Abstract
The ribosome-recycling factor (RRF) and elongation factor-G (EF-G) disassemble the 70S post-termination complex (PoTC) into mRNA, tRNA, and two ribosomal subunits. We have determined cryo-electron microscopic structures of the PoTC·RRF complex, with and without EF-G. We find that domain II of RRF initially interacts with universally conserved residues of the 23S rRNA helices 43 and 95, and protein L11 within the 50S ribosomal subunit. Upon EF-G binding, both RRF and tRNA are driven towards the tRNA-exit (E) site, with a large rotational movement of domain II of RRF towards the 30S ribosomal subunit. During this intermediate step of the recycling process, domain II of RRF and domain IV of EF-G adopt hitherto unknown conformations. Furthermore, binding of EF-G to the PoTC·RRF complex reverts the ribosome from ratcheted to unratcheted state. These results suggest that (i) the ribosomal intersubunit reorganizations upon RRF binding and subsequent EF-G binding could be instrumental in destabilizing the PoTC and (ii) the modes of action of EF-G during tRNA translocation and ribosome-recycling steps are markedly different.
Keywords: conformation of elongation factor G(EF-G) during ribosome recycling, reverse ratcheting of the ribosome, ribosome-recycling factor (RRF), RRF–EF-G interactions
Introduction
After the termination step of translation, deacylated tRNA and mRNA remain bound to the bacterial 70S ribosome. This complex is called the post-termination complex (PoTC), which must be disassembled so that each of its components can participate in the next round of protein synthesis. In bacteria, disassembly of PoTC is carried out by the reaction that requires the binding of the ribosome-recycling factor (RRF) to PoTC, subsequent binding of the elongation factor-G (EF-G) in complex with guanosine 5′-triphosphate (GTP), followed by GTP hydrolysis (Hirokawa et al, 2006). This reaction drives the dissociation of two ribosomal subunits (Karimi et al, 1999; Hirokawa et al, 2005; Peske et al, 2005; Zavialov et al, 2005) and the release of mRNA and deacylated tRNA (Hirokawa et al, 2005). The initiation factor 3 (IF3) has also been implicated (Seshadri and Varshney, 2006) to play a role in the splitting of the 70S ribosome or in keeping the split subunits separated (Hirokawa et al, 2005). IF3 was also suggested to be involved in the release of deacylated tRNA from the 30S subunit (Karimi et al, 1999; Peske et al, 2005) (but see Hirokawa et al, 2006).
Atomic structures of RRF from several bacterial species (Selmer et al, 1999; Kim et al, 2000; Toyoda et al, 2000; Yoshida et al, 2001; Nakano et al, 2003; Saikrishnan et al, 2005) are known. Each of these structures shows two distinct domains: domain I has a three-helix bundle, while domain II is a three-layer β/α/β sandwich, which is connected to domain I through highly flexible linkers. Relative positions of the two domains among these atomic structures are not the same. Therefore, an inherent interdomain flexibility of RRF has been proposed to be important for its function (Selmer et al, 1999; Yoshida et al, 2001). The binding position of RRF on the ribosome has been studied by using a variety of biochemical and structural biology techniques, such as the hydroxyl radical probing (HRP) (Lancaster et al, 2002), cryo-electron microscopy (cryo-EM) (Agrawal et al, 2004; Gao et al, 2005; Barat et al, 2007) and X-ray crystallography (Wilson et al, 2005; Borovinskaya et al, 2007; Weixlbaumer et al, 2007; Pai et al, 2008; Dunkle et al, 2011). These studies have revealed a predominant binding position of RRF on the ribosome, henceforth referred to as position 1 (or P1). Another weak and transient binding position, referred to as position 2 (or P2), was captured on a small fraction of the dissociated large (50S) ribosomal subunits in one of the cryo-EM studies (Barat et al, 2007). In its position 1, domain I of RRF resides on the 50S ribosomal subunit, stretching over the ribosomal aminoacyl (A)- and peptidyl (P)-tRNA-binding sites (Hirokawa et al, 2002; Lancaster et al, 2002; Agrawal et al, 2004), while domain II of RRF flexibly occupies the intersubunit space between ribosomal protein S12 of the small (30S) ribosomal subunit and the L11 stalk-base (Sb) region of the 50S ribosomal subunit (Agrawal et al, 2004; Gao et al, 2005; Barat et al, 2007; Borovinskaya et al, 2007; Weixlbaumer et al, 2007; Pai et al, 2008; Dunkle et al, 2011).
In order to describe the binding positions of RRF on the ribosome in conjunction with interdomain flexibility of the two RRF domains, we introduced a nomenclature in our previous study (Barat et al, 2007). Accordingly, RRF binds to the ribosome in position 1, or position P1/IIa, where ‘P1’ and ‘IIa’ refer to binding position of domain I and relative orientation of domain II, respectively. Subsequently, domain II of RRF adopts a different orientation, referred to as position P1/IIb, suggesting that the domain I of RRF stayed in the same position with an altered relative orientation of domain II. In all previous structural studies of the RRF-bound ribosome complexes, domain II adopts slightly different versions of P1/IIb on the ribosome. In position 2, or P2/IIb, RRF is situated exclusively in the P-site region of the 50S ribosomal subunit such that domain II and the interdomain elbow region of the bound RRF would make a significant steric clash with the 30S ribosomal subunit's neck and head regions. This implies that RRF could achieve this position during, or immediately after, the dissociation of the two ribosomal subunits (Barat et al, 2007). Cryo-EM structures of the 50S·RRF·EF-G·GDPNP complex have also been studied, using the pre-dissociated 50S ribosomal subunit for the formation of the complex (Gao et al, 2005, 2007). However, capturing both RRF and EF-G simultaneously on the PoTC, or on the vacant 70S ribosome, for structural studies has been technically challenging, due to rapid disassembly of the PoTC complex in the presence of both RRF and EF-G.
To understand the molecular details of the ribosome-recycling process, it is essential to study the interaction between RRF and EF-G on the PoTC. In order to capture the structure of the PoTC·RRF·EF-G complex, together with mRNA and tRNA, we exploited the fact that such a complex can be stabilized by using RRF and EF-G from two different bacterial species, that is, when Thermus thermophilus RRF (ttRRF) and Escherichia coli EF-G (henceforth referred to as EF-G) in conjunction with E. coli PoTC (PoTC) (Raj et al, 2005) are used. The ttRRF·EF-G combination slows down the recycling reaction (Raj et al, 2005). However, increasing the concentration of both factors results in rapid disassembly of the PoTC (NI, TY, AK, RKA and HK, in preparation). Furthermore, the same species combination of the two factors, from either T. thermophilus or E. coli, readily disassembles the PoTC (Raj et al, 2005). These results suggest that the mode and site of binding of ttRRF on the PoTC are the same as that of an ecRRF and that the two-species combination is biochemically active. We prepared the PoTC·ttRRF complex and then incubated it with EF-G·GTP in the presence of fusidic acid (FA) to obtain the PoTC·ttRRF·EF-G·GDP·FA complex. FA stabilizes EF-G on the 70S ribosome in a post-GTP hydrolysis state (Agrawal et al, 1998; Valle et al, 2003; Datta et al, 2005; Gao et al, 2009; Ratje et al, 2010), and its antimicrobial activity is primarily due to inhibition of the ribosome-recycling step (Savelsbergh et al, 2009). The PoTC·ttRRF and the PoTC·ttRRF·EF-G·GDP·FA complexes were analysed by the three-dimensional (3D) cryo-EM technique (Frank et al, 2000). The cryo-EM map of the PoTC·ttRRF·EF-G·GDP·FA complex shows densities for both RRF and EF-G on the PoTC and reveals an intermediate binding position of RRF that lies between previously described binding positions P1 and P2. Molecular analysis of the complex revealed interactions between domain II of RRF and domains III–V of EF-G on the PoTC. In addition, both RRF and EF-G adopt hitherto unknown configurations on the ribosome. These findings provide important insights into the mechanism of ribosome recycling, and suggest that the mode of action of EF-G in the ribosome-recycling step is different from that in the tRNA translocation step.
