Abstract
Inhibitor of apoptosis (IAP) proteins cIAP1, cIAP2, and XIAP (X-linked IAP) regulate apoptosis and cytokine receptor signalling, but their overlapping functions make it difficult to distinguish their individual roles. To do so, we deleted the genes for IAPs separately and in combination. While lack of any one of the IAPs produced no overt phenotype in mice, deletion of cIap1 with cIap2 or Xiap resulted in mid-embryonic lethality. In contrast, Xiap−/−cIap2−/− mice were viable. The death of cIap2−/−cIap1−/− double mutants was rescued to birth by deletion of tumour necrosis factor (TNF) receptor 1, but not TNFR2 genes. Remarkably, hemizygosity for receptor-interacting protein kinase 1 (Ripk1) allowed Xiap−/−cIap1−/− double mutants to survive past birth, and prolonged cIap2−/−cIap1−/− embryonic survival. Similarly, deletion of Ripk3 was able to rescue the mid-gestation defect of cIap2−/−cIap1−/− embryos, as these embryos survived to E15.5. cIAPs are therefore required during development to limit activity of RIP kinases in the TNF receptor 1 signalling pathway.
Keywords: apoptosis, IAP, NF-κB, RIP kinase, TNF
Introduction
The cellular inhibitor of apoptosis (IAP) proteins cIAP1 (Birc2), cIAP2 (Birc3), and XIAP (X-linked IAP, Birc4) were initially identified by their similarity to baculoviral IAP genes, and, in the case of cIAP1 and cIAP2, also because they bound to tumour necrosis factor (TNF) receptor-associated proteins (TRAFs) 1 and 2 (Rothe et al, 1995; Duckett et al, 1996; Liston et al, 1996; Uren et al, 1996). Cellular IAPs and XIAP are very similar proteins and belong to the BIRC (baculoviral IAP repeat containing) protein family, as they all bear three BIR domains, a ubiquitin-associated domain and a carboxy-terminal RING domain, which functions as an E3 ligase (Birnbaum et al, 1994; Gyrd-Hansen et al, 2008). Cellular IAP1 and IAP2 also both bear a caspase activation and recruitment domain, and appear to have arisen from a recent gene duplication event. They are therefore likely to have similar functions. Only XIAP is a direct inhibitor of caspase activity, and can potently inhibit caspase 3, 7, and 9 (Tenev et al, 2005; Eckelman et al, 2006).
Cellular IAPs 1 and 2 regulate various signalling pathways that promote activation of canonical NF-κB, but reduce activation of non-canonical NF-κB transcription factors (Vince et al, 2007; Hayden and Ghosh, 2008; Gyrd-Hansen and Meier, 2010). For example, in cells that have not been exposed to cytokines cIAPs reduce levels of the NF-κB inducing kinase, NIK (Varfolomeev et al, 2007; Vince et al, 2007), but binding of the TNF superfamily member TWEAK to its receptor leads to degradation of TRAF2 and cIAPs, so that NIK is stabilized, and activates non-canonical NF-κB (Vince et al, 2008). On the other hand, ligation of TNFR1 by TNF triggers formation of a multiprotein complex containing TRADD (TNFR1-associated death domain protein), TRAF2, and RIPK1 (receptor-interacting protein kinase 1). IAP-mediated ubiquitylation within the TNF receptor signalling complex leads to activation of TGF-β-activated kinase 1 (TAK1) and NF-κB essential modulator (NEMO/IKKγ) complexes, which in turn cause phosphorylation and degradation of IκB, and activation of p65/RelA canonical NF-κB (Micheau and Tschopp, 2003; Takaesu et al, 2003; Bertrand et al, 2008). However, the overlapping functions of cIAP1 and cIAP2 make it difficult to distinguish the precise role of each.
Deletion of genes in mice can often reveal their roles in development and in disease, and these roles can be distinguished by analysis of compound mutants. However, because the mouse cIap2 and cIap1 genes are only ∼15 kb apart on chromosome 9A2 (Liston et al, 1997), they operate as a single locus, so it has not been possible to generate cIap2/cIap1 double-knockout (DKO) mice by crossing single-KO strains. In the light of the similar and partially overlapping roles and functions of cIAP1, cIAP2, and XIAP, we sequentially targeted the same chromosome in ES cells to make conditional alleles of both cIap1 and cIap2. We crossed these mice with Xiap mutant mice to generate mice lacking combinations of two genes, and studied their development in vivo. We isolated single and compound IAP mutant cell lines to study the redundant and individual functions of IAPs in vitro.
Results
Generation of a double IAPs mutant allele
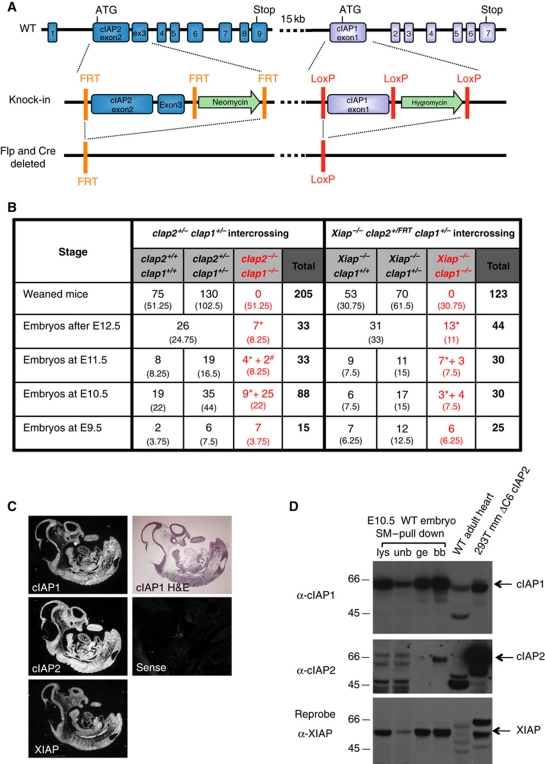
The mouse cIap2 and cIap1 genes are so close that it is impractical to delete them both by crossing individual KO lines. By re-targeting the same chromosome in ES cells, we have generated mice that bear a loxP-flanked cIap1 exon 1 as well as frt-flanked cIap2 exons 2 and 3, so that both genes can be conditionally deleted either separately or together (Gardam et al, 2011) (Figure 1A). These loxP–frt-flanked mice (cIap2FRT/FRTcIap1lox/lox) are viable and fertile and have a normal phenotype (Supplementary Figure S1A). FlpE (Rodriguez et al, 2000) and Cre-deleter (Schwenk et al, 1995) mice were used to generate germline deletions of the cIap1 and cIap2 loci either individually or together, and some were subsequently crossed with mice bearing Xiap mutant alleles. Both the cIap2FRT/FRTcIap1−/− and cIap2−/−cIap1lox/lox single mutant mice were viable and showed no reproductive or developmental defects (Supplementary Figure S1A), consistent with previous reports of independently generated cIap2 or cIap1-deficient mice (Conze et al, 2005; Conte et al, 2006). Among >200 progeny of 40 cIap2+/−cIap1+/− intercrosses, no homozygous cIap2−/−cIap1−/− mice were found, whereas double heterozygote and wild-type (WT) genotypes were present in a ratio of 2 to 1 (Figure 1B; Supplementary Figure S1B). This indicates that the absence of both cIAP1 and cIAP2 results in lethality at or before birth. Therefore, at least for development, cIAP1 and cIAP2 are redundant, but presence of at least one allele of either gene is required and sufficient for viability. Similarly, mice bearing one or two copies of the gene for cIap1 (i.e., Xiap−/−cIap1+/− and Xiap−/−cIap1+/+ mice, respectively) were found in a ratio of 2 to 1, indicating lethality of embryos lacking both cIAP1 and XIAP (Figure 1B). In contrast, mice that lacked genes for both XIAP and cIAP2 were viable and fertile without any obvious phenotype (Figure 8, Supplementary Figure S1A, and data not shown). Together, these results show that cIAP1 is sufficient for development in the absence of cIAP2 and XIAP, but that presence of both cIAP2 and XIAP is needed if cIAP1 is absent.
