Abstract
Bacillus subtilis 6S-1 RNA binds to the housekeeping RNA polymerase (σA-RNAP) and directs transcription of short ‘product’ RNAs (pRNAs). Here, we demonstrate that once newly synthesized pRNAs form a sufficiently stable duplex with 6S-1 RNA, a structural rearrangement is induced in cis, which involves base-pairing between sequences in the 5′-portion of the central bulge and nucleotides that become available as a result of pRNA invasion. The rearrangement decreases 6S-1 RNA affinity for σA-RNAP. Among the pRNA length variants synthesized by σA-RNAP (up to ∼14 nt), only the longer ones, such as 12–14-mers, form a duplex with 6S-1 RNA that is sufficiently long-lived to induce the rearrangement. Yet, an LNA (locked nucleic acid) 8-mer can induce the same rearrangement due to conferring increased duplex stability. We propose that an interplay of rate constants for polymerization (kpol), for pRNA:6S-1 RNA hybrid duplex dissociation (koff) and for the rearrangement (kconf) determines whether pRNAs dissociate or rearrange 6S-1 structure to trigger 6S-1 RNA release from σA-RNAP. A bioinformatic screen suggests that essentially all bacterial 6S RNAs have the potential to undergo a pRNA-induced structural rearrangement.
Keywords: non-coding RNA, RNAP, 6S RNA
Introduction
The discovery and investigation of bacterial small non-coding RNAs (ncRNAs) has become an emerging field in recent years. Beside roles in processing, translation and genome defence, many of these ncRNAs act as regulators, for example, by masking binding sites or inducing structural changes in their specific target RNA (Waters and Storz, 2009). A unique ncRNA is 6S RNA (Willkomm and Hartmann, 2005; Wassarman, 2007), which was identified as an abundant RNA species some 40 years ago (Brownlee, 1971), but whose function remained enigmatic for 30 years (Wassarman and Storz, 2000). 6S RNA, ∼200 nt in length, has now been identified in all bacterial branches (Barrick et al, 2005). In stationary phase Escherichia coli cells, 6S RNA competes with DNA promoters for binding to the housekeeping RNA polymerase (σ70-RNAP), resulting in transcriptional repression of a variety of σ70-dependent genes (Wassarman and Storz, 2000). The conserved secondary structure of 6S RNA, thought to resemble an open DNA promoter complex (Barrick et al, 2005; Trotochaud and Wassarman, 2005), enables stable binding to RNAP. During outgrowth from stationary phase, 6S RNA itself serves as a template for transcription of small ‘product’ RNAs (pRNAs), which results in the dissociation of the 6S RNA:RNAP complex by a so far unknown mechanism (Wassarman and Saecker, 2006; Gildehaus et al, 2007). In contrast to most bacteria, Bacillus subtilis harbours two 6S RNA homologues, termed 6S-1 and 6S-2, which differ in their expression profiles (Ando et al, 2002; Suzuma et al, 2002; Barrick et al, 2005; Trotochaud and Wassarman, 2005; Beckmann et al, 2011). In a previous study (Beckmann et al, 2011), we were able to demonstrate that 6S-1 RNA indeed serves as a template for pRNA synthesis, particularly during outgrowth from stationary phase, similar to the prototypic 6S RNA of E. coli. Deep sequencing and in vitro transcription revealed that pRNA synthesis is always initiated at nucleotide C40 of 6S-1 RNA, yielding primarily pRNAs with a length of ⩽5, ∼8 and ∼14 nt (Beckmann et al, 2011). In the present study, we have analysed the mechanism of 6S-1 RNA:RNAP release. We show that dissociation of 6S-1 RNA:RNAP complexes is the result of a structural rearrangement of 6S-1 RNA induced by pRNA transcripts that have a minimal length (⩾12nt) to confer sufficient stability to pRNA:6S-1 RNA hybrid duplexes. Furthermore, in silico analysis predicts that pRNA-induced rearrangements in 6S RNA structure occur in all known bacterial 6S RNAs. Our findings provide a basis towards understanding the escape of RNAP from its 6S RNA-mediated activity block. The unique mechanism interdigitates RNA-dependent RNAP activity with a pRNA length-dependent switch in 6S RNA structure that disrupts 6S-1 RNA:RNAP interactions.