Results
Structures of the PoTC·ttRRF complex and the PoTC·ttRRF·EF-G·GDP·FA complex
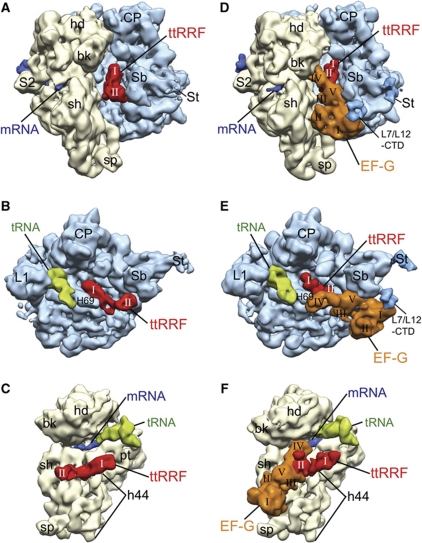
We isolated polysomes from growing E. coli cells and removed the peptidyl moiety, by treating with puromycin, to obtain the model PoTC. As described in the Introduction section, to capture both RRF and EF-G simultaneously on the PoTC, we used the combination of ttRRF, EF-G and PoTC. In order to establish that the binding position of ttRRF on PoTC is identical to the binding position of ecRRF, we first obtained a cryo-EM map of the PoTC·ttRRF complex (henceforth referred to as complex 1). The map shows distinct mass of densities corresponding to ttRRF, a tRNA at the P/E hybrid site (Moazed and Noller, 1989; Agrawal et al, 1999b) and the mRNA (Figure 1A–C; Supplementary Figure S1A for a stereo viewing), when compared with the structure of a vacant 70S ribosome (Schuwirth et al, 2005). The overall location of the density corresponding to domain I of ttRRF appears to be similar to those derived in previous studies (Lancaster et al, 2002; Agrawal et al, 2004; Gao et al, 2005; Barat et al, 2007; Borovinskaya et al, 2007; Weixlbaumer et al, 2007; Pai et al, 2008; Dunkle et al, 2011). However, domain II of RRF is predominantly oriented towards the 50S ribosomal subunit, making contacts with the stalk base (Sb) region (Figure 1A and B). This orientation of domain II of RRF on the 70S ribosome was first inferred from a hydroxyl radical probing (HRP) study (Lancaster et al, 2002) and subsequently observed in our previous cryo-EM studies (Agrawal et al, 2004; Barat et al, 2007), and was referred to as position P1/IIa (Barat et al, 2007). However, position P1/IIa has eluded all X-ray crystallographic studies (Borovinskaya et al, 2007; Weixlbaumer et al, 2007; Pai et al, 2008; Dunkle et al, 2011). These observations suggest that RRF in position P1/IIa is attained only in physiologically relevant solution conditions, such as hydroxyl radical probing (HRP) (Lancaster et al, 2002) and cryo-EM (Agrawal et al, 2004; Barat et al, 2007) conditions but not in crystallographic conditions, as all X-ray crystallographic structures of the 70S·RRF complexes invariably find RRF in the position P1/IIb (Borovinskaya et al, 2007; Weixlbaumer et al, 2007; Pai et al, 2008; Dunkle et al, 2011) with domain II of RRF oriented towards the 30S ribosomal subunit protein S12. In the complexes formed between the 70S ribosome and RRF in our previous cryo-EM studies, density for domain II was split between positions P1/IIa and P1/IIb (Agrawal et al, 2004; Barat et al, 2007). Since the interspecies combination of PoTC and ttRRF, allows us to capture the RRF predominantly in position P1/IIa, density corresponding to domain II is the strongest among all ribosome·RRF structures determined thus far. It is therefore possible to accurately dock all atom coordinates of domain II of ttRRF (Toyoda et al, 2000) into the cryo-EM density (Supplementary Figure S2A). A very weak density is also observed in position P1/IIb, suggesting that in a small fraction (∼10%) of the total population of complex 1 domain II of ttRRF is oriented towards the 30S ribosomal subunit (Supplementary Figure S2B and C). Since the binding position of domain I of ttRRF closely matches with that in E. coli 70S·RRF complexes (Agrawal et al, 2004; Barat et al, 2007; Pai et al, 2008) and the fact that ttRRF is capable of disassembling the PoTC with ttEF-G (Raj et al, 2005), the predominant P1/IIa position attained by the domain II of ttRRF on the PoTC must represent a functionally relevant binding state.
Figure 1.
Segmented cryo-EM maps of the PoTC·ttRRF and PoTC·ttRRF·EF-G·GDP·FA complexes. (A–C) The PoTC·ttRRF complex (complex 1): (A) The 70S ribosome (yellow, 30S subunit; and blue, 50S subunit) is viewed from the tRNA entry side, with strong densities for both domains (I and II) of ttRRF (red) clearly visible in the intersubunit space (see also Supplementary Figure S1A for a stereo viewing); (B) the same map is shown from the 50S interface side, where the 30S subunit has been computationally removed to reveal densities corresponding to ttRRF (in position P1/IIa) and deacylated tRNA (green, in the P/E site); and (C) the same complex is shown along with density corresponding to mRNA (dark blue) from the 30S interface side, without the 50S subunit. (D–F) The PoTC·ttRRF·EF-G·GDP·FA complex (complex 2), shown in matching views with (A–C): (D) Densities for both ttRRF and ecEF-G can be seen in the intersubunit space, with both domains of ttRRF and all five domains (I–V) of EF-G (orange) readily identifiable (see also Supplementary Figure S1B for a stereo viewing). Position occupied by domain II of ttRRF in (A–C) is now occupied by domain V of EF-G; (E) the same complex is shown from the 50S interface side (without 30S subunit) to reveal densities corresponding to ttRRF (in intermediate position Pi), EF-G, and deacylated tRNA (in the P/E and E sites); and (F) the same complex shown along with density corresponding to mRNA (dark blue; see Supplementary Figure S3 for complete mRNA density) from the 30S interface side, without the 50S subunit. A marked shift in RRF position can be seen when compared with the RRF position in (C). Landmarks of the 30S subunit: bk, beak; hd, head; pt, platform; sh, shoulder; sp, spur; h44, 16S rRNA helix 44; and S2, protein S2. Landmarks of the 50S subunit: CP, central protuberance; L1, protein L1 stalk; L7/L12-CTD, C-terminal domain of protein L7/L12; Sb, stalk base (protein L11 region); St, L7/L12 stalk; and H69, 23S rRNA helix 69. In (D, E), densities corresponding to extended St and L7/L12-CTD are shown at slightly lower threshold values than the threshold value used for displaying the rest of the 50S subunit.
By incubating complex 1 with EF-G, GTP and FA, we obtained the PoTC·ttRRF·EF-G·GDP·FA complex (henceforth referred to as complex 2). The computed cryo-EM map of complex 2 clearly shows distinct mass of densities for RRF, EF-G, tRNA and mRNA (Figure 1D–F; see also Supplementary Figure S1B for stereo viewing), when compared with the map of a vacant 70S ribosome (Schuwirth et al, 2005). Both domains of RRF (Selmer et al, 1999), all five domains of EF-G (Czworkowski et al, 1994; Ævarsson et al, 1994) and a tRNA can be readily identified in this map. However, densities of RRF and EF-G appear to be partially fused, suggesting a direct interaction between these two protein factors.