Figure 1.
Deletion of cIap1 plus cIap2, or cIap1 plus Xiap, results in embryonic lethality at ∼E10. (A) Generation of a genetically modified cIap locus. Exons 2 and 3 of cIap2 were flanked by frt sites and a neomycin phosphotransferase expression cassette and exon 1 of cIap1 was flanked by loxP sites with an hygromycin cassette. (B) Incidence of genotypes of weaned mice or embryos derived from indicated intercrossed mice; * represents embryos without heartbeat or reabsorbed and # represents embryos with minimal heart contractile activity. Expected numbers of each group shown in brackets (based on Mendelian ratios). (C) Detection of cIap1, cIap2 and Xiap expression during development (E10.5) by in situ hybridization as compared with sense control probe. (D) Expression of IAPs during mouse development detected by western blotting. A WT E10.5 embryo was lysed in DISC lysis buffer supplemented with protease inhibitors, NEM and Pefabloc. IAPs were trapped using biotinylated SM and precipitated with streptavidin beads. Lysates (lys), unbound fractions (unb), glycine elutions (ge), and boiled beads (bb) were separated using SDS/PAGE and blotted with antibodies to cIAP1, cIAP2, and XIAP. 293T cells transfected with a plasmid encoding mouse cIAP2ΔC6 were used as a positive control for detection of cIAP2. See also Supplementary Figure S1. Figure source data can be found in Supplementary data.
All three IAPs are expressed during early mouse development
To determine the expression and distribution of the three IAPs during development, we analysed cIAP1, cIAP2, and XIAP embryonic expression by in situ hybridization using sections of WT embryos from E9.5 to E15.5. Dark field photographs are shown in Figure 1C. Ubiquitous expression of cIAP1, cIAP2, and XIAP was observed at all stages, with the strongest expression being from E9.5 to E12.5. We also determined levels of IAP proteins by lysing whole E10.5 embryos, and immunoprecipitating using a biotinylated IAP antagonist/Smac mimetic (SM) (compound A, TetraLogic Pharmaceuticals) (Vince et al, 2007). Consistent with the in situ hybridization data, all three IAPs could be pulled down with the biotinylated compound at E10.5 (Figure 1D and data not shown). These results showed that cIAP1, cIAP2, and XIAP are all expressed during mouse development, and of these three proteins, only cIAP1 is sufficient to act alone to allow normal development.
Cardiovascular defect in cIap2−/−cIap1−/− and Xiap−/−cIap1−/− DKO embryos
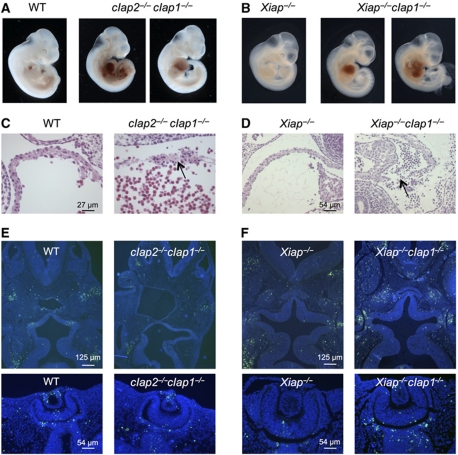
To determine the cause of death of the double IAP-deficient embryos, we examined them from E9.5 to E14.5. At each stage, no differences were observed between WT, double heterozygote cIap2+/−cIap1+/−, Xiap−/−cIap1+/− and Xiap−/− embryos. At E9.5, all cIap2−/−cIap1−/− and Xiap−/−cIap1−/− embryos were viable and could not be distinguished by overall morphology from WT or Xiap−/− embryos. However, between E10.5 and E11.5, we observed that the pericardial cavities of double homozygous mutant embryos were frequently swollen and filled with blood (Figure 2A and B). In most cases, mutant embryos were found with minimal or no contractile activity of the heart. Placentas appeared normal by gross morphology and histology (data not shown). Transverse serial sections of the mutant embryos showed that most tissues appeared normal, except for the heart, which showed sporadic discontinuities, usually in the atrial chamber. These discontinuities were often in association with pyknotic nuclei, suggestive of apoptosis (Figure 2C and D). However, elsewhere in the embryos we saw no major differences in the overall pattern of apoptosis between WT, Xiap−/−, cIap2−/−cIap1−/− and Xiap−/−cIap1−/− embryos (head and eye sections are shown as examples in Figure 2E and F). We conclude that the lethal phenotype of cIap2−/−cIap1−/− and Xiap−/−cIap1−/− embryos is not due to generalized growth retardation or widespread failure to control cell death, but is due to haemorrhages and cardiovascular failure at ∼E10.5.
Figure 2.
Embryonic lethality of cIap2−/−cIap1−/− and Xiap−/−cIap1−/− DKO embryos. (A, B) Whole view of E10.5 and E11.5 embryos derived from intercrosses of cIap2+/−cIap1+/− and Xiap−/−cIap1+/− mice, respectively. In the DKO embryos, blood has accumulated in the pericardial cavities, but not in those of WT or Xiap−/− littermates. (C, D) Histological analysis of the atria from a WT and cIap2−/−cIap1−/− DKO E10.5 embryo and Xiap−/− and Xiap−/−cIap1−/− DKO E11.5 embryos. The arrow shows discontinuity in the wall. (E, F) TUNEL (green) staining indicating fragmented DNA and nuclear (DAPI, blue) staining of E10.5–E11.5 transverse sections of embryos with genotypes as indicated. Heads are shown in the top panels and eyes are shown in the bottom panels. No gross differences in patterns of TUNEL-stained cells were observed.
In mouse embryonic fibroblasts, only cIAP1 is necessary, and provides sufficient IAP function, for normal induction of canonical NF-κB by TNF
Previous studies using cIap−/− mouse embryonic fibroblasts (MEFs) and siRNA have shown that cIAPs are required for TNF to efficiently activate canonical p65/RelA NF-κB in cultured cells (Mahoney et al, 2008; Varfolomeev et al, 2008). To further investigate the roles of the IAPs in NF-κB activation, we generated MEFs from cIap2−/−cIap1−/−, Xiap−/−cIap1−/− and Xiap−/−cIap2−/− DKO embryos and immortalized them with SV40 large T antigen. We used western blotting to determine the levels of the three IAPs in each DKO MEF line. As expected, cIAP1 was absent from the cIap2−/−cIap1−/− and Xiap−/−cIap1−/− lines, and was present at similar levels in WT and Xiap−/−cIap2−/− lines (Figure 3A). Similarly, XIAP was absent from the Xiap−/−cIap1−/− and Xiap−/−cIap2−/− lines, and present in the cIap2−/−cIap1−/− line. In contrast, cIAP2 was not only absent from cIap2−/− lines, but was also not detectable in WT MEFs or Xiap−/−cIap1−/− MEFs, presumably reflecting either low levels of transcription in unstimulated MEFs, protein instability, or low sensitivity of the antibody. Because the same antibody readily detected cIAP2 in embryos that was immunoprecipitated by a biotinylated SM compound, as well as overexpressed cIAP2 (Figure 1D), we believe unstimulated MEFs normally contain very low levels of cIAP2.