Results
Release of RNAP is controlled via stable pRNA binding
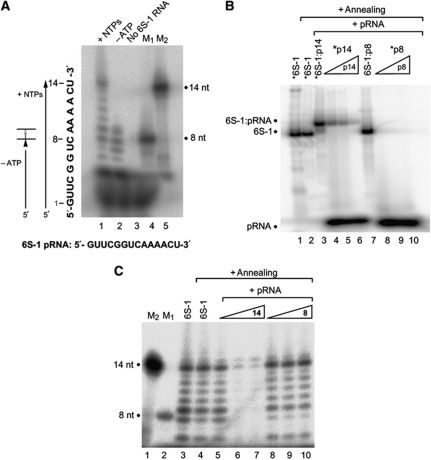
The major pRNA species transcribed from 6S-1 RNA as template are ⩽5, 8/9 and ∼14 nt in length (Figure 1A). (Beckmann et al, 2011). We analysed by native PAGE whether pRNA 8- and 14-mers (both chemically synthesized) are able to form gel-resolvable complexes with 6S-1 RNA. These experiments revealed that only annealing of pRNA 14-mer, but not 8-mer, to 5′-endlabelled 6S-1 RNA caused a shift in 6S-1 RNA mobility (Figure 1B, lane 3 versus lane 7). Reciprocally, we attempted to anneal 5′-endlabelled pRNA 8- or 14-mer to unlabelled 6S-1 RNA (Figure 1B, lanes 4 and 8). Again, formation of a stable hybrid was restricted to the 14-mer. Here, complexes between 6S-1 RNA and radiolabelled pRNA 14-mer could be outcompeted by the presence of excess amounts of unlabelled 14-mer (Figure 1B, lanes 4–6). We then tested if the B. subtilis σA-RNAP is able to synthesize pRNA transcripts from 6S-1 RNA pre-annealed to the pRNA 14-mer. As illustrated in Figure 1C (lanes 6 and 7), 6S-1 RNA:pRNA 14-mer complexes prevent de novo pRNA transcription. Replacement of the pRNA 14-mer with the 8-mer in the pre-annealing step abrogated the inhibitory effect on in vitro transcription (Figure 1C, lanes 9 and 10), consistent with the 8-mer being unable to stably anneal to 6S-1 RNA.
Figure 1.
B. subtilis 6S-1 RNA forms stable hybrids with pRNA in vitro. (A) In vitro transcription (2 μM B. subtilis RNAP holoenzyme) of pRNA using 3.2 μM 6S-1 RNA as template, either in the presence of all four NTPs (lane 1; each NTP 200 μM) or CTP, GTP and UTP only (lane 2, –ATP). Control lanes: lane 3, as in lane 1 but omission of the 6S-1 RNA template; lanes 4 and 5, chemically synthesized 5′-endlabelled pRNA 8-mer (5′-GUU CGG UC; lane M1) and 14-mer (5′-GUU CGG UCA AAA CU; lane M2) used as size markers. Omission of ATP (lane 2) results in synthesis of short pRNA (8-mers) because 6S-1 RNA encodes four consecutive A residues at pRNA positions 9–12 (see sequence at the bottom); our observation of 9- in addition to 8-mers despite the absence of ATP can be explained by RNAP adding a non-templated residue to the 3′-end. RNA products were separated by 20% denaturing PAGE. (B) Annealing of chemically synthesized pRNAs (8- or 14-mers) to 6S-1 RNA. Asterisks denote the radiolabelled RNA species (6S-1 RNA, 14-mer or 8-mer). Lanes 1 and 2: 