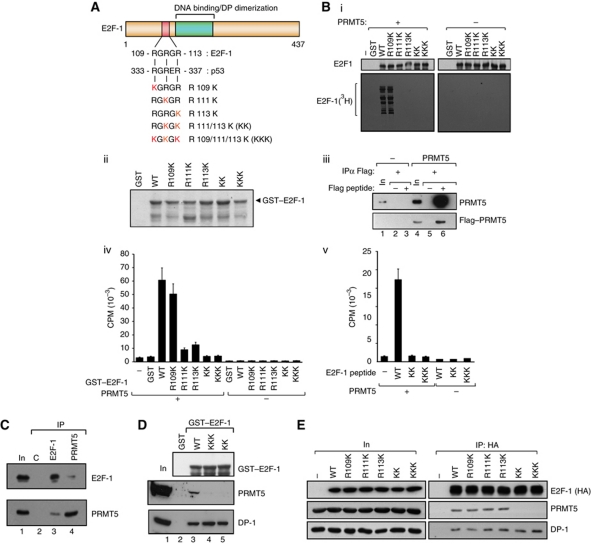
A short sequence motif exists in E2F-1, RGRGR, which is similar to the site of arginine methylation in p53, namely RGRER (Jansson et al, 2008; Figure 1A), and wild-type (WT) E2F-1 could be methylated in vitro by the arginine methyltransferase PRMT5 (Figure 1B). To assess whether the RGRGR motif was a direct target for PRMT5 methylation, a series of mutations were made in which each arginine (R) residue was substituted with a lysine (K) residue (Figure 1A), and the ability of each mutant derivative to be methylated by PRMT5 in vitro then measured. Both R111K and R113K exhibited a greater level of reduction compared with the minimal effect of mutating R109 (Figure 1B). The double R111/113K (referred to as KK) and triple R109/111/113K (KKK) mutants could not be methylated (Figure 1B), and when a short E2F-1 peptide containing the RGRGR sequence was methylated, the same residues (R111 and R113) were required (Figure 1B). Thus, R111 and R113 are the predominant sites of methylation by PRMT5 in vitro.
Figure 1.
E2F-1 undergoes arginine methylation by PRMT5. (A) Location of the RGRGR sequence motif in E2F-1, highlighting its similarity with the region in p53 targeted by PRMT5 (Jansson et al, 2008). The arginine (R) residues mutated to lysines (K) in the E2F-1 mutants are indicated (in red), together with the designation of the different mutant derivatives. (B) Each of the indicated GST–E2F-1 proteins (about 1 μg; ii) was used in the methylation reaction (i). In vitro methylated samples were analysed by SDS–PAGE as described. Quantitation of either GST–E2F-1 (iv) and associated mutant derivatives (A) or a 20-residue peptide (from residue 100 to 120) together with the equivalent KK and KKK peptides (v) after in vitro methylation by PRMT5. Incorporation of 3H-methyl groups was measured as DPM. Coomassie stain of the GST–E2F-1 proteins is shown in (ii), and the Flag–PRMT5 immunoprecipitated from transfected cells in (iii), which was used in (i, iv, v). (C) Endogenous E2F-1 or PRMT5 was immunoprecipitated with either anti-E2F-1 or anti-PRMT5 from untransfected U2OS cells and subsequently immunoblotted with anti-PRMT5 or anti-E2F-1 as indicated. The input (In) and control (C) immunoprecipitations (IPs) are indicated. (D) Binding of E2F-1 to PRMT5. The indicated GST–E2F-1 proteins were incubated with U2OS cell lysate, and bound proteins immunoblotted using anti-DP-1 or PRMT5 antibodies. The level of GST–E2F-1 input protein (top anti-GST immunoblot) is indicated. (E) Either WT E2F-1 or the indicated mutant derivative expression vectors were transfected into U2OS cells and HA11 antibody was used for IP followed by immunoblotting with HA11 (E2F-1), PRMT5 or DP-1 as indicated. The level of ectopic E2F-1 input (In) protein is shown, and (−) indicates empty vector control transfected cells. Figure source data can be found with the Supplementary data
We assessed whether E2F-1 could interact with PRMT5 and thereafter the role of the arginine residues in mediating the interaction. E2F-1 and PRMT5 exist in a complex in cells (e.g., U2OS cells; Figure 1C), and GST–E2F-1 bound to PRMT5 in cell extracts (Figure 1D). When expressed ectopically in cells, each of R109K, R111K and R113K could interact with PRMT5, although the KK and KKK mutants failed to bind to PRMT5 (Figure 1E). Binding to DP-1, the major heterodimeric partner for E2F-1 (Girling et al, 1993; Stevens and La Thangue, 2003), was not affected since it bound equally well to each mutant E2F-1 protein (Figure 1E). These results show that E2F-1 and PRMT5 interact in cells, and that R111 and R113 are the principal sites modified by PRMT5 and required for E2F-1 to interact with PRMT5.
E2F-1 is methylated in cells
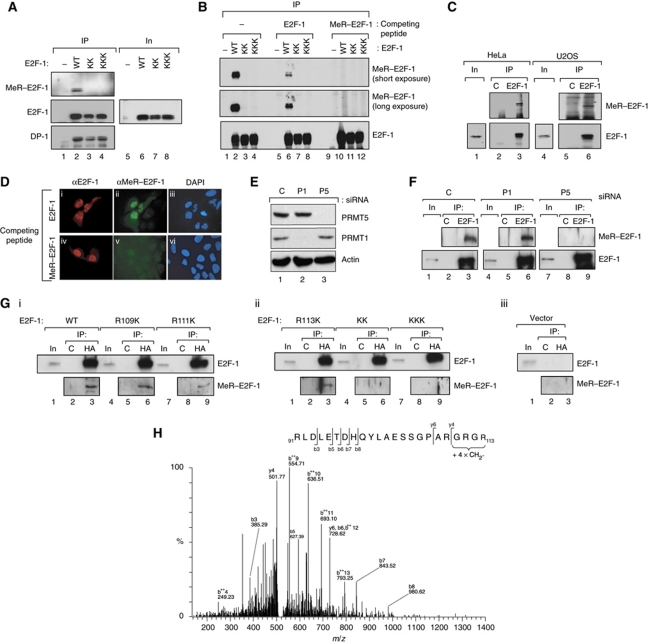
We investigated whether E2F-1 is methylated in cells using a modification-specific peptide antibody that we prepared against a methylated RGRGR peptide derived from E2F-1, in which R111 and R113 were symmetrically methylated. The anti-MeR–E2F-1 antibody recognized ectopic WT E2F-1 immunoprecipitated from cells, but failed to react with either the KK or KKK mutants (Figure 2A), indicating that the antibody recognized the relevant methylated region in E2F-1. Each ectopic protein was immunoprecipitated at an equivalent level and bound equally well to the DP-1 subunit control (Figure 2A).
Figure 2.