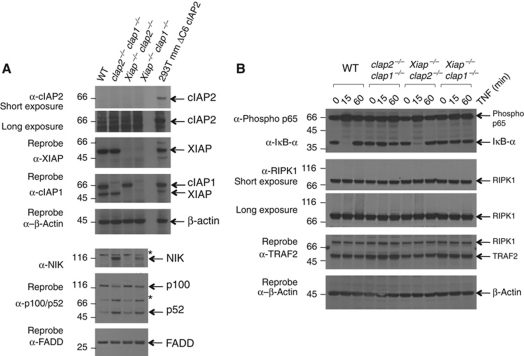
Figure 3.
Defects in NF-κB signalling in the absence of cIAP1. (A) Levels of NIK, and spontaneous processing of p100 are increased in MEFs lacking cIAP1. SV40 large T antigen-immortalized MEFs derived from WT, cIap2−/−cIap1−/− DKO, Xiap−/−cIap2−/− DKO, and Xiap−/−cIap1−/− DKO embryos, were lysed then separated using SDS/PAGE and probed with antibodies to cIAP1, cIAP2, XIAP, NIK, p100/p52, and FADD (* indicates non-specific bands due to the antibody to NIK). Lysate from 293T cells transfected with a plasmid encoding murine cIAP2 ΔC6 was used as a positive control for detection of cIAP2. (B) TNF does not efficiently trigger IκB degradation in cIap1 deleted cells. Cells described in (A) were incubated with or without 100 ng/ml Fc-TNF for the indicated times. Lysates were separated using SDS/PAGE and probed for phosphorylated p65, IκBα, RIPK1, and TRAF2 and β-actin. Figure source data can be found in Supplementary data.
Previously, we have found elevated levels of NIK, and processing of NF-κB2 to p52, in unstimulated cIap1−/− and cIap2−/−cIap1−/− MEFs, suggesting that a normal function of cIAP1 is to promote degradation of NIK in cells that have not been treated with cytokine (Vince et al, 2007; Feltham et al, 2010). Consistent with these observations, we found high levels of NIK and activation of non-canonical NF-κB and processed p100 NF-κB2 in untreated Xiap−/−cIap1−/− MEFs (Figure 3A). In contrast, the amounts of NIK, p100 and p52 in Xiap−/−cIap2−/− MEFs were similar to those in WT MEFs. Therefore, the presence of cIAP1 is both necessary and sufficient to cause degradation of NIK and prevent spontaneous activation of non-canonical NF-κB in untreated cells. Furthermore, the presence of cIAP1 was also both necessary and sufficient for TNF to promote phosphorylation of canonical p65/RelA NF-κB, and trigger degradation of IκBα (Figure 3B). These results also show that, on its own, XIAP is not able to activate canonical NF-κB following TNF stimulation of MEFs, and that cIAP2 is also not able, or is not present at high enough levels, to signal activation of canonical NF-κB, confirming that cIAP1 is the most important IAP for regulation of canonical and non-canonical NF-κB both in MEFs and for development.
Lack of cIAP1 sensitizes MEFs to TNF-induced apoptosis, but loss of both cIAP2 and XIAP does not
Low levels of cIAP1, whether due to gene deletion or treatment with SM, sensitized MEFs to TNF-induced cell death (Varfolomeev et al, 2007; Vince et al, 2007). Consistent with this, WT MEFs were not killed by TNF, but the cells died when TNF was combined with SM (Figure 4A). cIap2−/−cIap1−/− and Xiap−/−cIap1−/− MEFS were very sensitive to induction of cell death by TNF alone (Figure 4A). Compared with WT MEFs, those lacking both cIAP1 and cIAP2, or both cIAP1 and XIAP, were also much more sensitive to killing by CD95L (Fas ligand), or the topoisomerase II inhibitor etoposide, but not the DNA alkylating agent cisplatin (Figure 4A). Surprisingly, Xiap−/−cIap2−/− MEFs were similar to WT MEFs, because they were not killed by TNF alone, but could be significantly sensitized by addition of SM, which depletes the cells of cIAP1.
Figure 4.
Endogenous levels of cIAP1, and ectopic expression of cIAP2, protect MEFs from killing by TNF much more than XIAP. (A) Sensitivity of gene deleted lines to induction of cell death. Cells described in Figure 3A were incubated for 24 h with 100 ng/ml Fc-TNF or 20 ng Fc-CD95L, 500 nM SM, 1 μM etoposide or 5 μg/ml cisplatin where indicated. Cells were stained with PI and analysed by flow cytometry. The mean+s.e.m. of 3–11 independent experiments is shown. (B, C) Ability of ectopically expressed IAPs to protect cIap2−/−cIap1−/− DKO cells. cIap2−/−cIap1−/− MEFs were complemented with inducible mouse cIAP1, FLAG–cIAP2, or mouse XIAP and induced with 10 nM 4-HT for 24 h, and then incubated with or without 100 ng/ml Fc-TNF for a further 24 h. Cells were stained with PI and analysed by flow cytometry. The mean values+s.e.m. of 5–14 independent experiments are shown (* indicates a non-specific band). (D, E) Ability of ectopically expressed IAPs to protect Xiap−/−cIap1−/− DKO cells. Xiap−/−cIap1−/− MEFs were complemented with inducible mouse cIAP1 or EGFP–XIAP, induced with 10 nM 4-HT for 24 h and then treated with 100 ng/ml Fc-TNF or 20 ng/ml Fc-CD95L and 500 nM SM. The mean values+s.e.m. of three to six independent experiments are shown. See also Supplementary Figure S2. Figure source data can be found in Supplementary data.
To investigate further the ability of cIAPs to regulate TNF-induced cell death, we complemented the cIap2−/−cIap1−/− and Xiap−/−cIap1−/− MEFs with WT cIAP1, EGFP-tagged cIAP1, Flag–cIAP2, or XIAP, expressed from a tamoxifen-inducible lentiviral vector. EGFP-tagged cIAP1 and XIAP proteins were functional because they bound TRAF2 and Smac/DIABLO, respectively (Supplementary Figure S2). Induction of cIAP1 or Flag–cIAP2 by 4-hydroxy-tamoxifen (4-HT) in cIap2−/−cIap1−/− MEFs, and induction of EGFP–cIAP1 in Xiap−/−cIap1−/− MEFs made cells almost as resistant to TNF-induced death as WT MEFs (Figure 4B and D). Even the low background levels of cIAPs expressed from the uninduced lentiviral vector (Figure 4C and E) gave partial protection against TNF killing (Figure 4B and D). Moreover, reconstitution with cIAP1 or XIAP was able to give partial protection to Xiap−/−cIap1−/− MEFs against CD95L-induced cell death, but cIAP1 seemed to be more potent than XIAP (Figure 4D). Similarly, overexpression of XIAP in cIap2−/−cIap1−/− MEFs provided no protection, and reconstitution of XIAP into Xiap−/−cIap1−/− MEFs provided only modest protection from killing by TNF (Figure 4B and D). These data demonstrate that unlike cIAP1, XIAP is neither necessary nor sufficient to block apoptosis pathways activated by TNF in MEFs.