5′-endlabelled 6S-1 RNA; in lane 2, the 6S-1 RNA was subjected to the pRNA annealing procedure (for details, see Materials and methods); lane 3: 5′-endlabelled 6S-1 RNA (100 nM unlabelled 6S-1 RNA and trace amounts, <1 nM, of 5′-endlabelled 6S-1 RNA) with pRNA 14-mer (1 μM) pre-annealed as just mentioned; lanes 4–6: annealing of 100 nM unlabelled 6S-1 RNA, trace amounts (<1 nM) of 5′-endlabelled pRNA 14-mer, plus 100 nM (lane 4), 200 nM (lane 5) or 500 nM (lane 6) unlabelled 14-mer; lane 7, as lane 3, but using the pRNA 8-mer instead of the 14-mer; lanes 8–10: as lanes 4–6, but using the pRNA 8-mer. Samples were analysed by 9% native PAGE. (C) In vitro transcription of pRNA from 6S-1 RNA (1 μM) as template using 1 μM RNAP holoenzyme. Lanes 1–3: as lanes 5, 4 and 1, respectively, in (A); lane 4: as lane 3, but subjecting 6S-1 RNA to the annealing procedure before transcription; lanes 5–10: 2 μM (lane 5), 4 μM (lane 6), 20 μM (lane 7) pRNA 14-mer or 2 μM (lane 8), 4 μM (lane 9), 20 μM (lane 10) pRNA 8-mer were subjected to the annealing procedure in the presence of 2 μM 6S-1 RNA in a final volume of 5 μl 1 × TE buffer; then, 2 μl of 5 × activity buffer and 0.5 μl RNAP were added and samples were incubated for 30 min at 37°C before addition of nucleotides and transcription for 1 h at 37°C (final volume 10 μl). RNA products were separated by 20% denaturing PAGE.
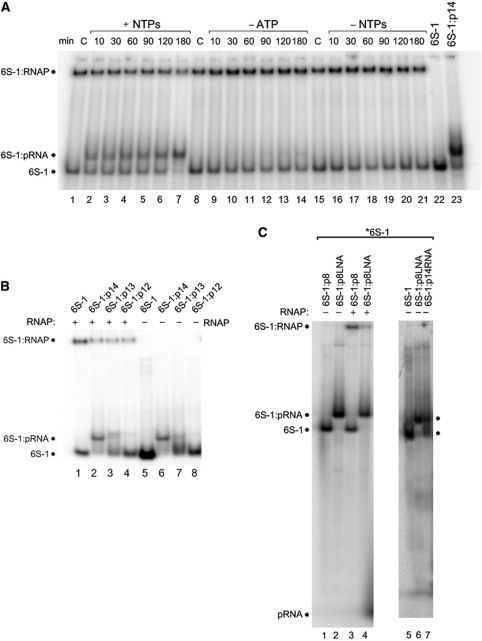
Next, we combined in vitro transcription by σA-RNAP with native PAGE for the analysis of 6S-1 RNA:RNAP complex formation, utilizing 5′-endlabelled 6S-1 RNA as transcription template. Binding to RNAP and transcription were started at different time points before all samples were concertedly loaded onto a native PAA gel. In addition to the formation of 6S-1 RNA:RNAP complexes, a 6S-1 RNA:pRNA hybrid band of retarded mobility appeared already at the first time point (10 min; Figure 2A, lanes 1–7). At the latest time point (180 min), very little free 6S-1 RNA remained and the fraction of 6S-1 RNA in complex with RNAP was substantially reduced (lane 7). The idiosyncratic feature of 6S-1 RNA to encode the first A residue at pRNA position 9 enabled us to transcribe only shorter pRNAs up to 8-mers through omission of ATP during in vitro transcription (Figure 1A, lane 2; the fact that we observed also 9-mers despite the absence of ATP can be explained by RNAP adding a non-templated residue to the 3′-end).