A comparison between the maps of complex 1 and complex 2 reveals a striking movement of RRF towards the tRNA-exit (E) site (or towards L1 stalk) upon EF-G binding (Figure 1; also described later), suggesting that a direct interaction between ttRRF and EF-G shifts the ttRRF from position P1/IIa (and position P1/IIb). We designate this new binding position of RRF as an intermediate position (or position Pi). During this shift, domain II of ttRRF severs its interaction with the L11 Sb region of the 50S ribosomal subunit and is rotated towards the 30S ribosomal subunit. The same Sb region makes contact with domain V of EF-G in complex 2 (Figure 1, compare panels D and E with panels A and B, respectively). In addition, we observe a strong mass of well-defined density corresponding to the C-terminal domain (CTD) of protein L7/L12 (Figure 1D and E) that forms a bridge (or the previously described ‘arc-like connection’ (Agrawal et al, 1998; Datta et al, 2005)) between EF-G and the Sb region of the 50S subunit. The map of complex 2 contains densities for both the mRNA and a tRNA (Figure 1D–F; also see Supplementary Figures S1 and S3). However, as compared with complex 1, the density corresponding to the anticodon end of the tRNA is significantly broadened (Figure 1, compare panel E with panel B) and partially bifurcated between P and E sites, suggesting that the codon–anticodon interaction is affected significantly upon EF-G binding (also described later).
Interactions between the PoTC and ttRRF in complex 1
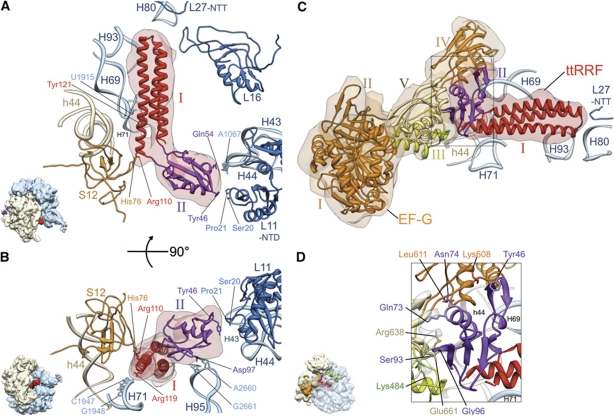
In order to determine molecular interactions between the PoTC and ttRRF, we docked the atomic coordinates of the ribosome (Schuwirth et al, 2005; Gao et al, 2009) and ttRRF (Toyoda et al, 2000) into corresponding densities within the cryo-EM map, using the MDFF program (Trabuco et al, 2008, 2009). This fitting reveals that domain I of ttRRF is situated within the cavity formed by helices 69, 71, 80 and 93 of the 23S rRNA (Figure 2A, see also Supplementary Movie S1), while its domain II interacts with the Sb of the 50S ribosomal subunit, involving helix 43 of the 23S rRNA and the N-terminal domain (NTD) of the ribosomal protein L11. Two amino-acid residues (Tyr46 and Gln54), located at the tip of domain II of RRF, appear to make direct contacts with the ribosome components. Tyr46 (E. coli # Tyr45 (or ecTyr45); see Supplementary Figure S4 for aligned sequences) is in close vicinity to Ser20 and Pro21 of protein L11, while Gln54 (ecGln53) would make a protein–RNA contact with the highly conserved nucleotide residue A1067 within helix 43 of the 23S rRNA (Figure 2A). Arg110 (ecArg109) in the elbow region of ttRRF lies in close proximity to amino-acid residue His76 of protein S12. Tyr121 (ecGlu120) of domain I of ttRRF would contact U1915 of helix 69 (H69) of the 23S rRNA. In addition, Asp97 (ecSer96) of domain II of ttRRF lies in close proximity to two other conserved nucleotides, A2660 and G2661, within the α-sarcin-ricin stem-loop (H95) of the 23S rRNA (Figure 2B). Arg119 (ecArg118) lies close to C1947 and G1948 of helix 71 (H71) of the 23S rRNA. Thus, while the interactions between domain I of RRF and the ribosome are essentially the same as derived in previous X-ray crystallographic studies (Borovinskaya et al, 2007; Weixlbaumer et al, 2007; Pai et al, 2008; Dunkle et al, 2011), interactions between domain II of RRF and the ribosome are found to be dramatically different from what has been reported in those studies. This is due to the fact that we captured the ttRRF predominantly in P1/IIa configuration, while P1/IIb is the preferred configuration in crystallographic studies. It is likely that the P1/IIa configuration precedes the P1/IIb configuration on the PoTC (see Discussion).
Figure 2.
Interactions between the ribosome and ttRRF in complex 1 (A, B), and between ttRRF and EF-G in complex 2 (C, D). (A) Flexibly fitted atomic structure of ttRRF (red and purple ribbons, PDB ID; 1EH1) into the corresponding RRF density (semitransparent pink). Contacts and proximities between the amino-acid residues of ttRRF and ribosomal components, such as amino-acid residues of proteins L11 and S12 and nucleotides of the 23S rRNA helices 43 (H43) and 69 (H69) are indicated. Domains I and II of the fitted ttRRF coordinates are depicted in red and purple, respectively. (B) Same as in (A), but rotated around a horizontal axis by ∼90°, to reveal interactions between an amino-acid residue of domain II of ttRRF and conserved nucleotides of the 23S rRNA helix 95 (H95), and between domain I of ttRRF and helix 71 (H71) of the 23S rRNA. (C) Flexibly fitted atomic structures of ttRRF and the homology model of E. coli EF-G into the corresponding cryo-EM densities of ttRRF (semitransparent red, in the intermediate position Pi) and EF-G (semitransparent orange), respectively, are shown with the ribosomal components present in the immediate vicinity of RRF. EF-G domains I–V are shown in distinctive colours, I (orange), II (brown), III (green), IV (orange) and V (yellow). L27-NTT refers to the N-terminal tail of protein L27. (D) Enlarged boxed area in (C) to reveal ribosomal neighbourhood of ttRRF in the position Pi and interaction of domain II of ttRRF with EF-G domains III–V. Five select pairs of amino-acid interactions are labelled (see also Supplementary Table S1B for the complete list of amino-acid interactions between ttRRF and EF-G). All helices of the 16S and 23S rRNAs are identified with h and H, respectively; while ribosomal proteins of the small and large subunits are prefixed by S and L, respectively. Thumbnails to the lower left of (A, B, D) depict overall orientations of the ribosome.
Interactions among the ribosome, ttRRF and EF-G in complex 2
Similarly to complex 1, we flexibly docked the atomic structures of the ribosome (Schuwirth et al, 2005; Gao et al, 2009), ttRRF (Toyoda et al, 2000) and a homology model of the E. coli EF-G, which was built using the structural templates of ttEF-G (Czworkowski et al, 1994; Ævarsson et al, 1994), into corresponding densities within the cryo-EM map of complex 2 (Figure 2C; Supplementary Figure S2D for a stereo viewing; and Supplementary Movie S2). Based on these fitting results, we examined the interactions among ttRRF, EF-G, and the ribosomal components. Similarly to complex 1, domain I of ttRRF is situated within the cavity formed by helices 69, 71, 80 and 93 of 23S rRNA (Figure 2C). However, upon EF-G binding, specific contacts between RRF domains and the ribosome change significantly (Supplementary Table S1A).