Deletion of Tnfr1, but not Tnfr2, allows cIap2−/−cIap1−/− embryos to develop until birth
Previous studies have shown that deletion of genes for Tnfr1 prevents death of p65/RelA NF-κB KO and Traf2 KO mice during embryonic development, and allows them to survive until shortly after birth (Nguyen et al, 1999; Rosenfeld et al, 2000). Because MEFs derived from p65/RelA KO and Traf2 KO mice resemble those derived from cIap1 KO mice, in that they are all very sensitive to induction of apoptosis by TNF alone, and all show deficits in activation of canonical (p65/RelA) NF-κB in response to TNF, we hypothesized that deletion of Tnfr1 would allow cIap2−/−cIap1−/− embryos to survive until birth. We therefore generated cIap2−/−cIap1−/−Tnfr1−/− triple KO mice. Unlike the cIap2−/−cIap1−/− embryos, most of the cIap2−/−cIap1−/−Tnfr1−/− pups were born alive (Figure 5A and B). Some of them survived until day 2 with no overt phenotype, but none of the cIap2−/−cIap1−/−Tnfr1−/− mice were found at weaning. In contrast, deletion of TNFR2 was not able to prolong survival of cIap2−/−cIap1−/− embryos (Figure 5A). To further analyse the relative contributions of the two different TNF receptors in death induced by TNF, we generated MEFs from cIap2−/−cIap1−/−Tnfr1−/− and cIap2−/−cIap1−/−Tnfr2−/− embryos and immortalized them with SV40 large T antigen. For comparison, we generated WT, Tnfr1 KO, and Tnfr2 KO MEFs. As expected, none except the cIap2−/−cIap1−/− Tnfr2−/− MEFs was killed by TNF alone (Figure 5C). Moreover, all MEF lines lacking Tnfr1 were resistant to TNF even in the presence of SM. These results demonstrate that death of cIap2−/−cIap1−/− MEFs in response to TNF, and death at E10.5 of cIap2−/−cIap1−/− embryos, are both dependent on the presence of TNFR1 but not TNFR2.
Figure 5.
Deletion of Tnfr1, but not Tnfr2 allows cIap2−/−cIap1−/− embryos to develop until birth. (A) Incidence of genotypes of weaned mice or embryos derived from intercrosses of genotypes as indicated; * represents embryos without heartbeat or reabsorbed. Expected numbers of each group are shown in brackets. (B) Representative photographs of three 1-day-old pups from a litter from a cIap2+/−cIap1+/−Tnfr1−/− intercross. (C) Sensitivity to TNF is conferred by TNFR1 not TNFR2. SV40 large T antigen transformed MEFs derived from WT embryos or mutant embryos as indicated were incubated for 24 h in the presence or absence of 100 ng/ml Fc-TNF and 500 nM SM. Cells were stained with PI and analysed by flow cytometry. The mean values+s.e.m. of three to seven independent experiments are shown.
TNF can activate both caspase-dependent and caspase-independent, RIPK1 kinase-dependent cell death mechanisms
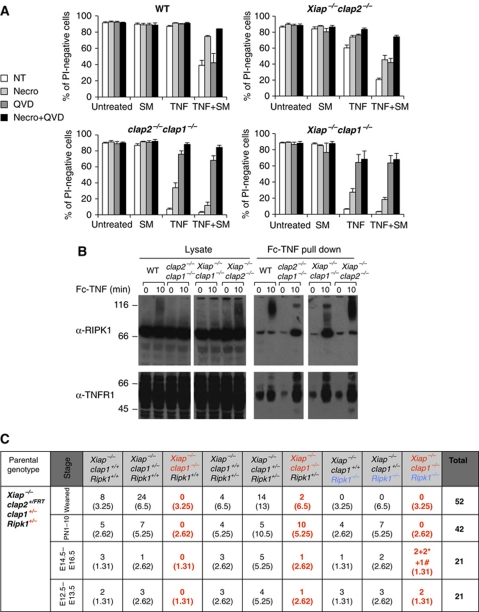
The amount of cell death induced by signalling through TNFR1 is greatly increased when IAPs are absent, in cIap2−/−cIap1−/− embryos as well as in MEFs, whether cIAPs are absent due to gene deletion or treatment with SM. To determine if TNF causes cell death via a caspase- or a RIPK1-dependent mechanism, we analysed the effect of adding a pan-caspase inhibitor, QVD-OPh (QVD) (Caserta et al, 2003), and a specific RIPK1 kinase inhibitor, necrostatin (Degterev et al, 2008). In all types of cells tested (WT, Xiap−/−cIap2−/−, cIap2−/−cIap1−/−, Xiap−/−cIap1−/−), TNF caused death when cIAP1 was genetically deleted, or was reduced by treatment with SM (Figure 6A). Moreover, in every case, TNF-induced cell death could be almost entirely blocked when QVD and necrostatin were added together (Figure 6A, black columns). When added alone, the degree of protection provided by necrostatin (Figure 6A, light grey columns) appeared to correlate with the amount of remaining cIAP1. In contrast, the extent of protection by QVD was greater when cIAP1 was genetically deleted than when it was reduced by SM treatment. These results suggest a larger role for caspases, but a lesser role for RIPK1 kinase activity, in TNF-induced death in the absence of cIAP1. Furthermore, in agreement with previous results (Haas et al, 2009), ubiquitylation of RIPK1 was reduced in the absence of both cIAP1 and cIAP2, but the amount of unmodified RIPK1 bound to the TNFR1 signalling complex was increased (Figure 6B). Findings were similar in Xiap−/−cIap1−/− cells, whereas normal levels of RIPK1 ubiquitylation were observed when only cIAP1 was present (i.e., in Xiap−/−cIap2−/− MEFs) (Figure 6B). These experiments indicate that in MEFs, TNF mainly activates a caspase-dependent death process, but provide some evidence, suggesting that it can also activate a caspase-independent, RIPK1 kinase-dependent death mechanism, which is more apparent when cIAP1 levels are reduced by SM, than when cIAP1 is eliminated by gene deletion.
Figure 6.
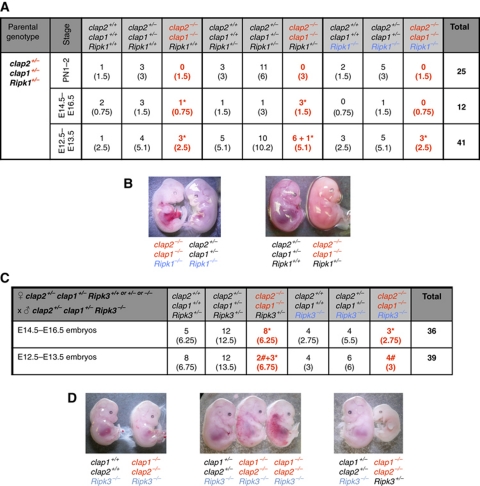
In the absence of IAPs, both TNF-induced death of MEFs, and embryonic death during development, involves RIPK1. (A) Together, the RIPK1 kinase inhibitor necrostatin and the caspase inhibitor QVD protect IAP gene deleted MEFs from killing by TNF. MEFs derived from WT and compound mutant embryos were incubated for 24 h with or without 100 ng/ml Fc-TNF and 500 nM SM in the presence or absence of 50 μM necrostatin or 10 μM QVD. Cells were stained with PI and analysed by flow cytometry. The mean values+s.e.m. of 3–11 independent experiments are shown. (B) TNF-induced RIPK1 modification fails, and receptor-associated RIPK1 is increased when cIap1 genes are deleted. MEFs were stimulated with 1 μg/ml Fc-TNF prior to Fc pull down, and analysed by western blotting. (C) Incidence genotypes of offspring from Xiap−/−cIap1+/−Ripk1+/− intercrosses. *Represents embryos without heartbeat or reabsorbed and # represents embryos alive but with a small liver. See also Supplementary Figure S3. Figure source data can be found in Supplementary data.