E2F-1 is methylated on arginine residues under physiological conditions. (A) Anti-MeR–E2F-1 recognizes WT E2F-1 but not the KK or KKK mutants. Either WT E2F-1, KK or KKK expression vectors were transfected into HeLa cells and immunoprecipitated (IP) with HA11, followed by immunoblotting (IB) with anti-MeR–E2F-1, HA11 (E2F-1) or DP-1 antibodies. The level of input (In) protein is indicated. (B) Anti-MeR–E2F-1 recognizes arginine methylated E2F-1. Either WT E2F-1, KK or KKK expression vectors were transfected to HeLa cells and IP with HA11, followed by immunoblotting with anti-MeR–E2F-1 or HA11 (E2F-1), either in the presence of methylated (MeR–E2F-1) or unmethylated E2F-1 peptide (1 μg), as indicated. Two different (short and long) exposures of the HA11 (E2F-1) blot are shown. (C) Endogenous E2F-1 is methylated in cells. HeLa or U2OS cell lysates were harvested and IP with anti-E2F-1 (KH95) or control (C) antibody, followed by immunoblotting with anti-MeR–E2F-1 or anti-E2F-1 antibodies. The level of input (In) E2F-1 protein is shown. (D) U2OS cells transfected with E2F-1 expression vector were immunostained with anti-E2F-1 or anti-MeR–E2F-1 in the presence or absence of competing methylated E2F-1 peptide as indicated. DAPI shows the location of nuclei. (E) U2OS cells were transfected with PRMT5 (P5), PRMT1 (P1) or control (C) non-targeting siRNA (50 nM) and harvested at 72 h. Extracts were immunoblotted as indicated. Actin served as the loading control. (F) The extracts described in (E) were IP with anti-E2F-1 (KH95) or control (C) antibody, followed by immunoblotting with anti-MeR–E2F-1 or anti-E2F-1 antibodies as indicated. The level of input (In) E2F-1 protein is shown. (G) U2OS cells were transfected with the indicated E2F-1 mutant derivatives (i, ii) and immunoprecipitated with anti-HA or control (C) antibody, followed by immunoblotting with anti-MeR–E2F-1 or anti-E2F-1 antibodies as indicated. The level of input (In) E2F-1 protein is shown; (iii) shows the vector transfected control. (H) E2F-1 expressed in MCF7 cells was immunopurified as described and subjected to analysis by tandem mass spectrometry. Analysis by LC-MS/MS revealed the presence of four methyl groups present in the peptide fragment GRGR (110–113) in E2F-1 (Sprot Acc Nr: Q01094/ IPI00005630). The MS/MS spectrum of the modified tryptic peptide RLDLETDHQYLAESSGPARGRGR + 4 x CH2-[M+5H]5+ 528.8783 Da (MW 2639.3551 Da) is shown. Fragment ions are indicated as b and y ions. ++ represents loss doubly charged ions. Figure source data can be found with the Supplementary data
The specificity of the antibody was established by immunoprecipitating ectopic E2F-1, followed by immunoblotting with anti-MeR–E2F-1 in the presence of competing E2F-1 peptides that differed only in the methylation status of R111 and R113. Anti-MeR–E2F-1 recognized the WT protein, and its binding activity was competed by the methylated but not the unmodified E2F-1 peptide (Figure 2B, compare tracks 6 and 10). As expected, neither the KK nor KKK mutants were recognized by the anti-MeR–E2F-1 antibody (Figure 2B). These results establish that the anti-MeR–E2F-1 peptide antibody detects methylated E2F-1 and, further, that E2F-1 expressed ectopically in cells undergoes arginine methylation.
We used the anti-MeR–E2F-1 antibody to study the methylation of endogenous E2F-1 by immunoprecipitation of E2F-1 followed by immunoblotting with anti-MeR–E2F-1. E2F-1 was methylated in different cancer cell lines, including HeLa and U2OS cells (Figure 2C), and further localized to nuclei (Figure 2D). These results indicate that endogenous E2F-1 undergoes arginine methylation at the RGRGR motif.
It was important to establish that PRMT5 is responsible for methylating E2F-1 in cells. To pursue this question, we depleted PRMT5 with siRNA (Figure 2E) and thereafter assessed the level of E2F-1 methylation. Methylated E2F-1 was readily detected when E2F-1 was immunoprecipitated from cells, which was reduced when a parallel immunoprecipitation was performed from PRMT5 depleted cells (Figure 2F). The level of E2F-1 methylation was not affected upon depleting another member of the PRMT family, the asymmetric arginine methyltransferase PRMT1 (Figure 2E), as methylation remained at a similar level to the control-treated cells (Figure 2F). Further, methylation of R111K and R113K, in addition to KK and KKK, was reduced compared with WT E2F-1 (Figure 2A and G), thus establishing the methylation of R111 and R113 in cells. Additional analysis by tandem mass spectrometry (Taylor and Goodlett, 2005; Batycka et al, 2006; Fischer et al, 2011) provided further support for di-methylation at R111 and R113 upon in vitro methylation of E2F-1 (Supplementary Figure S1G), and an analysis of E2F-1 immunopurified from MCF7 cells further substantiated methylation at R111 and R113 (Figure 2H). Altogether, these results indicate that E2F-1 undergoes symmetrical arginine methylation under physiological conditions and identify PRMT5 as the enzyme involved.
Arginine methylation regulates E2F-1 stability and target gene expression
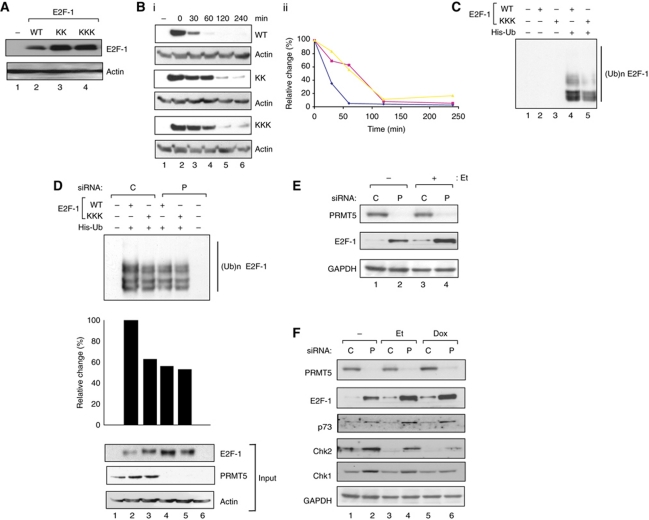
We considered that arginine methylation may play an important role in controlling E2F-1 in DNA-damaged cells, where E2F-1 levels increase (Stevens et al, 2003). When the level of ectopic protein was studied under conditions where each protein underwent equivalent nuclear accumulation (Supplementary Figure S1B), KK and KKK were expressed at increased levels compared with WT E2F-1 (Figure 3A). In part, the difference in protein level was caused by the shorter half-life of WT E2F-1 compared with the mutant derivatives (25–75 min respectively; Figure 3B), which corresponded to the increased steady-state level of KK and KKK compared with E2F-1 (Figure 3A). The R111K and R113K mutant also exhibited an extended half-life relative to WT E2F-1 (Supplementary Figure S2C). One type of modification that was affected by PRMT5, and which might account for different stabilities, was ubiquitination because the mutants exhibited a lower level of ubiquitination relative to WT E2F-1 (Figure 3C). In support of this idea, the level of ubiquitination that occurred on WT E2F-1 was reduced upon PRMT5 depletion, to a level similar to that seen with the KKK mutant (Figure 3D).