Several amino-acid residues within domains III–V of EF-G interact with domain II of RRF (Figure 2D; Supplementary Table S1B). Lys484 on domain III of EF-G appears to contact Ser93 (ecAsn92) of ttRRF. Lys608 and Leu611 of EF-G domain IV, near the linker region between domains IV and V, would contact Tyr46 (ecTyr45) and Asn74 (ecSer73) of ttRRF, respectively. Glu661 and Arg638 on domain V of EF-G is in close vicinity to amino-acid residues Gly96 (ecGly95) and Gln73 (ecArg72) of ttRRF. Domain II of ttRRF also contacts H69 of the 23S rRNA. Thus, domain II of RRF is somewhat sandwiched between EF-G and the ribosome components. In addition, Tyr121 (ecGlu120) in domain I of RRF appears to switch partner from H69 nucleotide U1915 in position P1 to A1916 in position Pi (Supplementary Table S1A).
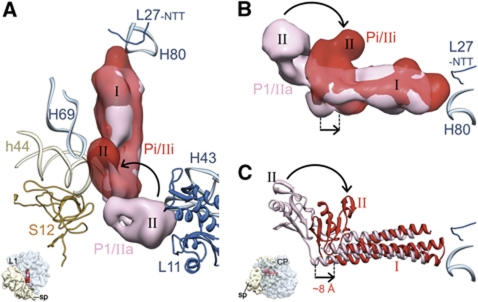
As pointed out earlier, there is a marked shift in overall position of ttRRF (Figure 3A), as can be inferred by comparing the densities corresponding to the elbow region of ttRRF, before and after EF-G binding (Figure 3B). Domain I of ttRRF is shifted towards the E site. Flexible fitting shows that domain I of ttRRF is shifted by ∼8 Å such that the long axis of domain I of ttRRF is not perfectly parallel to that in position P1 (Figure 3C). Furthermore, the relative orientation of domain II of RRF is different from previously described IIa and IIb configurations (Barat et al, 2007) (Supplementary Figure S5). Therefore, this intermediate RRF-binding position is referred to as position Pi/IIi, where IIi refers to a unique relative orientation of domain II of ttRRF (Supplementary Figures S5 and S6). Thus, in addition to pushing the RRF towards the E site, the binding of EF-G to complex 1 also induces a rotational shift in domain II of ttRRF, from the 50S-subunit side to the 30S-subunit side (curved arrows in Figure 3).
Figure 3.
Comparison of ttRRF-binding positions before and after EF-G binding. (A, B) Cryo-EM densities and (C) flexibly fitted atomic coordinates corresponding to ttRRF in complex 1 (pink) and complex 2 (red) are superimposed. The overall direction and magnitude of ttRRF movement is depicted by straight arrows. The direction of rotational shift in domain II of ttRRF, upon EF-G binding, is indicated with curved arrows (see also Supplementary Figure S6). Thumbnails to the lower left in (A, C) depict the orientation of the 70S ribosome: In (A), the ribosome is in top view, whereas in (B, C) it is shown from the L7/L12 stalk side of the 50S subunit to reveal the shift in the elbow region of RRF.
Because of a pre-bound ttRRF, ecEF-G is unable to attain its previously known binding position on the ribosome in the 70S·EF-G·GDP·FA complex (Agrawal et al, 1998; Datta et al, 2005; Gao et al, 2009; Ratje et al, 2010). Domains IV and V of EF-G are situated more towards the shoulder of the 30S subunit within the intersubunit space, when compared with their positions in the ec70S·ecEF-G·GDP·FA complex (Datta et al, 2005) (Figure 4). Domain IV of EF-G adopts a skewed conformation, where both its tip and the proximal end, that is, the end continuing with domains III and V, are shifted by 8–9 Å towards the shoulder of the 30S ribosomal subunit (Figure 4C). In addition, the flexible docking, places the C-terminal α-helix of EF-G in a markedly different position. However, there are no appreciable changes of domain I and II of EF-G as compared with what has been observed during EF-G-dependent tRNA translocation on the ribosome. Nevertheless, these observations suggest that EF-G can adopt more than one conformational state on the ribosome in the post-GTP hydrolysis state and that the mechanism of interaction of EF-G with the RRF-bound PoTC is significantly different from that with the pre-translocational ribosome complex.
Figure 4.
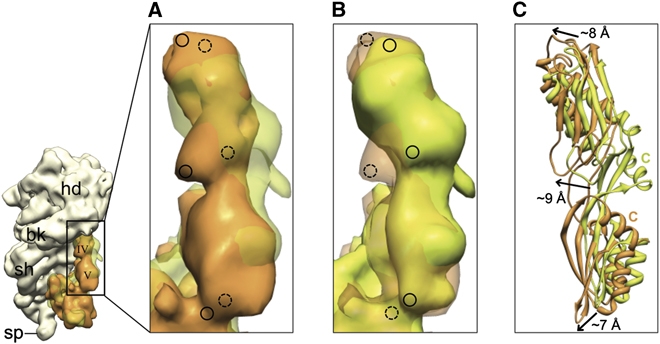
Positions of domains IV and V of EF-G in the presence and absence of ttRRF on the ribosome. (A) Corresponding cryo-EM densities of domains IV and V of EF-G in complex 2 (orange) and the 70S·EF-G·GDP·FA complex (semitransparent green (Datta et al, 2005)) are superimposed. (B) Same as in panel A, but the transparency has been switched to reveal marked shifts in the position of EF-G domains. Dashed and solid circles in (A, B) highlight the equivalent positions in semitransparent and solid densities. (C) Flexibly fitted coordinates of EF-G domains IV and V into the maps shown in (A) are superimposed. Arrows depict the direction and magnitude of shifts in EF-G domains as compared with their positions in the absence of ttRRF. C, C-terminal α-helix.
Dynamic movements of domain II of RRF
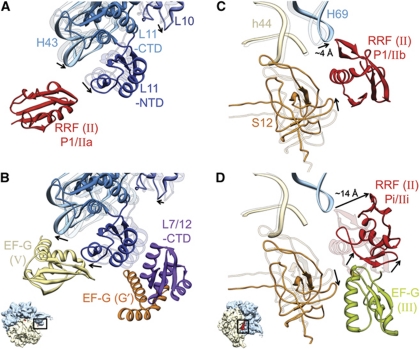
Domain II of RRF has been suggested to be the main functional component of the RRF structure (Guo et al, 2006). We have indentified three configurations of domain II on the ribosome, namely IIa, IIb and IIi. The binding positions of domain I of RRF on the ribosome are essentially the same in P1/IIa and P1/IIb configurations. It is possible that the domain II exists in a dynamic equilibrium between these two configurations. We find that specific ribosomal components switch their binding partners from domain II of RRF (in both positions P1/IIa and P1/IIb) to specific domains of the subsequently bound EF-G. The cleft region between tip of the 23S rRNA helix 43 and NTD of protein L11 in the Sb region of the 50S subunit, previously occupied by domain II of ttRRF in position P1/IIa within complex 1 (Figure 5A), is subsequently occupied by domain V of EF-G in complex 2 (Figure 5B). Similarly, the interaction between protein S12 of the 30S ribosomal subunit and domain II of RRF in position P1/IIb (Agrawal et al, 2004; Gao et al, 2005; Barat et al, 2007; Borovinskaya et al, 2007; Weixlbaumer et al, 2007; Pai et al, 2008; Dunkle et al, 2011) would be severed upon EF-G binding. The domain II of RRF in position Pi/IIi has moved away from protein S12. The space previously occupied by domain II of RRF in its P1/IIb position (Figure 5C) is partially occupied by domain III of ecEF-G in complex 2 (Figure 5D). Furthermore, the interactions between the tip of the 23S rRNA helix 95 (the SRL) and domain II of RRF in position P1/IIa (Figure 2B) would be disrupted upon its transition to P1/IIb as well as upon EF-G binding. These results clearly show that RRF has moved slightly towards the E site upon EF-G binding.