cIap2−/−cIap1−/− E10.5 cardiac defects are absent if Ripk1 or Ripk3 genes are deleted, and heterozygosity for Ripk1 allows Xiap−/−cIap1−/− mice to survive embryogenesis
The ability of necrostatin to increase survival of MEFs with reduced levels of IAPs when exposed to TNF raised the possibility that the death of cIap2−/−cIap1−/− and Xiap−/−cIap1−/− DKO embryos at day E10.5 might also involve RIPK1 or RIPK3. To test this, we generated Xiap−/−cIap1−/−Ripk1−/−, cIap2−/−cIap1−/−Ripk1−/−, and cIap2−/−cIap1−/−Ripk3−/− mice. Xiap−/−cIap1−/−Ripk1−/− embryos were viable to E14.5, but by E15.5, they had died. Surprisingly, Xiap−/−cIap1−/−Ripk1+/− mice were born alive (Figure 6C). While most Xiap−/−cIap1−/−Ripk1+/− mice died within the first 10 days of birth, two runted males even survived for 6 weeks (Figure 6C; Supplementary Figure S3B and C). Unlike the cIap2−/−cIap1−/− embryos that never survive past E10.5, cIap2−/−cIap1−/−Ripk1−/− and cIap2−/−cIap1−/−Ripk3+/− embryos survived to E12.5, and cIap2−/−cIap1−/−Ripk1+/− and cIap2−/−cIap1−/−Ripk3−/− embryos survived to E15.5 (Figure 7A and C). Six out of seven cIap2−/−cIap1−/−Ripk1+/− embryos were viable at E12.5 and three embryos were found dead at E15.5 (Figure 7B and D). Four cIap2−/−cIap1−/−Ripk3−/− embryos were viable at E13.5 and three embryos were found dead around E15.5 (Figure 7C). These results show that the absence of RIPK3 or loss of only one allele of Ripk1 can overcome the mid-embryonic lethality in cIap2−/−cIap1−/− or Xiap−/−cIap1−/− mutant mice, respectively. Moreover, the presence of cIap2 allowed the further development of Xiap−/−cIap1−/−Ripk1+/− to birth, but Xiap was not sufficient for cIap2−/−cIap1−/−Ripk1+/− embryos to survive beyond E15.5. Together with the data presented above, these results indicate that of the IAPs, only cIAP1 is sufficient on its own to prevent RIP kinases from acting in the TNF receptor signalling pathway to cause mid-embryonic lethality.
Figure 7.
Prevention of E10.5 lethality of cIap2−/−cIap1−/− embryos by deletion of Ripk1 or Ripk3 genes. (A) Incidence of genotypes of born pups or embryos derived from intercrosses of cIap2+/−cIap1+/−Ripk1+/− mice. (B) Representative photographs of E12.5 embryos from a cIap2+/−cIap1+/−Ripk1+/− intercross. (C) Incidence of genotypes of embryos derived from crosses of cIap2+/−cIap1+/−Ripk3+/+or+/−or−/− females with cIap2+/−cIap1+/−Ripk3−/− males. In (A, C), * represents embryos without heartbeat or reabsorbed and # represents embryos alive but with a small liver. (D) Representative photographs of E13.5 and E14.5 embryos from a cIap2+/−cIap1+/−Ripk3+/− female with cIap2+/−cIap1+/−Ripk3−/− male cross.
Discussion
Consistent with earlier reports, we found that mice lacking a single IAP, either cIap1, cIap2, or Xiap, are viable and fertile and exhibit relatively subtle phenotypes (Harlin et al, 2001; Conze et al, 2005; Conte et al, 2006). To study the functional redundancy of the IAPs, we examined compound IAP mutant mice. While those lacking both cIAP2 and XIAP were viable and fertile, without any obvious developmental phenotype, in marked contrast, cIap2−/−cIap1−/− and Xiap−/−cIap1−/− DKO mice died in utero after 10 days of development.
The phenotype of the compound mutants implied that all three IAPs are expressed during early embryogenesis. However, Shafey et al (2006), who used cIap2 null, lacZ reporter mice, concluded that cIAP2 was not expressed before E11.5. To resolve this, we performed immunoprecipitations using biotinylated SM, and analysed them by western blot. Using this technique, we were able to detect cIAP2 from E10.5, which suggests that at this stage of embryogenesis all three IAPs are expressed, but cIAP2 protein is present at comparatively low levels. These low levels of cIAP2 protein, together with the easily detectable mRNA revealed by in situ hybridization, are consistent with cIAP2 having a shorter half-life than cIAP1, which might be due to an increased propensity for cIAP2 than cIAP1 to spontaneously dimerize, autoubiquitylate, and be degraded (Feltham et al, 2011).
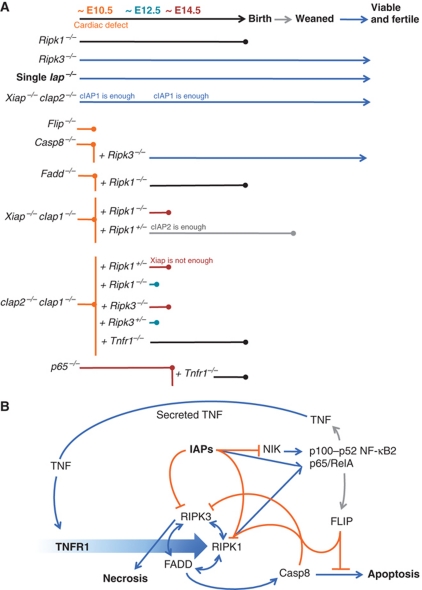
The double IAPs-deficient embryos and placentas appeared normal with the exception of the heart. The appearance of these embryos bore similarities to zebrafish mutant for their single cIAP gene (Santoro et al, 2007). These embryos exhibit the tomato phenotype, characterized by severe haemorrhage and vascular regression during development. A similar phenotype is also seen in mouse embryos lacking other genes in the death receptor signalling pathway (Figure 8A), such as Fadd, Casp8, and Flip mutants (Varfolomeev et al, 1998; Yeh et al, 1998, 2000). This raised the possibility that there might be common mechanisms for the developmental deaths of the embryos, even though some of the proteins (e.g., FADD and caspase 8) are generally thought to be pro-apoptotic, whereas others (e.g., FLIP and IAPs) are generally thought to promote survival or inhibit apoptosis. Interestingly, apoptosis (as revealed by TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labelling) staining) was unremarkable in the cIap2−/−cIap1−/− embryos, just as it was in the Fadd−/−, Casp8−/−, and Flip−/− mutants, raising the possibility that some other cell death process, such as necroptosis (Hitomi et al, 2008) might be involved.
Figure 8.