Figure 3.
Properties of arginine methylated E2F-1. (A) Levels of WT E2F-1, KK and KKK after transfection of the indicated expression vectors (1 μg) into U2OS cells and immunoblotting with HA11; – indicates untransfected cells. (B) Stability of WT E2F-1, and the KK and KKK mutants. Expression vectors (1 μg) encoding WT E2F-1, KK and KKK mutants were transfected into U2OS cells for 48 h. Cells were treated with 100 μg/ml of cyclohexamide and then harvested at 0, 2, 4, 6 h post-treatment time points as indicated for subsequent immunoblotting (i), and further quantitated (ii). The HA11 antibody was used for immunoblotting and actin served as protein loading control; n=2. Both the KKK (red) and KK (yellow) mutants had similarly increased half-life compared with WT (blue) E2F-1 (from 75 to 25 min, respectively). (C) Ubiquitination of WT E2F-1 and the KKK mutant. Cells were transiently transfected with expression vectors encoding WT E2F-1 or KKK (2 μg), together with His6-ubiquitin (4 μg) as indicated, and treated with MG132 (20 nM) for 4 h before harvesting. Cell lysates and Ni2+ pull-down eluates were analysed as described. (D) Effect of PRMT5 on E2F-1 ubiquitination: U2OS cells were transfected with PRMT5 (P) or control (C) siRNA. After 24 h, cells were transfected with expression vectors encoding WT E2F-1 or the KKK mutant (2 μg) and His6-ubiquitin (4 μg) as indicated. Cells were treated with MG132 (20 nM) for 4 h before being collected. Cell lysates and Ni2+ pull-down eluates were analysed as described. Graphical presentation of quantitation of ubiquitin signals was performed using ImageJ 1.43u, and the input protein levels are shown underneath. (E) PRMT5 siRNA increases E2F-1 protein levels. PRMT5 (P) or control (C) non-targeting siRNA was transfected into U2OS cells, and cells harvested 72 h post-transfection with or without etoposide (Et) treatment (10 μM) in the last 16 h. Extracts were immunoblotted with anti-PRMT5 and E2F-1, and GAPDH levels served as a loading control. (F) Protein levels of E2F-1 target genes in PRMT5 siRNA-treated cells. PRMT5 (P) or control (C) non-targeting siRNA was transfected into U2OS cells and cells were harvested 72 h post-transfection with or without etoposide (Et; 10 μM) or doxorubicin (Dox; 2 μM) treatment in the last 16 h. Extracts were immunoblotted with anti-PRMT5, E2F-1, p73, Chk2 and Chk1 as indicated. Levels of GAPDH served as the loading control.
To explore this possibility in greater detail and establish whether PRMT5 had an effect on endogenous E2F-1, we depleted PRMT5 in cells using siRNA and thereafter measured E2F-1 protein levels. Depleting PRMT5 under conditions that reduced arginine methylation of E2F-1 (Figure 2E and F) caused a coincident increase in E2F-1 protein (about three-fold; Figure 3E), a result that is compatible with the increased stability observed for each of the E2F-1 mutants (Figure 3B; Supplementary Figure S2C). Significantly, the increased E2F-1 levels that occurred upon PRMT5 depletion also coincided with an increase in the expression of a variety of E2F target genes, including p73, Chk2 and Chk1 (Figure 3F). Together, these results establish that PRMT5 regulates the level of E2F-1 protein through altered stability.
Functional consequences of arginine methylation
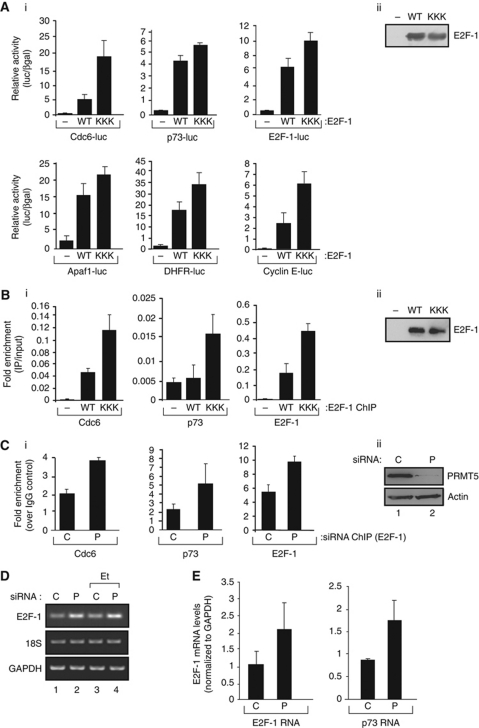
We reasoned that the transcriptional activity of E2F target genes might be affected by PRMT5, and tested this idea on a small group of genes in a reporter-based transfection assay. When the WT E2F-1 and KKK mutant were compared on E2F responsive promoters under conditions of equivalent protein expression (Figure 4Aii), the KKK mutant consistently exhibited increased transcriptional activity compared with WT E2F-1 (Figure 4Ai); the R111K, R113K and KK mutants behaved in a similar fashion (Supplementary Figure S2E). Furthermore, when endogenous PRMT5 was depleted, E2F responsive promoters were similarly more active compared with the control treatment (Supplementary Figure S1B). These results suggest, therefore, that arginine methylation also impacts on the ability of E2F-1 to activate transcription.
Figure 4.
Functional properties of arginine methylated E2F-1. (A) Transcription properties of WT E2F-1 and the KKK mutant. U2OS cells were transfected with expression vectors encoding WT E2F-1 or the KKK mutant, together with p73-luciferase, Cdc6-luciferase, E2F-1-luciferase, Apaf1-luciferase, DHFR-luciferase or cyclin E-luciferase for 48 h as indicated, and pCMV-βgal to monitor transfection efficiency. Relative luciferase activity (luciferase/βgal) is shown together with the expression level of the ectopic proteins underneath; n=3. (B) ChIP of U2OS cells transfected with expression vectors encoding HA-tagged WT E2F-1 or the KKK mutant as described, followed by immunoprecipitation with anti-HA on the E2F target genes, and quantitation by real-time PCR shown in (ii); n=4. (C) ChIP of U2OS cells transfected with PRMT5 (P) or control (C) siRNA followed by immunoprecipitation with anti-E2F-1 antibodies on the indicated E2F target genes, and quantitation by real-time PCR (i); the level of the input protein is shown below (ii); n=2. (D) Effect of PRMT5 siRNA on E2F-1 RNA: PRMT5 (P) or control (C) siRNA was transfected into U2OS cells in the presence or absence of etoposide (Et) and cells harvested at 72 h post-transfection. RNA levels for E2F-1, 18S and GAPDH were assayed as indicated. (E) Effect of PRMT5 (P) or control (C) siRNA on E2F-1 and p73 RNA at 72 h post-transfection and quantitation by real-time PCR.