Figure 5.
Movements of domain II of RRF, and conformational changes of ribosomal components that interact with RRF and EF-G. Fitted atomic structures of the ribosomal components in two functionally relevant regions are shown. (A, B) The L11 Sb region; and (C, D) the bridge B2a region, shown along with protein S12. In each panel, fitted structures of two functional states are shown. In (A, C), the ribosomal components of the PoTC complex are shown as semitransparent ribbons, while those of complex 1 are shown as solid ribbons. Similarly, in (B, D), ribosomal components of complex 1 are shown as semitransparent ribbons, whereas those of complex 2 are shown as solid ribbons. Arrows point to movements in the ribosomal components and RRF. Domain II of RRF in specified positions (A, C, D), domains V and G’ of EF-G (B), and domain III of EF-G (D) are also shown. Since only a weak density for domain II of RRF in P1/IIb configuration was observed in this study, its corresponding position in (C) is based on previous studies (Agrawal et al, 2004; Pai et al, 2008). The position of L7/L12-CTD shown in (B) is similar to that derived in previous cryo-EM (Datta et al, 2005) and X-ray crystallographic (Gao et al, 2009) studies (also see Supplementary Figure S11). Thumbnails to lower left of (B, D) depict the orientations of the 70S ribosome in (A, B) and (C, D), respectively.
Conformational changes of ribosomal components that interact with RRF and EF-G
The ribosomal components that interact with domain II of RRF also undergo large conformational changes during the transition from P1/IIa (or P1/IIb) to Pi/IIi. For example, H43 in the Sb region moves towards the solvent side by ∼5 Å upon binding of ttRRF alone (Figure 5A). Upon subsequent binding of EF-G, the Sb shifts back towards the intersubunit space by ∼5 Å and the CTD of protein L7/L12 forms a bridge between the NTD of L11 and the G′ domain of EF-G (Figure 5B), corroborating our previous study (Datta et al, 2005) and indicating that the CTD of L7/12 adopts this relatively stable position on the ribosome in a post-GTP hydrolysis state, both during the tRNA translocation and recycling steps. In addition, we observe a distinct movement in density corresponding to protein S12 on the intersubunit face of the 30S-subunit body. Upon ttRRF binding (in both P1/IIa and P1/IIb configurations), S12 moves towards the 16S rRNA helix 44 (Figure 5C), and reverts back, close to its original position, upon subsequent binding of EF-G (Figure 5D).
Upon RRF binding to the PoTC, helix 69 (H69) of the 23S rRNA undergoes a conformational change. The tip of H69 is shifted by ∼4 Å towards the rest of the 50S ribosomal subunit, apparently severing its contact with helix 44 of the 16S rRNA (Figure 5C) to disrupt the bridge B2a (Yusupov et al, 2001). This movement in H69 is consistent with some of the previous cryo-EM (Agrawal et al, 2004; Barat et al, 2007) and X-ray crystallographic (Borovinskaya et al, 2007; Pai et al, 2008) studies; however, the overall movement is less pronounced than what was reported in those X-ray studies. The altered position of tip of H69 remains unchanged between P1/IIa and P1/IIb states and upon subsequent binding of ecEF-G to complex 1 (Figure 5D). In other words, the position of H69 is found to be the same in both complexes 1 and 2. Except for the helix 69, no other rRNA helices that are known to interact with domain I of RRF (e.g., H71, H80 and H93) show any noticeable movement in either of our complexes, when compared with their respective positions in the map of control PoTC.
Global conformational changes of the ribosome and tRNA movement
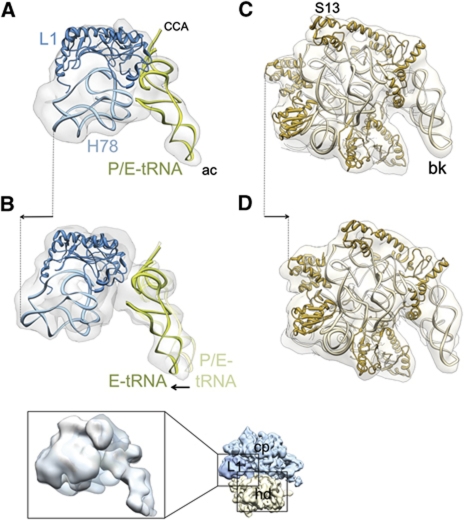
In addition to the conformational changes described in the previous section, there are global conformational changes of the ribosome in both complexes 1 and 2 (Supplementary Figure S7; Supplementary Movie S3). This includes a marked movement of the L1 stalk due to RRF binding and upon subsequent binding of EF-G (Figure 6A and B). While RRF binding moves the L1 stalk towards the intersubunit space, the subsequent binding of EF-G moves it in the opposite direction, that is, towards the solvent side. The deacylated tRNA, which is exclusively situated in the P/E state in complex 1 (Figure 6A), is present in both P/E and E/E states in complex 2 (Figure 6B, also see Supplementary Figure S8), suggesting that the anticodon end of the tRNA has been pulled into the E site in a significant population of complex 2, along with outward movement of the L1 stalk upon EF-G binding. In addition, the ribosomal components corresponding to intersubunit bridges B1a-c and B3 undergo large conformational changes upon ttRRF binding (complex 1), corroborating our previous study (Barat et al, 2007), and upon EF-G binding (complex 2) (Supplementary Figure S7). Furthermore, the 70S ribosome in complex 1 is in the ratcheted state (Supplementary Figure S7), where the 30S subunit is found rotated in an anticlockwise direction relative to the 50S subunit (Agrawal et al, 1999a; Frank and Agrawal, 2000). This observation is consistent with previous studies (Gao et al, 2005; Barat et al, 2007). However, in complex 2, the 30S subunit is not rotated in an anticlockwise direction relative to the 50S subunit. In other words, subsequent binding of EF-G to complex 1 brings back the 30S subunit to its unratcheted state (Figure 6C and D; Supplementary Movie S3), which is unusual for the EF-G-bound 70S ribosome. It should be noted that the movement of the 30S subunit head is in a direction opposite to the earlier described L1 stalk movement that takes place upon binding of EF-G (indicated with arrows in panels B and D of Figure 6). This observation also implies that modes of action of EF-G are markedly different between tRNA translocation and ribosome-recycling steps of protein synthesis.
Figure 6.
Conformational changes of the ribosome and tRNA movement due to binding of RRF and EF-G. Each structure was aligned, using the core portion of the 50S subunit of the cryo-EM maps as the main guide. (A, B) Movement of the L1 stalk (light blue, rRNA helices 77 and 78; dark blue, protein L1) and tRNA (green) during transition from (A) complex 1 to (B) complex 2. (C, D) Movement of the 30S subunit head (light brown, 16S rRNA; dark brown, 30S subunit head proteins) during transition from (C) complex 1 to (D) complex 2. Cryo-EM densities are shown as semitransparent grey. In all panels, the ribosomes are viewed from the top, in an overall orientation depicted in the thumbnail at the lower right. In (A, B), only the L1 stalk region (corresponding to left boxed area on the thumbnail) of the 50S subunits, and in (C, D), only the head portion (corresponding to lower right boxed area on the thumbnail) of the 30S subunits are shown (see also Supplementary Figure S7 for the overall conformational changes). The inset shown below (B) depicts the split densities for tRNA anticodons between the P (semitransparent) and E (solid) sites in complex 2. Arrows indicate major movements (see Supplementary Figure S8 for stereo viewing of the tRNA movement). Landmarks: ac, anticodon end; CCA, acceptor end; H78, helix 78 of the 23S rRNA; S13, small ribosomal subunit protein S13. The rest of the landmarks of the ribosomal subunits are the same as in Figure 1.