Figure 2.
Role of pRNA length and pRNA:6S-1 RNA duplex stability. (A) Trace amounts (<1 nM) of 5′-endlabelled 6S-1 RNA and 2.5 μM unlabelled 6S-1 RNA were subjected to the annealing procedure in a volume of 4 μl (see Materials and methods); then, 1 μl of a heparin solution (400 ng/ μl) and 2 μl 5 × activity buffer were added and samples were kept at 37°C; then 1.06 μl RNAP holoenzyme (8 μg/μl) were added and samples were incubated for 30 min at 37°C, followed by addition of 2 μl nucleotide solution (all four NTPs: lanes 2–7; only CTP, GTP and UTP: lanes 9–14) or 2 μl of double-distilled water instead (no NTPs; lanes 15–21) (final volume 10 μl; f.c. RNAP: 2 μM; f.c. 6S-1 RNA: 1 μM; f.c. NTPs: 200 μM each); lanes C: samples incubated for 180 min at 37°C in the absence of NTPs. After transcription at 37°C for the time period indicated above each lane, samples were analysed by 7.5% native PAGE (1 × TBE). (B) Trace amounts (<1 nM) of 5′-endlabelled 6S-1 RNA and 1.7 μM unlabelled 6S-1 RNA, either alone (lane 5) or in the presence of 17 μM pRNA 14-mer (lane 6), 13-mer (5′-GUU CGG UCA AAA C, lane 7) or 12-mer (5′-GUU CGG UCA AAA, lane 8) were annealed in 6 μl 1 × TE buffer and then loaded on a 7.5% native PAA gel (1 × TBE); lanes 1–4: as lanes 5–8, but before gel loading 1 μl of a heparin solution (400 ng/μl) and 2 μl 5 × activity buffer were added and samples were kept at 37°C; then 1.06 μl RNAP holoenzyme (8 μg/μl) was added and samples were incubated for 30 min at 37°C followed by gel loading. (C) Test for annealing of the pRNA 8-mer (lane 1) or an isosequential all-LNA 8-mer (lane 2) to 6S-1 RNA; lanes 3 and 4: as lanes 1 and 2, but after the annealing procedure samples were further incubated with RNAP holoenzyme; for details, see (B) and Materials and methods. Lanes 5–7 illustrate that 6S-1 RNA hybrid complexes with the LNA 8-mer cause the same mobility shift as obtained with the RNA 14-mer.
When ATP or all four NTPs were omitted during the transcription step (Figure 2A, lanes 8–21), neither a signal corresponding to the 6S-1 RNA:pRNA hybrid nor a time-dependent reduction in the amount of 6S-1 RNA:RNAP complexes were observed.
We conclude that pRNA 7–9-mers, shown to be synthesized in the absence of ATP (Figure 1A, lane 2) under the conditions of the experiment illustrated in Figure 2A, are unable to form stable hybrids with 6S-1 RNA and thus do not affect 6S-1 RNA:RNAP complex formation (Figure 2A, lanes 8–14 versus 1–7).