Significantly, by chromatin immunoprecipitation (ChIP), increased binding of the methylation defective mutants compared with WT E2F-1 was evident on the promoter region of E2F target genes when each protein was expressed at an equivalent level (Figure 4B). Upon PRMT5 depletion, increased E2F-1 DNA binding was also apparent on a variety of target genes (Figure 4C). The level of histone H3 K4 acetylation, reflecting an active chromatin environment, also occurred upon PRMT5 depletion (Supplementary Figure S1C). Importantly, the enhanced binding of E2F-1 to endogenous promoters reflected, as expected, increased levels of E2F target gene RNA (Figure 4D and E). These results indicate that arginine methylation regulates E2F-1 DNA-binding activity, which represents another level of control that impacts on E2F-1 transcriptional activity. Overall, therefore, PRMT5 regulates several aspects of E2F-1 activity.
Arginine methylation regulates E2F-1-dependent cell growth and apoptosis
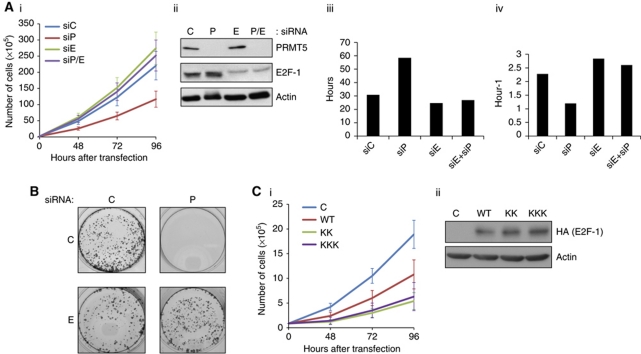
Because arginine methylation by PRMT5 affects on the biochemical and transcription properties of E2F-1, we considered that PRMT5 might influence cellular growth. Consequently, we evaluated the effect of PRMT5 and E2F-1 in growth control. When an analysis of cell growth was performed, there was a significant decrease in cell number over 96 h after PRMT5 depletion (Figure 5A). This effect of reduced growth was rescued upon co-depleting E2F-1 with PRMT5 (Figure 5A). The decreased cell number reflected an increase in cell doubling time and a decreased growth rate (Figure 5A). The inhibitory effect of PRMT5 was also evident when cell growth was measured in the context of a colony assay, where PRMT5 siRNA caused a dramatic reduction in growth after 10 days, which again was rescued upon co-depletion of E2F-1 (Figure 5B). Significantly, the methylation defective KK and KKK mutants were able to hinder cell growth more effectively than WT E2F-1 (Figure 5C). These results indicate that PRMT5 regulates the ability of E2F-1 to negatively impact on cell growth.
Figure 5.
PRMT5 and growth control. (A) Effect of PRMT5 and E2F-1 siRNA on cell growth. PRMT5 (P), E2F-1 (E) or control (C) siRNA (25 nM) was transfected as indicated into U2OS cells and cells counted (in triplicate) after 48, 72 and 96 h (i). The level of PRMT5 and E2F-1 in siRNA-treated cells is shown (ii) and the doubling time (iii) and growth rate (iv) was calculated from the results in (i). (B) U2OS cells were transfected with PRMT5 (P), E2F-1 (E) or control (C) or (25 nM) as indicated, and colony growth measured at 10 days after staining with crystal violet. (C) U2OS cells were transfected with expression vectors encoding WT E2F-1 (WT) or the KK and KKK mutant derivatives as indicated, and cells counted (in triplicate) at 48, 72 and 96 h (i). The level of ectopic E2F-1 protein is shown in (ii), which was adjusted for equivalent expression.
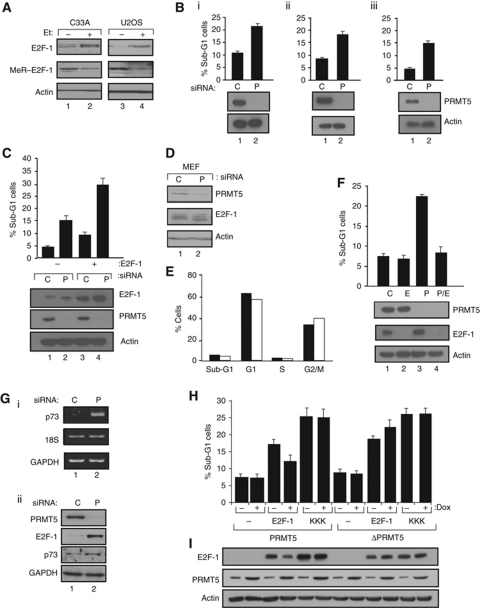
E2F-1 is stabilized and able to induce apoptosis in cells treated with DNA damaging agents (Stevens et al, 2003; Stevens and La Thangue, 2004; Polager and Ginsberg, 2008). We reasoned therefore that arginine methylation of E2F-1 could be regulated during DNA damage, and considered that such regulation might also be responsible for the increased level of E2F-1 and cessation of cell growth and associated apoptosis. In tumour cell lines (such as C33A and U2OS), the higher level of E2F-1 that occurred upon DNA damage (Stevens et al, 2003) coincided with reduced E2F-1 arginine methylation compared with untreated cells (Figure 6A). This is consistent with the increased stability of the KK and KKK mutants, the enhanced level of E2F-1 protein in PRMT5 depleted cells (Figure 3B and E) and the reduced level of PRMT5 that occurs under DNA damage conditions (Supplementary Figure S2). In addition to the decreased growth rate (Figure 5), an increase in apoptosis was evident when PRMT5 was depleted in different tumour cell lines (e.g., U2OS, HCT116 and SAOS2; Figure 6B and C), which occurred independently of p53 status since apoptosis was apparent upon PRMT5 depletion in HCT116 (p53+/+ or p53−/− background; Figure 6B) and SAOS2 (p53−/−) cells (Figure 6C). In contrast, the effect of PRMT5 on apoptosis was less evident in normal cells; thus, upon depleting PRMT5 in mouse embryonic fibroblasts (MEFs), there was an insignificant change in the overall level of apoptosis (Figure 6D and E).
Figure 6.