Discussion
Capturing both RRF and EF-G simultaneously on the PoTC is essential for understanding the mechanism of the ribosome-recycling step. Since the mRNA and deacylated tRNA remain bound to the ribosome (Figure 1D–F) and the ttRRF has advanced slightly (by ∼8 Å) towards the tRNA-exit (E) site (Figure 3), the PoTC·ttRRF·EF-G·GDP·FA complex (complex 2) represents an intermediate state of the ribosome recycling that is stalled by FA immediately after EF-G-dependent GTP hydrolysis. The Pi/IIi position of ttRRF, captured in this study in conjunction with ecEF-G, is significantly different from both previously identified positions P1/IIa and P1/IIb. In position Pi/IIi, the tip of domain 1 of ttRRF is shifted slightly upward (Figure 3B and C) such that it is situated between the tip of domain I at positions P1 and the previously described short-lived position P2 (Barat et al, 2007), indicating the path of RRF during its transition from positions P1 to P2 on the ribosome. A comparison of mRNA densities between the PoTC·ttRRF complex (complex 1) and complex 2, along entire mRNA path (Yusupova et al, 2006; Supplementary Figure S3), shows that the contact points between mRNA and the ribosome remain largely unaffected at this stage. However, there is a marked shift in the tRNA's anticodon from the P site to the E site in a subpopulation of complex 2 (Figure 6B; Supplementary Figure S8). This shift in the tRNA's anticodon appears to be coupled to the movement of the L1 stalk towards the solvent side, which could facilitate the ultimate tRNA release from the PoTC. Since the codon–anticodon interaction is expected to be relatively weak at the E site (Jenner et al, 2007), it is possible that the mRNA and tRNA releases from the PoTC are triggered during this intermediate state.
The intersubunit ratchet-like rotation (Agrawal et al, 1999a; Frank and Agrawal, 2000) is essential for the tRNA translocation (Horan and Noller, 2007). Normally, during EF-G-dependent tRNA translocation, EF-G binding stabilizes the ribosome in ratcheted state (Agrawal et al, 1999a; Frank and Agrawal, 2000). In sharp contrast, we find that EF-G binding to the complex 1 reverts the ribosome to its unratcheted state (Supplementary Figure S9). It is possible that during its initial binding to the complex 1, the tip of domain IV of EF-G instantaneously pushes the 30S subunit head towards the E site, facilitating the movement of tRNA anticodon from P to E site. However, due to the presence of ttRRF, the rest of domain IV, along with the insertion domain V, of EF-G is not able to occupy its usual binding position on the ribosome (Figure 4). This steric constraint would force the tip of domain IV to retract back, and in the process, pull back the 30S subunit head to an unratcheted state (Figure 6C and D). Furthermore, there are marked differences in contact points of EF-G with tRNA and RRF between EF-G-dependent tRNA translocation and recycling processes. During tRNA translocation, the tip of EF-G interacts with the anticodon of tRNA (Gao et al, 2009), whereas during recycling, there is no direct interaction between EF-G and tRNA, instead, the junction between domains III–V of EF-G interacts with domain II of RRF (Figure 2C and D). Thus, modes of EF-G action are strikingly different during tRNA translocation and recycling steps of protein synthesis. In fact, in mammalian mitochondria, a separate isoform of EF-G is recruited exclusively for the recycling step (Tsuboi et al, 2009). Similarly, existence of two separate isoforms of EF-G, one each for the tRNA translocation and recycling steps, has also been reported in a bacterial species (Suematsu et al, 2010).
Previous cryo-EM (Agrawal et al, 2004; Barat et al, 2007) and X-ray crystallographic studies (Borovinskaya et al, 2007; Pai et al, 2008) have shown a large movement in the 23S rRNA helix 69 (H69) upon RRF binding to the 70S ribosome. The tip of H69 has been found to be moving away from the 16S rRNA helix 44 (h44) of the 30S subunit, implying disruption of an intersubunit bridge B2a to facilitate the subunit dissociation during the recycling process. These findings corroborate previous studies showing that the rate of ribosomal subunits association is directly affected by deletion (Ali et al, 2006) or mutations (Hirabayashi et al, 2006) of H69. In addition, several conformational changes associated with components of other intersubunit bridges, such as B1a-c, and B3 were also noted (Barat et al, 2007). We observe similar conformational changes involving bridges B1a-c and B2a in the maps of both complexes 1 and 2 (Supplementary Figure S7). These changes might contribute to the dissociation of ribosomal subunits during the recycling steps. Interestingly, one of the X-ray crystallographic studies of the 70S·RRF complex (Weixlbaumer et al, 2007) such conformational changes, including those previously described for H69, were not reported. In that study, an anticodon stem-loop (ASL) was pre-bound in the ribosomal P site. Since an intact tRNA molecule, including the CCA end, is required for tRNA to adopt the P/E state (Joseph and Noller, 1998), it is likely that ASL stabilized the bridge B2a in that 70S·ASL·RRF complex by maintaining a P/P-like state.
Previous cryo-EM studies of a complex of RRF and EF-G with the pre-dissociated 50S subunit (Gao et al, 2005, 2007) placed the highly flexible domain II of RRF in a position that is firmly occupied by the 16S rRNA helix 44 of the 30S subunit in its natural substrate, PoTC. Since a pre-dissociated 50S subunit is not a substrate for ribosome recycling, and since RRF bound to the pre-dissociated 50S subunit cannot be removed by EF-G while RRF bound to the 70S ribosome is removed by EF-G (Kiel et al, 2003), the binding of RRF and EF-G to pre-dissociated 50S subunit is less likely to represent an actual step of the process of recycling.
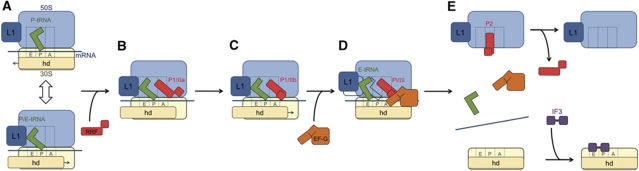
Based on the results presented here and previous studies (Agrawal et al, 2004; Barat et al, 2007), we outline the possible sequence of events of the ribosome recycling (Figure 7). The PoTC carries mRNA and a deacylated tRNA, which fluctuates between P/P and P/E states. This fluctuation is coupled to the unratcheted and ratcheted states, respectively, of the ribosome (Figure 7A; also see Dunkle et al, 2011). Upon RRF binding, the PoTC is locked in a ratcheted state, accompanied with distortion (or partial destabilization) of an intersubunit bridge, B2a. Since the ratcheted state of ribosome is observed irrespective of the relative orientation of domain II (Gao et al, 2005; Barat et al, 2007; Dunkle et al, 2011), interaction with domain I must be responsible for locking the ribosome in the ratcheted state. It is likely that RRF binds to the PoTC in P1/IIa configuration, as domain II in this case is extended more to the solvent side with interaction with the functionally important and flexible Sb region of the 50S subunit (Figure 7B). This initial interaction may trigger a movement of RRF's flexible domain II to the P1/IIb configuration such that tip of domain is reoriented into the intersubunit space and towards protein S12 of the 30S subunit (Figure 7C), as also proposed in our previous study using ecRRF on the ecPoTC (Barat et al, 2007). While the possibility that RRF is in a dynamic equilibrium between P1/IIa and P1/IIb configurations cannot be ruled out and the question that which of the two configurations present the substrate for the subsequent binding of EF-G to complex 1 remains unresolved, a spontaneous P1/IIa to P1/IIb transition presents a logical sequence of events as position of domain II of RRF in P1/IIb is close to that attained after EF-G binding (Supplementary Figures S5 and S6). Upon EF-G binding, RRF advances to an intermediate position Pi/IIi and the anticodon of the deacylated tRNA from P to E site, with mRNA staying firmly bound to the ribosome, while the 30S subunit reverts back to an unratcheted state (Figure 7D). Since the reverse ratchet (or the unratcheted state) is not an energetically favoured state for the EF-G-bound ribosome (see Munro et al, 2009), complex 2 represents a high-energy state that ultimately leads to disassembly of the PoTC by pushing RRF to previously observed position P2 (Barat et al, 2007; Figure 7E). As previously suggested (Barat et al, 2007), RRF at position P2 has an extremely low affinity for the dissociated 50S subunit, and is spontaneously released after the ribosomal subunit dissociation.