Roles of cIAPs in signalling and cell death during development. (A) Diagram of extent of viability of single, double, and triple gene deleted mice. (B) Speculative model to account for the phenotypes of gene deleted mice. Binding of TNF to TNFR1 triggers (large blue arrow) formation of complexes that can culminate in cell death by apoptosis or necroptosis, or lead to cell survival. Activating interactions are indicated with blue arrows; inhibitory interactions are indicated with orange lines; transcriptional induction is indicated with grey arrows. Merging of lines (such as those from Casp8 and FLIP to RIPK1, or IAPs and RIPK1 to p65/RelA) indicate proteins that can act together. cIAPs limit levels of NIK, and inhibit cell death mediated by RIPK1 and RIPK3, but cooperate with RIPK1 to activate p65/RelA. RIPK1 has both pro-death and pro-survival functions, by promoting necroptosis via RIPK3, apoptosis via FADD and Casp8, and cell survival via p65/RelA. Casp8 inhibits cell death by necrosis, but promotes cell death by apoptosis. FLIP inhibits cell death by both pathways. This model is not complete, and does not include other important proteins, such as TRADD, TRAF2, CYLD, A20, TAK1, HOL, HOIP, or Sharpin, just to name a few. For further details, see Discussion.
Depending on the cell type and environment, TNFR1 signalling can result in a variety of responses in living cells, or death of cells by apoptosis or necroptosis. We found that in untreated cells, cIAP1 played a much more important role than cIAP2 and XIAP in limiting levels of NIK, and cIAP1 was the most critical IAP for activating canonical p65/RelA NF-κB to prevent TNF-induced cell death. In MEFs, only cIAP1 was necessary and sufficient to stop cells dying when they were exposed to TNF, or to allow TNF to rapidly activate p65/RelA NF-κB. cIAP1 was necessary and sufficient to cause degradation of NIK, and prevent the spontaneous activation of the non-canonical p100/p52 NF-κB2 pathway. In contrast, in the absence of cIAP1, neither cIAP2 nor XIAP allowed TNF to activate NF-κB. Our results confirm that cIAP1 is a crucial regulator of both canonical and non-canonical NF-κB, in both resting and TNF-stimulated cells.
In MEFs, TNF mainly activated a caspase-dependent death process, but it could also activate a caspase-independent, RIPK1 kinase-dependent death mechanism. The differences in the ability of QVD and necrostatin to block TNF-induced cell death in cIAP1 gene deleted MEFs compared with MEFs depleted of cIAP1 by SM suggest a complex interaction between cIAP1 and RIPK1. One possibility is that the amount of ubiquitylation of RIPK1 by cIAP1 determines the outcome following TNF treatment. In normal cells, TNF triggers cIAP1 to poly-ubiquitylate RIPK1, which efficiently activates p65/RelA NF-κB, but RIPK1 is then degraded and no cell death occurs; if cIAP1 levels are reduced by SM, RIPK1 is ubiquitylated to a small extent, which increases its ability to cause cell death by a necrostatin blockable, caspase-independent mechanism; and if no cIAP1 is present, there is no RIPK1 ubiquitylation, and most of RIPK1 remains within the TNFR1 signalling complex to help recruit and activate caspase 8.
Recently, deletion of RIPK1 was shown to rescue the embryonic lethality of Fadd null embryos to birth (Zhang et al, 2011), and deletion of RIPK3, which is thought to act downstream of RIPK1 in the necroptotic pathway, was shown to allow Casp8 KO mice to develop into normal, fertile adults (Ch’en et al, 2011; Kaiser et al, 2011; Oberst et al, 2011) (Figure 8A). However, later in life, mice lacking both RIPK3 and caspase 8 develop a progressive lymphoaccumulative disease, a phenotype similar to Fas deficiency (Kaiser et al, 2011; Oberst et al, 2011). These experiments imply that FADD and caspase 8 are required to inhibit RIPK1 and/or RIPK3, which, when left uncontrolled, cause lethal developmental defects at E10.5. Our observations of the cIap2−/−cIap1−/−Ripk1+/− and cIap2−/−cIap1−/−Ripk3−/− mice suggest that cIAPs inhibit RIPK activity during development, presumably through ubiquitylation (Bertrand et al, 2008), whereas FADD and caspase 8 limit RIPKs by proteolysis (Pastorino et al, 1999; Feng et al, 2007). Furthermore, inhibition of RIPKs by FADD/caspase 8 or cIAP1/2 alone is not sufficient to prevent RIPK-dependent embryonic lethality, rather, both FADD/caspase 8 and cIAP1/2 are required to control RIP kinases.
Together, these results show that mouse development is relatively normal in the absence of cIap1 alone, or the absence of both cIap2 and Xiap. Therefore, the critical IAP functions can be fulfilled by just cIAP1, or by XIAP and cIAP2 acting together. Perhaps, cIAP2 on its own might not be present at high enough levels for normal development to proceed, but when XIAP is also present, it supports cIAP2's function, either by inhibiting caspases, by binding to TAB1, or by sequestering BIR-binding proteins (such as Smac/DIABLO).
Remarkably, because deletion of Tnfr1 rescued development of cIap2−/−cIap1−/− embryos to birth, either TNF signalling occurs as early as E10.5, and this will lead to embryonic lethality unless it is controlled by IAPs, or, IAPs are needed to prevent aberrant production of TNF, just as depletion of IAPs by SM provokes autocrine production of TNF by some tumour cell lines (Vince et al, 2007). Regardless, the pathway activated by TNF during development of cIap2−/−cIap1−/− embryos must involve RIPK1 and RIPK3, because rather than dying at E10.5, cIap2−/−cIap1−/− embryos that lacked both copies of Ripk3, or Xiap−/−cIap1−/− and cIap2−/−cIap1−/− embryos hemizygous for Ripk1, were able to survive to E15.5 or longer, sometimes surviving beyond birth.
A possible model to account for these observations is shown in Figure 8B. According to this model, ligation of TNFR1 can trigger formation of a complex (sometimes termed the ‘ripoptosome’) in which RIPK1, RIPK3, and FADD interact in complex ways. When activated, RIPK1 does three things: it can activate RIPK3 (which can lead to necroptosis); together with FADD, it can activate caspase 8 (which can lead to apoptosis); and together with cIAPs, it can promote activation of canonical (p65/RelA) NF-κB. However, RIPK1 and RIPK3 are both subject to negative regulation, both by cIAP-dependent ubiquitylation, and by FADD/FLIP/caspase 8-mediated cleavage. cIAPs inhibit cell death by ubiquitylating RIPK1 and RIPK3, by promoting activation of canonical NF-κB, and by reducing activation of non-canonical (NIK-dependent) NF-κB. When genes for either FADD, FLIP, caspase 8, or cIAPs are deleted, RIPK1 and RIPK3 are no longer restrained, and cause death of cells in the developing heart at E10. This phenotype can be suppressed by also deleting genes for RIPK1 or RIPK3. Early activation of RIPK1 and RIPK3 in the cIap DKOs might be in response to normal production of TNF, or aberrant production of TNF due to elevated levels of NIK and p100/p52 NF-κB. According to this model, deletion of one or both copies of Ripk1 in cIap DKO mice is able to reduce RIPK1 levels enough to allow development past E10, but RIPK1 heterozygous mice might survive better than RIPK1 null mice after birth because in the absence of cIAPs, RIPK1 heterozygotes probably retain some ability to activate canonical p65/RelA NF-κB.
Materials and methods
Generation of cIap2FRT/FRT and cIap1lox/lox mice by gene targeting and timed mating
Animal experimental procedures were approved by the Animal Ethics Committee of La Trobe University. cIap1lox/lox and cIap2FRT/FRT constructs are described in Gardam et al (2011). In order to generate the cIap2FRT/FRTcIap1lox/lox mice, both constructs were inserted into BRUCE embryonic stem cells, which were derived from C57BL6 mice (Koentgen et al, 1993; Hughes et al, 2007). The cIap2FRT/FRTcIap1lox/lox mice were crossed to transgenic mice expressing FLPe recombinase to generate the cIap2−/−cIap1lox/lox line, and to mice expressing Cre recombinase to generate the cIap2FRT/FRTcIap1−/− line. The cIap2FRT/FRTcIap1−/− mice were subsequently crossed to FLPe mice to generate the cIap2+/−cIap1+/− mice then used for timed matings. For all timed matings, a single male was housed with 2–5 females. The females were checked each morning for vaginal plugs. When a vaginal plug was detected, the female was separated from the male. The embryonic days were counted starting at E0.5 on the day the plug was detected. The embryos were taken on the embryonic days described throughout.