Arginine methylation and apoptosis. (A) E2F-1 arginine methylation is regulated upon DNA damage treatment. C33A or U2OS cells were treated with etoposide (Et+; 10 μM for 16 h) or left untreated (−), extracts prepared and subsequently immunoblotted with anti-E2F-1, anti-MeR–E2F-1 or actin (loading control) as indicated. (B) Apoptosis upon PRMT5 depletion. PRMT5 (P) or control (C) non-targeting siRNA (25 nM) was transfected into HCT116 p53+/+ (i), HCT116 p53−/− (ii) or U2OS (iii) cells and harvested 72 h post-transfection and analysed by FACS. The graph represents the actual percentage of sub-G1 cells in the indicated conditions in the total cell population, and the immunoblot shows the level of PRMT5 and actin control; n=3. (C) Apoptosis upon PRMT5 depletion: PRMT5 (P) or control (C) non-targeting siRNA was transfected into SAOS2 together with an expression vector encoding WT E2F-1 (1 μg) as indicated, harvested at 72 h and processed as described in (A); n=3. (D) Effect of PRMT5 siRNA in MEFs. PRMT5 (P) or control (C) non-targeting siRNA was transfected into MEFs and cells harvested at 72 h post-transfection as indicated. Cell extracts were immunoblotted with anti-PRMT5 and anti-E2F-1. Actin levels served as the loading control. (E) MEFs were treated as described in (C) and thereafter analysed by FACS. The black bar represents the effect of C siRNA, and clear bar the effect of P siRNA; n=3. (F) Apoptosis upon PRMT5 and E2F-1 depletion. PRMT5 (P), E2F-1 (E) or control (C) non-targeting siRNA was transfected into U2OS cells and harvested 72 h post-transfection and analysed by FACS as described. The graph shows the percentage of sub-G1 cells compared with the control treatment and the level of endogenous PRMT5 and E2F-1 is shown underneath; n=3. (G) Effect of PRMT5 siRNA on p73. PRMT5 (P) or control (C) siRNA was transfected into U2OS cells and cells harvested at 72 h post-transfection. Both RNA (i) and protein (ii) levels were measured, as indicated, for p73, 18S and GAPDH (i) and PRMT5, E2F-1, p73 and GAPDH (ii). (H) Regulation of apoptosis by PRMT5. Stable Tet-On cell lines expressing either WT PRMT5 or catalytically inactive ΔPRMT5 were treated with doxycycline (+; 1 μg/ml) as indicated and transfected with expression vectors encoding either WT E2F-1 or the KKK mutant derivative, or empty vector (2 μg). After 72 h, cells were harvested and analysed by FACS. The graph represents the actual percentage of sub-G1 cells in the indicated conditions (each treatment in triplicate); n=3. (I) The immunoblot shows the level of PRMT5 and E2F-1 for the experiment described above (H). Actin levels served as a loading control. Figure source data can be found with the Supplementary data
The enhanced level of E2F-1 upon PRMT5 depletion and increased apoptosis is consistent with the idea that E2F-1 activity is required for the apoptosis. We tested this by co-depleting PRMT5 and E2F-1, and monitoring any effect on apoptosis. The increased level of apoptosis upon PRMT5 depletion was substantially reduced when E2F-1 was co-depleted with PRMT5 (Figure 6F) and, conversely, apoptosis was enhanced when E2F-1 levels were increased by expressing ectopic E2F-1 in PRMT5 siRNA-treated cells (Figure 6C). Thereafter, we considered the role of p73 in apoptosis induction by PRMT5 siRNA, since p73 is an established E2F target gene involved in mediating E2F-1-dependent apoptosis (Irwin et al, 2000), and activated at the transcriptional level upon PRMT5 depletion (Figures 4E and 6G). Co-depleting PRMT5 with p73 caused a decrease in the level of apoptosis (Supplementary Figure S1F).
To gather further evidence that arginine methylation is important in regulating E2F-1-dependent apoptosis, we evaluated the effect of PRMT5 on apoptosis induced by E2F-1 and the methylation defective KKK mutant. Under conditions of apoptosis induced by WT E2F-1, ectopic PRMT5 suppressed apoptosis (Figure 6H and I). In contrast, the effect of PRMT5 on apoptosis induced by the KKK mutant was minimal (Figure 6I). As a control, we compared the effect of WT PRMT5 to ΔPRMT5, which lacks catalytic activity (Branscombe et al, 2001; Jansson et al, 2008). The apoptosis induced by WT E2F-1 was not affected by ΔPRMT5, contrasting with the effect of WT PRMT5 (Figure 6H); in fact, a modest increase in apoptosis occurred, most probably reflecting the ability of ΔPRMT5 to act in a dominant-negative fashion (Rho et al, 2001; Jansson et al, 2008). These results suggest that PRMT5, specifically its arginine methyltransferase activity, suppresses negative growth control and apoptosis driven by E2F-1.
PRMT5 and E2F-1 in tumour biopsies
Since E2F-1-dependent growth control and apoptosis is influenced by PRMT5, we reasoned that the mechanism may be relevant to tumourigenesis in situ, where a pathway that controls negative growth control could provide a survival advantage for tumour cells. To explore this possibility in greater detail, we studied the level of E2F-1 and PRMT5 in human tumour biopsies. In a preparatory study, we assessed PRMT5 and E2F-1 levels by immunohistochemistry (IHC) in biopsies taken from murine xenografts derived from human MDA-MB-231 tumour cells, where both PRMT5 and E2F-1 were readily detectable; there was a heterogeneous staining pattern, with intensely stained PRMT5 and E2F-1 cells being evident throughout the tumour biopsy (Supplementary Figure S1D). We then assessed PRMT5 expression in a diverse collection of human tumours, including follicular B-cell lymphoma, where interestingly the highest PRMT5 expression was restricted to the follicular areas of concentrated malignant cell growth (Supplementary Figure S2A).
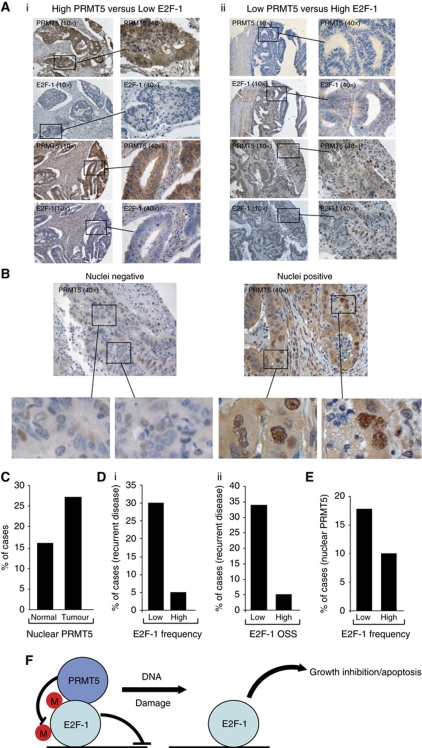
To evaluate the relationship between PRMT5 and E2F-1 during clinical disease, we studied PRMT5 and E2F-1 levels in a collection of biopsies taken from patients suffering from CRC, where clinical outcome and disease history had been thoroughly annotated for each biopsy (Midgley et al, 2010; Supplementary Table S1). We found that PRMT5 was present at high levels, and further its expression was mostly confined to malignant cells (Figure 7A). We observed however in a subgroup of tumours that high-level PRMT5 expression occurred coincidently with low levels of E2F-1 and, in another subgroup, low PRMT5 expression occurred coincidently with high levels of E2F-1 (Figure 7A). In a proportion of the biopsies, PRMT5 was clearly present in the nuclei of tumour cells (Figure 7B), which was apparent in a greater proportion of tumour biopsies compared with normal tissue biopsies (Figure 7C). At a general level therefore, the observations made on tumour cell lines, where high PRMT5 expression coincides with low E2F-1 expression, reflect the expression profile seen in a subgroup of human CRC.