Deep sequencing revealed that 6S-1 RNA-derived pRNA 14–15-mers represent the second most abundant length variant identifiable in vivo beyond the shorter 8–12-mers, whereas surprisingly few 13-meric pRNA reads were detected (Beckmann et al, 2011). This raised the question of the minimal pRNA length that confers sufficient stability to pRNA:6S-1 RNA hybrids in order to be able to induce the mobility shift of 6S-1 RNA as seen in Figure 2A. We addressed this by using chemically synthesized length variants of the pRNA 14-mer lacking one or two nucleotides at the 3′-end (13-mer and 12-mer). The pRNA oligomers were annealed to 5′-endlabelled 6S-1 RNA and loaded onto the native PAA gel either directly (Figure 2B, lanes 6–8) or after pre-incubation with σA-RNAP (lanes 2–4). This experiment revealed that the 14-mer is required for persistent gel-resolvable binding of pRNA to 6S-1 RNA, as inferred from the prevalence of the shifted 6S-1 RNA conformer (Figure 2B, lane 6). Some mobility shift was still detectable with the 13-mer, but barely with the 12-mer (Figure 2B, lanes 7 and 8). In the case of the 13-mer, the free 6S-1 RNA band migrated more diffusely than in the lane representing the 12-mer, suggesting that 13-mer:6S-1 RNA hybrids underwent multiple cycles of dissociation and reassociation during electrophoresis, in line with the intermediate stability relative to 14-mer:6S-1 RNA and 12-mer:6S-1 RNA complexes. When pRNA/6S-1 RNA mixtures were incubated with σA-RNAP after the pRNA pre-annealing step and before gel loading, we observed a reduction in the amount of 6S-1 RNA:RNAP complexes for the 14-, 13-, as well as the 12-mer (Figure 2B, lanes 2–4). We conclude that hybrid structures of pRNA 13- and 12-mer with 6S-1 RNA are sufficiently long-lived to induce the conformational shift in 6S-1 RNA. However, the 6S-1 RNA hybrids containing the 13- or 12-mer have higher dissociation rate constants relative to the 14-mer, leading to increased irreversible dissociation of pRNA:6S-1 RNA complexes under the dilute conditions of native PAGE.
We further addressed the question whether an 8-mer, if able to form a more stable duplex with 6S-1 RNA, may be capable of inducing the mobility switch of 6S-1 RNA. For this purpose, we pre-annealed an isosequential all-LNA 8-mer (LNA, locked nucleic acid (Vester and Wengel, 2004; Grünweller and Hartmann, 2007); all-LNA, LNA analogues at every position) to 6S-1 RNA, exploiting the duplex-stabilising effect of LNA residues. In contrast to the pRNA 8-mer, the pLNA 8-mer was able to form a stable hybrid with 6S-1 RNA and to induce a mobility switch (Figure 2C, cf. lanes 1 and 2), as observed for the RNA 14-mer (Figure 2C, cf. lanes 6 and 7). We conclude that stability of pRNA:6S-1 RNA hybrids is the critical feature here, rather than pRNA length per se. Likewise, when pre-annealed pLNA 8-mer:6S-1 RNA complexes were incubated with σA-RNAP, 6S-1 RNA binding to RNAP was reduced relative to the control sample with the pRNA 8-mer (Figure 2C, cf. lanes 3 and 4). The same was seen with the pRNA 14-mer (Figure 2B, cf. lanes 1 and 2), suggesting that the decrease in affinity for σA-RNAP is similar for pLNA 8-mer:6S-1 RNA and pRNA 14-mer:6S-1 RNA complexes relative to free 6S-1 RNA.