Figure 7.
Schematic diagram showing steps of the recycling process. (A) The model PoTC, with 30S (yellow) and 50S (blue) subunit. The head (hd) of the 30S subunit is depicted in a darker-shade block to indicate its relative position with respect to the rest of the 30S subunit body. Three canonical tRNA-binding sites, A, P and E, are depicted by dotted lines on both ribosomal subunits. The deacylated tRNA (green) is shown to fluctuate between P/P (P-tRNA, upper panel) and P/E (P/E-tRNA, lower panel) sites between the two ribosomal populations in dynamic equilibrium. (B) Initial binding of RRF in position P1/IIa, where domain II of RRF is oriented towards the 50S subunit and tRNA is exclusively in the P/E site, with the 30S subunit rotated in an anticlockwise direction (complex 1). (C) Subsequent reorientation of domain II towards the 30S subunit to attain position P1/IIb. (D) Binding of EF-G shifts RRF to position Pi/IIi, tRNA anticodon moves from P to E site in a subpopulation of complex 2, while the head of the 30S subunit is fully rotated back to its original position. (E) Depiction of final disassembly of the PoTC through concomitant movement of RRF to P2/IIb, a position exclusively attained on the 50S subunit during the subunit dissociation (Barat et al, 2007). The precise sequence of events that leads to release of all bound components is not known, while binding of IF3 to the 30S subunit keeps the latter from re-associating with the 50S subunit to facilitate the translation re-initiation. Relative positions of the blocks representing 30S subunit body and head in (A–D) depict movements of those 30S-subunit domains with respect to the 50S subunit.
Materials and methods
Preparation of ribosomal complexes
The E. coli polysome, EF-G and ttRRF were prepared as described previously (Raj et al, 2005). The PoTC·ttRRF complex (complex 1) was obtained by incubating 32 nM polysome, 100 μM puromycin with 41 μM ttRRF in the buffer R, containing 50 mM Tris–HCl (pH 7.5), 10 mM Mg(OAc)2, 25 mM KCl for 20 min at 30°C. Binding of ttRRF to the PoTC was ascertained by passing a portion of the reaction mixture through NanoSep 300k Omega to remove unbound ttRRF and then the isolated complex 1 was subjected to quantitative western blot using anti-ttRRF antibody (Raj et al, 2005). The PoTC·ttRRF·EF-G·GDP·FA complex (complex 2) was prepared by incubating complex 1 with 4 μM EF-G and 1.5 mM GTP for 30 min at 30°C, followed by the addition of 200 μM FA and continued incubation for another 10 min. As described for complex 1, a portion of the reaction mixture was analysed by quantitative western blot for the presence of ttRRF and EF-G. The cryo-EM grids were prepared immediately after the preparation of the complexes.
Cryo-EM and image processing
Cryo-EM grids were prepared using standard protocol (Grassucci et al, 2007) with 32 nM ribosomal complexes, and data were collected on a Tecnai F20 field emission gun electron microscope (FEI) at liquid nitrogen temperature, using an Oxford cryo-holder (Oxford Instruments) at 200 kV and under low-dose conditions. Images were recorded on film (SO-163, Kodak) at magnification calibrated at × 51 282. Micrographs were scanned by a PhotoScan scanner (Z/I imaging) with a step size of 14 μm, which corresponds to 2.78 Å pixel size on the object scale. For complex 1, a total of 153 927 images were manually picked from 195 micrographs, covering a defocus range from −0.4 to −4.3 μm. After cross-correlation-based screening, 112 425 images were used in the final reconstruction. For complex 2, a total of 338 823 images were manually picked from 383 micrographs, covering a defocus range from −0.4 to −4.0 μm. After cross-correlation-based screening, 245 866 images were used in the final reconstruction. The methods of supervised classification (Valle et al, 2002), using vacant ratcheted- and unratcheted-state ribosome maps (Agirrezabala et al, 2008) as two references, and maximum-likelihood 3D classification (Scheres et al, 2007) were applied to separate possible conformational states within data sets of both complex 1 and complex 2. These analyses suggested that particle images used for both complexes were highly homogeneous. However, the possibility of existence of low levels of conformational heterogeneity in both data sets cannot be ruled out. The SPIDER (Shaikh et al, 2008) was used for all image processing, including particle selection, alignment and 3D reconstruction. Projection matching (Penczek et al, 1994) in conjunction with small-angle alignment (up to 0.2° angular spacing) was employed for the iterative refinements. Resolutions of the final cryo-EM maps of complex 1 and complex 2 were 11.1 Å (at 0.5 cutoff of FSC; or 9.4 Å at 0.143 cutoff of FSC (Rosenthal and Henderson, 2003)) and 9.9 Å (at 0.5 cutoff of FSC; or 8.4 Å at 0.143 cutoff of FSC), respectively (Supplementary Figure S10; Supplementary Table S2).
Molecular interpretations
For molecular interpretations, atomic structures of the ribosomes (Schuwirth et al, 2005) (PDB IDs; 2AVY and 2AW4), ttRRF (Toyoda et al, 2000) (PDB ID; 1EH1) and the homology model of ecEF-G, generated using a protein homology modelling server, SWISS-MODEL (Arnold et al, 2006), were used. Initially, coordinates of all domains of RRF and EF-G were individually aligned to the corresponding cryo-EM densities. The independently fitted domains were then linked together using LOOPY (Xiang et al, 2002). The manually fitted models were further refined by flexible fitting and energy minimization using MDFF (Trabuco et al, 2008, 2009). Both RRF and EF-G models fit extremely well into corresponding cryo-EM densities, as judged by high cross-correlation coefficient values of 0.87 for the ttRRF in complex 1, and 0.89 and 0.94 for the ttRRF and ecEF-G, respectively, in complex 2. Densities remained after attributing densities to the ribosome, tRNA, RRF and EF-G were assigned to mRNA, which matched closely with the previously determined path of mRNA (Yusupova et al, 2006; Schuette et al, 2009). The achievable accuracy of fittings of atomic models into cryo-EM maps has been estimated to be 6–10 times better than the resolution of the cryo-EM map (Rossmann, 2000), which would translate to an average value of ∼1.4 Å in our case. Chimera software (Pettersen et al, 2004) was used for all visualization work.