Figure 7.
PRMT5 and E2F-1 expression in clinical tumour biopsies. (A) Representative IHC images of PRMT5 and E2F-1 in CRC. Examples of tumours expressing high PRMT5/low E2F-1 (i), and low PRMT5/high E2F-1 (ii) are shown (× 10), together with fields of increased magnification (× 40). Two representative examples of different biopsies are shown for each subgroup. (B) Representative IHC images of biopsies from CRC exhibiting either negative or positive nuclear PRMT5 immunostaining (× 40), with enlarged images of nuclei shown underneath. (C) Graph representing the percentage of nuclear PRMT5 staining in normal colon versus tumour tissue; 165 CRC tumour and 63 normal biopsies were assessed. (D) Either E2F-1 staining frequency (i) or OSS (ii) was assessed in a total of 146 CRC biopsies, and related to the appearance of recurrent disease. (i) χ2 analysis (29.52 with 4 degrees of freedom) gave P<0.0001 (comparing low-frequency score (0) to high-frequency score (4) and (ii) χ2 analysis (43.94 with 4 degrees of freedom) gave P<0.0001 (comparing low OSS (0–2) with high OSS (6–7). (E) Comparison of E2F-1 staining frequency in CRC biopsies expressing nuclear PRMT5; a total of 165 CRC biopsies were assessed for nuclear PRMT5 and E2F-1 staining frequency. χ2 analysis (25.6 with 4 degrees of freedom) gave P<0.0001 (comparing low-frequency score (0) with high-frequency score (4)). (F) Model for the effect of PRMT5 on E2F-1 activity: in unperturbed cells PRMT5 methylates E2F-1, and retains cells in the proliferative cycle. In conditions where PRMT5 activity becomes limiting, for example, during DNA damage, E2F-1 levels increase through a mechanism that includes increased half-life, resulting in higher transcription and DNA-binding activity, thereby facilitating activation of diverse E2F target genes, reduced growth rate and apoptosis. Under DNA damage, regulated arginine methylation may work hand-in-hand with phosphorylation events on E2F-1, such as by Chk2. In tumour cells, high levels of PRMT5 were frequently found to coincide with low levels of E2F-1.
The in vitro cell line studies suggested that PRMT5 impacts on the ability of E2F-1 to regulate growth, by suppressing negative growth control and apoptosis (Figures 5 and 6), and that low levels of PRMT5 frequently relate to an E2F-1-dependent inhibitory effect on growth and apoptosis. It was of interest therefore to evaluate the level of E2F-1 and PRMT5 in the CRC biopsies and relate this information to clinical outcome. When a subgroup of tumour biopsies exhibiting either high or low E2F-1 levels where compared, there was a clear difference in the history of disease progression; high E2F-1 expressing tumours (either frequency of cells expressing E2F-1 or overall E2F-1 staining score; OSS) had a considerably better prognosis and disease history (measured as the appearance of recurrent disease) compared to biopsies with low E2F-1 (Figure 7Di and ii). Further, a greater proportion of tumours exhibiting low E2F-1 levels, namely with poor prognosis, exhibited increased levels of nuclear PRMT5 (Figure 7E). Given the evidence that PRMT5 favours growth by suppressing E2F-1 activity, these results suggest that the relationship between PRMT5 and E2F-1 observed in vitro is relevant to tumourigenesis in situ. Specifically, high-level PRMT5 expression might suppress growth inhibition by E2F-1 and thereby provide tumour cells with a survival advantage. In turn, this relationship could account for the poorer prognosis observed in tumours expressing high levels of PRMT5 and low E2F-1.
Discussion
Arginine methylation of E2F-1
One of the most important questions in the biology of E2F-1 relates to the mechanisms that give rise to the different physiological outcomes that have been ascribed to E2F-1 activity. While the role of E2F-1 in cell-cycle progression reflects its temporally regulated interaction with pRb (Stevens and La Thangue, 2003; Frolov and Dyson, 2004; van den Heuvel and Dyson, 2008), the mechanisms that are responsible for balancing its growth inhibitory activity, including apoptosis and cell-cycle progression, remain unclear. Phosphorylation of E2F-1 by kinases activated upon DNA damage, namely ATM/ATR and Chk1/Chk2, together with acetylation, play a positive role in E2F-1 activity under DNA damage conditions (Pediconi et al, 2003; Stevens and La Thangue, 2003; Stevens et al, 2003). However, whether mechanisms exist that act directly on E2F-1 in unperturbed cells, and allow cells to downregulate E2F-1 and continue to proliferate, has to date been unclear.
The results presented here strongly implicate arginine methylation with a key role in regulating E2F-1; thus, PRMT5 targets E2F-1 and influences a number of biochemical and functional properties, culminating in reduced activity. Further, and consistent with a role for arginine methylation in the control of E2F-1 activity, depleting PRMT5 caused a coincident increase in the expression of E2F target genes, reduced growth rate and heightened apoptosis. Interestingly, the E2F-1 gene was found to be a target for E2F-1 protein under conditions of reduced PRMT5 activity, which reflected increased E2F-1 RNA in treated cells. Such a positive auto-regulatory feedback mechanism might contribute to enhanced growth inhibition upon PRMT5 depletion by co-ordinating the increased expression of diverse E2F-1 target genes, such as p73 (Irwin et al, 2000), which was required for the apoptotic activity apparent upon PRMT5 depletion.
PRMT5 in malignant disease
Significantly, therefore, it is possible that high-level expression of PRMT5 facilitates oncogenesis by providing tumour cells with a survival advantage, in part by limiting growth inhibition by E2F-1 and thereby maintaining cells in a proliferative mode. In support of this idea, an analysis of PRMT5 expression in tumour biopsies showed that elevated levels of PRMT5 were frequently observed, and a subgroup of CRC tumours, which expressed high levels of nuclear PRMT5 with low E2F-1 expression had a poorer prognosis than the complementary subgroup that expressed high levels of E2F-1. At a general level, the clinical history and prognosis of disease expressing high E2F-1 was more favourable in comparison to disease expressing low E2F-1. Relating this information to the results derived from the in vitro studies would lead us to hypothesise therefore that tumours expressing high E2F-1 and low PRMT5 are more likely to exhibit decreased growth rate and increased apoptosis and, perhaps therefore be associated with a more favourable clinical outcome. Conversely, high PRMT5 levels might contribute to a mechanism that limits the growth inhibitory effect of E2F-1 activity, and thus the oncogenic process. Indeed, a number of reports have implicated E2F-1 with a role in negative growth control (Lee and Farnham, 2000; Wang et al, 2007) and a tumour cell sensitizer to chemotherapy, perhaps providing a biomarker for improved prognosis, which could reflect its ability to induce apoptosis and limit growth in tumour cells (Stanelle and Putzer, 2006; Evangelou et al, 2008; Lee et al, 2008; Kwon et al, 2010). The pilot study described here, evaluating PRMT5 and E2F-1 in tumour biopsies and relating this information follow-up clinical history, supports such an idea and, further, is consistent with the hypothesis that PRMT5 plays a role in regulating E2F-1 in tumours in situ.