In situ hybridization
In situ hybridization was performed as described previously (Thomas et al, 2000). Briefly, sections were dewaxed, rehydrated through graded concentrations of alcohol, incubated for 10 min at room temperature in 10 mg/ml proteinase K, fixed in 4% paraformaldehyde 10 min, then dehydrated through graded concentrations of alcohol. Sections were air dried, and hybridization solution containing 5 × 105 counts/min/μl of in vitro transcribed cRNA probe was placed over the section. Slides were incubated overnight at 56°C and then washed as described (Hogan et al, 1986).
Histological analysis
Embryos were dissected between E10.5 and E11.5, fixed in 4% paraformaldehyde, paraffin embedded and serially sectioned (7 μm). Sections were stained with haematoxylin and eosin or used unstained for TUNEL and immunofluorescence, which were conducted as described previously (Thomas et al, 2000).
Generation of MEFs and lentiviral particles
Generation of MEFs has been previously described in detail (Vince et al, 2007). Briefly, primary MEFs were generated from E9.5/E10.5 (cIap2−/−cIap1−/−), E10.5/E11.5 (Xiap−/−cIap1−/−), E10.5/E12.5 (cIap2−/−cIap1−/−Tnfr2−/−), E14.5 (cIap2−/−cIap1−/−Tnfr1−/−), E15.5 (WT, cIap2−/−cIap1−/−Tnfr1−/−, Tnfr2−/− and Xiap−/−cIap2−/−) embryos, using standard protocols, and then infected with SV40 large T antigen expressing lentivirus to generate immortal cell lines.
Generation of lentiviral particles has recently been described (Vince et al, 2007; Geserick et al, 2009). Briefly, to generate cIap2−/−cIap1−/− or Xiap−/−cIap1−/− MEFs expressing inducible mouse cIAP1, EGFP–cIAP1, Flag–cIAP2, mouse XIAP, or EGFP–XIAP, 293T cells were transfected with packaging constructs pCMV ðR8.2, VSVγ and the relevant lentiviral plasmid. DKO MEFs were infected with packaged lentivirus and polyclonal MEFs were obtained after puromycin (2–5 μg/ml: pF 5xUAS selection) and hygromycin selection (100–500 μg/ml: GEV16 selection). Cells were subsequently tested for expression of the respective proteins after 24 h of induction with 10 or 100 nM 4-HT.
Death assay
Cells were seeded on 12-well tissue culture plates at ∼40% confluency and were allowed to adhere for 24 h. Cells were incubated in the presence or absence of 10 nM 4-OH-tamoxifen (4-HT) for 24 h, then 100 ng/ml Fc-TNF or 20 ng/ml Fc-CD95L (Bossen et al, 2006) plus or minus 500 nM SM (compound A), and/or 10 μM QVD were added to cells for 24 h and cell viability measured by propidium iodide (PI) exclusion using flow cytometry. In each sample, 10 000 events were measured and the percentages of PI-negative (viable) cells are shown.
Western blotting
Samples were lysed in DISC lysis buffer containing 1% Triton X-100 supplemented with protease inhibitor cocktail (Roche), N-EthylMaleimide (NEM) and pefabloc on ice for 30 min and clarified by centrifugation. Samples were separated on precast 4–12% polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes for antibody detection. The blots were stained with Ponceau S to confirm the uniformity of protein loading in each lane. All membrane blocking steps and antibody dilutions were performed with 5% skim milk in PBS containing 0.1% Tween 20 (PBS-T). Antibodies were used as follows: monoclonal anti-β-actin (Sigma), polyclonal mouse anti-TRAF2 1:1000 (Santa Cruz), monoclonal anti-cIAP1 1:500 (Alexis), monoclonal anti-cIAP2 1:300 (in house), monoclonal anti-XIAP 1:1000 (MBL), anti-polyclonal RIPK1 1:1000 (BD Biosciences), monoclonal anti-Flag 1:2000 (Sigma), polyclonal anti-phospho p65 1:000 (Cell Signaling), polyclonal anti-p100–p52 1:1000 (Cell Signaling), polyclonal anti-IκBα 1:1000 (Cell Signaling), polyclonal anti-NIK 1:1000 (Cell Signaling). Washing steps were performed with PBS-T. After incubating with HRP-coupled secondary antibodies, western blots were visualized by ECL (GE Healthcare, Rydalmere, NSW, Australia).
Immunoprecipitation of EGFP–cIAP1 and EGFP–XIAP
Xiap−/−cIap1−/− MEFs reconstituted with inducible EGFP–cIAP1 or EGFP–XIAP were seeded on 10 cm tissue culture plate and induced for 24 h with 10 nM 4-HT. Cells were lysed in DISC lysis buffer and GFP fusion proteins were precipitated using GFP–Trap-A beads for 1 h (Chromotek). The beads were washed four times. Samples then were separated on 4–12% polyacrylamide gels and transferred to Hybond C nitrocellulose membrane as described in western blotting section (see above).
TNFR1 signalling complex precipitation
Cells were seeded on 15 cm tissue culture plates and treated for the indicated time with 1 μg/ml of Fc-TNF. Cells were lysed in DISC lysis buffer at 4°C for 30 min. The lysates were centrifuged for 30 min and the TNF-RSC was precipitated using protein A beads overnight. The beads were washed four times. Samples then were separated on 4–12% polyacrylamide gels and transferred to Hybond C nitrocellulose membrane as described in western blotting section (see above).
Supplementary Material
Acknowledgments
This work was funded by a Centre grant from the Leukemia and Lymphoma Society, and NHMRC grants and fellowships 433063, 461221, 541901, 575512, 1003435, and was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. We thank Michelle Kelliher for providing Ripk1+/− mice; Vishva Dixit for the Ripk3−/− mice; Heinrich Korner for the Tnfr1−/− and Tnfr2−/− mice; Mark McKinlay for biotinylated and non-biotinylated Smac mimetic compounds (TetraLogic Pharmaceuticals). We thank CAH staff at Latrobe University, especially Jose Ramos, Samantha Kelly, and Melinda Boulton for mouse husbandry, and Frank Koentgen (Ozgene) for generating the cIap2FRT/FRTcIap1lox/lox mice.
Author contributions: MM performed most of the experimental work; HA was responsible for mouse breeding, husbandry, and genotyping; AKV and TT analysed the embryos; WWW, AB, RF, DC, and WDC performed tissue culture experiments, made constructs and viruses, the anti-cIAP2 mAb, and performed western blot analysis. MM, AKV, TT, and DLV wrote the manuscript. WDC and WWW provided critical comments. MM, JS, and DLV designed and directed the experiments.
Footnotes
DLV and JS are on the Scientific Advisory Board of TetraLogic Pharmaceuticals.