Accession codes
The cryo-EM maps of both complexes 1 and 2 have been deposited in the EM database (http://emdatabank.org/) with accession codes EMD-1915 and EMD-1917, respectively. The extracted factor densities from both complexes have also been deposited with accession codes EMD-1916 and EMD-1918, respectively. The fitted coordinates of ttRRF, ecEF-G and relevant components of the ribosome structure corresponding to complex 1 and complex 2 have been deposited in the protein data bank (http://www.rcsb.org) under PDB ID codes 3J0D and 3J0E, respectively.
Supplementary Material
Acknowledgments
We thank Timothy Booth for some of the cryo-EM data collection, Kae Yokoyama for help with image processing, and Nilesh Banavali for help with the MDFF program. We also thank Aymen Yassin and Manjuli Sharma for helpful discussions. TY was in part and NI was fully supported through a Creative Biomedical Research Institute grant (to AK and HK). This work was supported by the NIH Grant R01 GM61576 (to RKA).
Author contributions: HK, AK, and RKA designed the research. TY prepared the complexes, collected cryo-EM data, performed image processing, flexible docking and molecular interpretation. TRS helped with image processing. TY and RKA analysed and interpreted the data. NI provided purified ttRRF, EF-G and polysomes and performed biochemical assays. TY, HK, AK and RKA wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ævarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J 13: 3669–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J (2008) Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 32: 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J (1999a) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol 6: 643–647 [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J (1999b) Effect of buffer conditions on the position of tRNA on the 70S ribosome as visualized by cryoelectron microscopy. J Biol Chem 274: 8723–8729 [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Frank J (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci USA 95: 6134–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J (2004) Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc Natl Acad Sci USA 101: 8900–8905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF (2006) Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell 23: 865–874 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK (2007) Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol Cell 27: 250–261 [DOI] [PubMed] [Google Scholar]
- Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH (2007) Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 14: 727–732 [DOI] [PubMed] [Google Scholar]
- Czworkowski J, Wang J, Steitz TA, Moore PB (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J 13: 3661–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK (2005) Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell 20: 723–731 [DOI] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH (2011) Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Agrawal RK (2000) A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 318–322 [DOI] [PubMed] [Google Scholar]
- Frank J, Penczek P, Agrawal RK, Grassucci RA, Heagle AB (2000) Three-dimensional cryoelectron microscopy of ribosomes. Methods Enzymol 317: 276–291 [DOI] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Ehrenberg M, Frank J (2007) Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits. J Mol Biol 374: 1345–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J (2005) Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell 18: 663–674 [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V (2009) The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Frank J (2007) Preparation of macromolecular complexes for cryo-electron microscopy. Nat Protoc 2: 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Zhang L, Zhang H, Feng Y, Jing G (2006) Domain II plays a crucial role in the function of ribosome recycling factor. Biochem J 393: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi N, Sato NS, Suzuki T (2006) Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J Biol Chem 281: 17203–17211 [DOI] [PubMed] [Google Scholar]
- Hirokawa G, Demeshkina N, Iwakura N, Kaji H, Kaji A (2006) The ribosome-recycling step: consensus or controversy? Trends Biochem Sci 31: 143–149 [DOI] [PubMed] [Google Scholar]
- Hirokawa G, Kiel MC, Muto A, Selmer M, Raj VS, Liljas A, Igarashi K, Kaji H, Kaji A (2002) Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J 21: 2272–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G, Nijman RM, Raj VS, Kaji H, Igarashi K, Kaji A (2005) The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA 11: 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan LH, Noller HF (2007) Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA 104: 4881–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Rees B, Yusupov M, Yusupova G (2007) Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Rep 8: 846–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S, Noller HF (1998) EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J 17: 3478–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell 3: 601–609 [DOI] [PubMed] [Google Scholar]
- Kiel MC, Raj VS, Kaji H, Kaji A (2003) Release of ribosome-bound ribosome recycling factor by elongation factor G. J Biol Chem 278: 48041–48050 [DOI] [PubMed] [Google Scholar]
- Kim KK, Min K, Suh SW (2000) Crystal structure of the ribosome recycling factor from Escherichia coli. EMBO J 19: 2362–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Kiel MC, Kaji A, Noller HF (2002) Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell 111: 129–140 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148 [DOI] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC (2009) Navigating the ribosome's metastable energy landscape. Trends Biochem Sci 34: 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H, Yoshida T, Uchiyama S, Kawachi M, Matsuo H, Kato T, Ohshima A, Yamaichi Y, Honda T, Kato H, Yamagata Y, Ohkubo T, Kobayashi Y (2003) Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium. J Biol Chem 278: 3427–3436 [DOI] [PubMed] [Google Scholar]
- Pai RD, Zhang W, Schuwirth BS, Hirokawa G, Kaji H, Kaji A, Cate JH (2008) Structural insights into ribosome recycling factor interactions with the 70S ribosome. J Mol Biol 376: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek PA, Grassucci RA, Frank J (1994) The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy 53: 251–270 [DOI] [PubMed] [Google Scholar]
- Peske F, Rodnina MV, Wintermeyer W (2005) Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell 18: 403–412 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Raj VS, Kaji H, Kaji A (2005) Interaction of RRF and EF-G from E. coli and T. thermophilus with ribosomes from both origins—insight into the mechanism of the ribosome recycling step. RNA 11: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CM (2010) Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468: 713–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R (2003) Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333: 721–745 [DOI] [PubMed] [Google Scholar]
- Rossmann MG (2000) Fitting atomic models into electron-microscopy maps. Acta Crystallogr D Biol Crystallogr 56: 1341–1349 [DOI] [PubMed] [Google Scholar]
- Saikrishnan K, Kalapala SK, Varshney U, Vijayan M (2005) X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications. J Mol Biol 345: 29–38 [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W (2009) Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 15: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM (2007) Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods 4: 27–29 [DOI] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM (2009) GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH (2005) Structures of the bacterial ribosome at 3.5 A resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Selmer M, Al-Karadaghi S, Hirokawa G, Kaji A, Liljas A (1999) Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science 286: 2349–2352 [DOI] [PubMed] [Google Scholar]
- Seshadri A, Varshney U (2006) Mechanism of recycling of post-termination ribosomal complexes in eubacteria: a new role of initiation factor 3. J Biosci 31: 281–289 [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J (2008) SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3: 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suematsu T, Yokobori S, Morita H, Yoshinari S, Ueda T, Kita K, Takeuchi N, Watanabe Y (2010) A bacterial elongation factor G homologue exclusively functions in ribosome recycling in the spirochaete Borrelia burgdorferi. Mol Microbiol 75: 1445–1454 [DOI] [PubMed] [Google Scholar]
- Toyoda T, Tin OF, Ito K, Fujiwara T, Kumasaka T, Yamamoto M, Garber MB, Nakamura Y (2000) Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch. RNA 6: 1432–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K (2008) Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K (2009) Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M, Morita H, Nozaki Y, Akama K, Ueda T, Ito K, Nierhaus KH, Takeuchi N (2009) EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol Cell 35: 502–510 [DOI] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003) Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V (2007) Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol 14: 733–737 [DOI] [PubMed] [Google Scholar]
- Wilson DN, Schluenzen F, Harms JM, Yoshida T, Ohkubo T, Albrecht R, Buerger J, Kobayashi Y, Fucini P (2005) X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J 24: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Soto CS, Honig B (2002) Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc Natl Acad Sci USA 99: 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Uchiyama S, Nakano H, Kashimori H, Kijima H, Ohshima T, Saihara Y, Ishino T, Shimahara H, T, Yokose K, T, A, Y (2001) Solution structure of the ribosome recycling factor from Aquifex aeolicus. Biochemistry 40: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M (2006) Structural basis for messenger RNA movement on the ribosome. Nature 444: 391–394 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Hauryliuk VV, Ehrenberg M (2005) Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell 18: 675–686 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.