Arginine methylation and the DNA damage response
Arginine methylation targets the p53 protein where it impacts on the physiological outcome of the p53 response. That arginine methylation of p53 prompts cell-cycle arrest (Jansson et al, 2008) is complementary to the observations described in the present study relating to the regulation apoptosis by E2F-1, and highlights a new level of cross-talk between the p53 and E2F pathways. Our results suggest that upon E2F-1 methylation, E2F-1-dependent growth inhibition and apoptotic activity is held in check (through low E2F-1 protein levels and reduced expression of target genes), enabling arginine methylated p53 (Jansson et al, 2008) to predominate and perhaps drive the p53 response. In contrast, under conditions that limit arginine methylation (e.g., certain types of DNA damage or, experimentally, treatment with PRMT5 siRNA), E2F-1 levels increase, which we suggest give rise to growth inhibition and apoptosis (Figure 7F). It would appear therefore that arginine methylation allows a level of interplay to occur between these two pathways; by providing a means for cross-talk between E2F-1 and p53, arginine methylation controls a key decision point in cell-cycle control. We suggest therefore that arginine methylation provides a signal, and PRMT5 the mechanism, through which E2F-1 and p53 activity is integrated.
Materials and methods
Cell culture
U2OS, C33A, MCF7, HCT116, SAOS2, HeLa, H1299 cells and MEFs were maintained in DMEM with 5% FCS. Stable Tet-On inducible cell lines were prepared as previously described (Jansson et al, 2008).
Antibodies
The following antibodies were used: anti-Flag peptide monoclonal antibody M2 (Sigma), anti-Flag peptide monoclonal antibody M2 coupled agarose beads (Sigma) and anti-HA11 monoclonal antibody (Covance). PRMT5 and acetylated H3K4 antibodies were from Upstate, and PRMT1 antibody from Cell Signaling. GST, E2F-1, Chk1 and p53 antibodies were monoclonal antibodies from Santa Cruz. DP-1, p73, Chk2 and GAPDH were polyclonal antibodies from Santa Cruz. β-Actin monoclonal antibody was from Sigma. PARP monoclonal antibody was from BD Pharmingen.
Transfection
Gene Juice transfection reagent (Novagen, San Diego, CA) was used to transfect cells with DNA. All transfections with the indicated plasmids included a pCMV-βGal internal control for gauging transfection efficiency. Empty pcDNA3.1a vector was used to equalize amounts of transfected DNA where appropriate.
siRNA
Oligofectamine transfection reagent (Invitrogen) was used to transfect cells with siRNAs. siRNA treatment of cells was carried out using siRNA non-targeting (Dharmacon) as control, or PRMT5 (5′CCGCUAUUGCACCUUGGAA3′) or PRMT1 (5′GGACAUGACAUCCAAAGA) siRNA. p73 Smart Pool siRNA was from Dharmacon. siRNA treatment was for 72 h at 50 nM.
Flow cytometry
Cells were transfected with the indicated expression vectors together with pBB14–GFP (for 48 h) to monitor transfection efficiency or siRNA (for 48 or 72 h) with or without treatment of the DNA damage agent for the last 16 h. Then, cells were fixed and stained with propidium iodide and analysed by flow cytometry. Tet-On stable cell lines expressing PRMT5 or ΔPRMT5 were prepared as previously described (Jansson et al, 2008).
Proliferation rate determination
To determine cell proliferation rate, 2 × 104 cells were seeded per well in a six-well plate. The cells were harvested at the indicated time points and stained with equal volume of 0.4% trypan blue. Viable cells were counted using a haemocytometer. Dead cells, which were stained blue, were excluded from the count.
Reporter assays
Cells were transfected with expression vectors (1 μg) encoding WT or the mutant E2F-1 proteins as indicated with E2F responsive luciferase reporters together with pCMV-βgal to monitor transfection efficiency. Cells were harvested, lysed 48 h post-transfection and luciferase activity measured as previously described (Jansson et al, 2008). RNA was extracted from U2OS cells using TRIzol reagent (Invitrogen). A total of 1 μg RNA was reverse transcribed and quantified by real-time PCR. Gene expression values were normalized to GAPDH.
Chromatin immunoprecipitation
U2OS cells were maintained in DMEM containing 5% fetal calf serum. Cells were transfected with 50 nM PRMT5 siRNA or a non-targeting control (C) siRNA for 72 h, or transfected with expression vectors for WT E2F-1 or the KKK derivative (1 μg; HA-tagged). Cells were cross-linked with formaldehyde (final concentration 1%). Chromatin was prepared as described previously (Zalmas et al, 2008). Antibodies used for immunoprecipitation were as follows: anti-E2F-1 (C-20, Santa Cruz), anti-acetyl-histone H3 K4 (Millipore) or anti-HA (Santa Cruz). The non-specific rabbit IgG used as a negative control in the ChIP assays was from Jackson ImmunoResearch. The recovered DNA was analysed by real-time PCR as described (Jansson et al, 2008) on the MX3005P QPCR system (Stratagene) using Brilliant II SYBR Green QPCR Master Mix (Stratagene) according to the manufacturer's instructions. Primer sequences are available upon request.
Cyclohexamide half-life assay
Cells were transfected with WT or mutant E2F-1 plasmids for 48 h as described. Cells were treated with 100 μg/ml of cyclohexamide (Fluka) and then harvested at different time points as indicated for immunoblotting.
Preparation of whole cell extracts, immunoprecipitation and immunoblot analysis
Cells were either left untransfected or transfected with expression vectors as indicated. At 48 h post-transfection, cells were harvested in TNN buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 0.5% Ipegal, 50 mM NaF, 0.2 mM sodium orthovanadate and protease inhibitor cocktail (1 mM PMSF, leupeptin (1 μg/ml), aprotinin (1 μg/ml) and pepstatin A (1 μg/ml))) and rotated at 4°C for 30 min to 1 h. The cell lysate was then homogenized, centrifuged at 11 000 r.p.m. for 10 min to remove cell debris and the supernatant collected. All lysates were normalized for protein concentration and, where appropriate, for transfection by β-galactosidase activity. Immunoprecipitation was carried out by incubation of lysates with the indicated antibody and protein-A/G Sepharose beads or agarose-antibody conjugated at 4°C for 4 h or overnight. The beads were collected and washed three times with TNN buffer before denaturation and SDS–PAGE. Protein was transferred to nitrocellulose and probed with the indicated antibody. Enhanced chemi-luminescence (Pierce Biotechnology, Rockford, IL) was used to visualize antibody binding.
Ubiquitination assays
Cells were transfected with pcDNA3.1a-ubiquitin (His-tagged) and treated as described. Briefly, cells were harvested into 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris–HCl and pH 8.0. Cell lysate was incubated with nickel (Ni2+-NTA)-agarose beads (Qiagen), 5 mM imidazole and 10 mM β-mercapthoethanol overnight at 4°C. The beads were collected and washed as described (Liu and Warbrick, 2006). Bound proteins were then eluted and assayed by immunoblotting.