References
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 30: 689–700 [DOI] [PubMed] [Google Scholar]
- Birnbaum MJ, Clem RJ, Miller LK (1994) An apoptosis inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with cys/his sequence motif. J Virol 68: 2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossen C, Ingold K, Tardivel A, Bodmer JL, Gaide O, Hertig S, Ambrose C, Tschopp J, Schneider P (2006) Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J Biol Chem 281: 13964–13971 [DOI] [PubMed] [Google Scholar]
- Caserta TM, Smith AN, Gultice AD, Reedy MA, Brown TL (2003) Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis 8: 345–352 [DOI] [PubMed] [Google Scholar]
- Ch’en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM (2011) Mechanisms of necroptosis in T cells. J Exp Med 208: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte D, Holcik M, Lefebvre CA, Lacasse E, Picketts DJ, Wright KE, Korneluk RG (2006) Inhibitor of apoptosis protein cIAP2 is essential for lipopolysaccharide-induced macrophage survival. Mol Cell Biol 26: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze DB, Albert L, Ferrick DA, Goeddel DV, Yeh WC, Mak T, Ashwell JD (2005) Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol Cell Biol 25: 3348–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, Hedrick SM, Gerber SA, Lugovskoy A, Yuan J (2008) Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 4: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett CS, Nava VE, Gedrich RW, Clem RJ, Vandongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB (1996) A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J 15: 2685–2694 [PMC free article] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL (2006) Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep 7: 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltham R, Bettjeman B, Budhidarmo R, Mace PD, Shirley S, Condon SM, Chunduru SK, McKinlay MA, Vaux DL, Silke J, Day CL (2011) Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J Biol Chem 286: 17015–17028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltham R, Moulin M, Vince JE, Mace PD, Wong WW, Anderton H, Day CL, Vaux DL, Silke J (2010) Tumor necrosis factor (TNF) signaling, but not TWEAK (TNF-like weak inducer of apoptosis)-triggered cIAP1 (cellular inhibitor of apoptosis protein 1) degradation, requires cIAP1 RING dimerization and E2 binding. J Biol Chem 285: 17525–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Castanares M, Wu M (2007) Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal 19: 2056–2067 [DOI] [PubMed] [Google Scholar]
- Gardam S, Turner VM, Carter H, Limaye S, Basten A, Koentgen F, Vaux DL, Silke J, Brink R (2011) Deletion of cIAP1 and cIAP2 in B lymphocytes constitutively activates cell survival pathways and inactivates the germinal center response. Blood 117: 4041–4051 [DOI] [PubMed] [Google Scholar]
- Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B, Gollnick H, Silke J, Leverkus M (2009) Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol 187: 1037–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Darding M, Miasari M, Santoro MM, Zender L, Xue W, Tenev T, da Fonseca PC, Zvelebil M, Bujnicki JM, Lowe S, Silke J, Meier P (2008) IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol 10: 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P (2010) IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer 10: 561–574 [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, Koschny R, Komander D, Silke J, Walczak H (2009) Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36: 831–844 [DOI] [PubMed] [Google Scholar]
- Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB (2001) Characterization of XIAP-deficient mice. Mol Cell Biol 21: 3604–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2008) Shared principles in NF-kappaB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J (2008) Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Costantini F, Lacy E (1986) Manipulating the mouse Embryo: A Laboratory Manual, 1st edn, pp 232–228, Plainview, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hughes ED, Qu YY, Genik SJ, Lyons RH, Pacheco CD, Lieberman AP, Samuelson LC, Nasonkin IO, Camper SA, Van Keuren ML, Saunders TL (2007) Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm Genome 18: 549–558 [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, Caspary T, Mocarski ES (2011) RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 471: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koentgen F, Suss G, Stewart C, Steinmetz M, Bluethmann H (1993) Targeted disruption of the Mhc class-Ii Aa gene in C57bl/6 mice. Int Immunol 5: 957–964 [DOI] [PubMed] [Google Scholar]
- Liston P, Lefebvre C, Fong WG, Xuan JY, Korneluk RG (1997) Genomic characterization of the mouse inhibitor of apoptosis protein 1 and 2 genes. Genomics 46: 495–503 [DOI] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Chertonhorvat G, Farahani R, Mclean M, Ikeda JE, Mackenzie A, Korneluk RG (1996) Suppression of apoptosis in mammalian cells by naip and a related family of iap genes. Nature 379: 349–353 [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, Arora V, Mak TW, Lacasse EC, Waring J, Korneluk RG (2008) Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA 105: 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Tschopp J (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114: 181–190 [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Duncan GS, Mirtsos C, Ng M, Speiser DE, Shahinian A, Marino MW, Mak TW, Ohashi PS, Yeh WC (1999) TRAF2 deficiency results in hyperactivity of certain TNFR1 signals and impairment of CD40-mediated responses. Immunity 11: 379–389 [DOI] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR (2011) Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino JG, Tafani M, Farber JL (1999) Tumor necrosis factor induces phosphorylation and translocation of BAD through a phosphatidylinositide-3-OH kinase-dependent pathway. J Biol Chem 274: 19411–19416 [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25: 139–140 [DOI] [PubMed] [Google Scholar]
- Rosenfeld ME, Prichard L, Shiojiri N, Fausto N (2000) Prevention of hepatic apoptosis and embryonic lethality in RelA/TNFR-1 double knockout mice. Am J Pathol 156: 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV (1995) The tnfr2-traf signaling complex contains two novel proteins related to baculoviral-inhibitor of apoptosis proteins. Cell 83: 1243–1252 [DOI] [PubMed] [Google Scholar]
- Santoro MM, Samuel T, Mitchell T, Reed JC, Stainier DY (2007) Birc2 (cIap1) regulates endothelial cell integrity and blood vessel homeostasis. Nat Genet 39: 1397–1402 [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxp-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23: 5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafey D, Korneluk R, Holcik M (2006) Distinct patterns of expression of the inhibitor of apoptosis protein cIAP2 during murine embryogenesis. Apoptosis 11: 1257–1259 [DOI] [PubMed] [Google Scholar]
- Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB (2003) TAK1 is critical for IkappaB kinase-mediated activation of the NF-kappaB pathway. J Mol Biol 326: 105–115 [DOI] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P (2005) IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol 7: 70–77 [DOI] [PubMed] [Google Scholar]
- Thomas T, Voss AK, Chowdhury K, Gruss P (2000) Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 127: 2537–2548 [DOI] [PubMed] [Google Scholar]
- Uren AG, Pakusch M, Hawkins CJ, Puls KL, Vaux DL (1996) Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci USA 93: 4974–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D (2007) IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131: 669–681 [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, Fairbrother WJ, Vucic D (2008) c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem 283: 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P, Wallach D (1998) Targeted disruption of the mouse caspase 8 gene ablates cell death induction by the TNF receptors, fas/apo1, and dr3 and is lethal prenatally. Immunity 9: 267–276 [DOI] [PubMed] [Google Scholar]
- Vince JE, Chau D, Callus B, Wong WW, Hawkins CJ, Schneider P, McKinlay M, Benetatos CA, Condon SM, Chunduru SK, Yeoh G, Brink R, Vaux DL, Silke J (2008) TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1-TRAF2 complex to sensitize tumor cells to TNFalpha. J Cell Biol 182: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, Schneider P, Callus BA, Koentgen F, Vaux DL, Silke J (2007) IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131: 682–693 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Delapompa JL, Mccurrach ME, Shu HB, Elia AJ, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, Eldeiry WS, Lowe SW, Goeddel DV, Mak TW (1998) Fadd—essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 279: 1954–1958 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A, Mirtsos C, Suzuki N, Bonnard M, Goeddel DV, Mak TW (2000) Requirement for casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity 12: 633–642 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J (2011) Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 471: 373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.