Supplementary Material
Acknowledgments
We thank the CRUK, MRC, LLR and EU for supporting this research and Rosemary Williams and Sarah Atkinson for help in preparing the manuscript. SZ was supported by the Agency for Science, Technology and Research of Singapore. We thank Dr Nicola Ternette and Dr Roman Fischer for their expert help with the mass spectrometry analysis. BK and RK are supported by the Biomedical Research Centre (NIHR), Oxford, UK.
Author contributions: E-CC, SZ, SM, GL, SC, JM, Y-CL, LS, OK and AS designed, performed and analysed the experiments. RK, JM and BK performed and interpreted the mass spectrometry. DJK and OK performed the analysis of clinical material. NBLT analysed the data, set the experimental strategy and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, Herlyn M, Hua X, Rustgi AK, McMahon SB, Diehl JA (2010) Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara LR, La Thangue NB (1991) Adenovirus E1a prevents the retinoblastoma gene product from complexing with a cellular transcription factor. Nature 351: 494–497 [DOI] [PubMed] [Google Scholar]
- Batycka M, Inglis NF, Cook K, Adam A, Fraser-Pitt D, Smith DG, Main L, Lubben A, Kessler BM (2006) Ultra-fast tandem mass spectrometry scanning combined with monolithic column liquid chromatography increases throughput in proteomic analysis. Rapid Commun Mass Spectrom 20: 2074–2080 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG (2009) Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Blattner C, Sparks A, Lane D (1999) Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol 19: 3704–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S (2001) PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem 276: 32971–32976 [DOI] [PubMed] [Google Scholar]
- Evangelou K, Kotsinas A, Mariolis-Sapsakos T, Giannopoulos A, Tsantoulis PK, Constantinides C, Troupis TG, Salmas M, Kyroudis A, Kittas C, Gorgoulis VG (2008) E2F-1 overexpression correlates with decreased proliferation and better prognosis in adenocarcinomas of Barrett oesophagus. J Clin Pathol 61: 601–605 [DOI] [PubMed] [Google Scholar]
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG Jr, Livingston DM, Orkin SH, Greenberg ME (1996) E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85: 549–561 [DOI] [PubMed] [Google Scholar]
- Fischer R, Trudgian D, Wright C, Thomas G, Bradbury L, Brown M, Bowness P, Kessler B (2011) Discovery of candidate serum proteomic and metabolomic biomarkers in Ankylosing Spondylitis. Mol Cell Proteomics, (advance online publication, 13 October 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ (2004) Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117: 2173–2181 [DOI] [PubMed] [Google Scholar]
- Girling R, Partridge JF, Bandara LR, Burden N, Totty NF, Hsuan JJ, La Thangue NB (1993) A new component of the transcription factor DRTF1/E2F. Nature 362: 83–87 [DOI] [PubMed] [Google Scholar]
- Hofferer M, Wirbelauer C, Humar B, Krek W (1999) Increased levels of E2F-1-dependent DNA binding activity after UV-or y-irradiation. Nucl Acids Res 27: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaquinta PJ, Lees JA (2007) Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol 19: 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG Jr (2000) Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407: 645–648 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB (2008) Arginine methylation regulates the p53 response. Nat Cell Biol 10: 1431–1439 [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A (2010) Crosstalk between C/EBPbeta phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J 29: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MJ, Nam ES, Cho SJ, Park HR, Shin HS, Park JH, Park CH, Lee WJ (2010) E2F1 expression predicts outcome in Korean women who undergo surgery for breast carcinoma. Ann Surg Oncol 17: 564–571 [DOI] [PubMed] [Google Scholar]
- Lee J, Park CK, Park JO, Lim T, Park YS, Lim HY, Lee I, Sohn TS, Noh JH, Heo JS, Kim S, Lim do H, Kim KM, Kang WK (2008) Impact of E2F-1 expression on clinical outcome of gastric adenocarcinoma patients with adjuvant chemoradiation therapy. Clin Cancer Res 14: 82–88 [DOI] [PubMed] [Google Scholar]
- Lee TA, Farnham PJ (2000) Exogenous E2F expression is growth inhibitory before, during, and after cellular transformation. Oncogene 19: 2257–2268 [DOI] [PubMed] [Google Scholar]
- Liu G, Warbrick E (2006) The p66 and p12 subunits of DNA polymerase delta are modified by ubiquitin and ubiquitin-like proteins. Biochem Biophys Res Commun 349: 360–366 [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U (2001) Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 11: 1990–1994 [DOI] [PubMed] [Google Scholar]
- Midgley RS, McConkey CC, Johnstone EC, Dunn JA, Smith JL, Grumett SA, Julier P, Iveson C, Yanagisawa Y, Warren B, Langman MJ, Kerr DJ (2010) Phase III randomized trial assessing rofecoxib in the adjuvant setting of colorectal cancer: final results of the VICTOR trial. J Clin Oncol 28: 4575–4580 [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24: 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, Gulino A, Levrero M (2003) Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol 5: 552–558 [DOI] [PubMed] [Google Scholar]
- Polager S, Ginsberg D (2008) E2F -- at the crossroads of life and death. Trends Cell Biol 18: 528–535 [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD (2002) E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS (2001) Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem 276: 11393–11401 [DOI] [PubMed] [Google Scholar]
- Rodicker F, Stiewe T, Zimmermann S, Putzer BM (2001) Therapeutic efficacy of E2F1 in pancreatic cancer correlates with TP73 induction. Cancer Res 61: 7052–7055 [PubMed] [Google Scholar]
- Stanelle J, Putzer BM (2006) E2F1-induced apoptosis: turning killers into therapeutics. Trends Mol Med 12: 177–185 [DOI] [PubMed] [Google Scholar]
- Stevens C, La Thangue NB (2003) E2F and cell cycle control: a double-edged sword. Arch Biochem Biophys 412: 157–169 [DOI] [PubMed] [Google Scholar]
- Stevens C, La Thangue NB (2004) The emerging role of E2F-1 in the DNA damage response and checkpoint control. DNA Repair (Amst) 3: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Stevens C, Smith L, La Thangue NB (2003) Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol 5: 401–409 [DOI] [PubMed] [Google Scholar]
- Taylor GK, Goodlett DR (2005) Rules governing protein identification by mass spectrometry. Rapid Commun Mass Spectrom 19: 3420. [DOI] [PubMed] [Google Scholar]
- Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T (1998) Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell 2: 293–304 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang R, Herrup K (2007) E2F-1 Works as a cell cycle suppressor in mature neurons. J Neuro Sci 27: 12555–12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T (1998) Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−)mice. Nat Genet 18: 360–364 [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ (1996) Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85: 537–548 [DOI] [PubMed] [Google Scholar]
- Zalmas LP, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, La Thangue NB (2008) DNA-damage response control of E2F7 and E2F8. EMBO Rep 9: 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M, La Thangue NB (1992) Adenovirus E1a prevents the retinoblastoma gene product from repressing the activity of a cellular transcription factor. EMBO J 11: 2603–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.