pRNA synthesis induces structural rearrangements of 6S-1 RNA
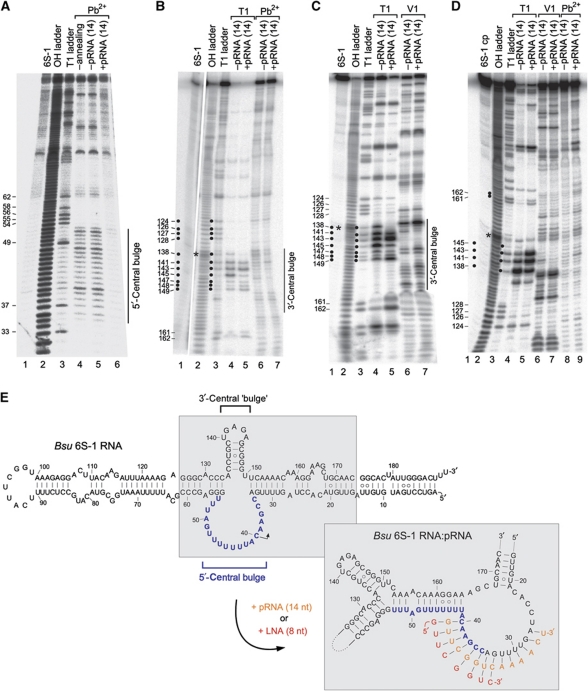
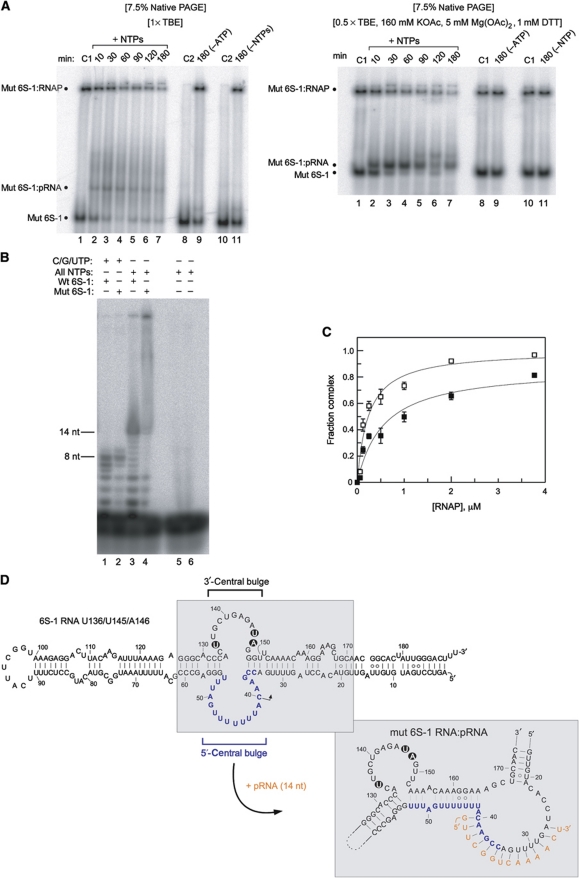
To understand the mobility shift induced by base-pairing of pRNA to 6S-1 RNA, we performed probing experiments with 6S-1 RNA in its free form or in complex with the pRNA 14-mer (Figure 3A–D). Using 5′- and 3′-endlabelled mature 6S-1 RNA (190 nt), we were able to identify structural differences particularly in the central bulge region of 6S-1 RNA. Pb2+-induced cleavage, preferentially occurring in single-stranded flexible RNA regions, decreased substantially in the 5′-part of the central bulge (nt 35–53) upon annealing of the pRNA 14-mer (Figure 3A, cf. lanes 5 and 6). Likewise, we observed pRNA-mediated protection from cleavage by RNase T1 (preference for single-stranded G residues) in the 3′-part of the central bulge at G138 and G145, and an enhanced signal at G143 (Figure 3B and C, cf. lanes 4 and 5); protection from Pb2+-induced cleavage was found in the region of ∼nt 135–149, except for G141 which showed enhanced accessibility upon pRNA binding (Figure 3B, cf. lanes 6 and 7). Since 3′-endlabelled 6S-1 RNA showed a band compression in the region of nt 131–137 (Figure 3B, lane 2), preventing a clear assignment of signals in this part of 6S-1 RNA, we employed a 5′-endlabelled circularly permuted 6S-1 RNA (with its new 5′-end in the apical loop; Supplementary Figure S1) for better resolution, which behaved as the wild-type (wt) 6S-1 RNA in terms of the pRNA-induced structural rearrangement and binding to σA-RNAP (Supplementary Figure S1). This RNA enabled us to resolve the region of nt 131–138, while the region of band compression was now shifted to around nt 147 (Figure 3D, lane 2). As for wt 6S-1 RNA (Figure 3B and C), partial protection from RNase T1 cleavage was identified at G138 and G145, and enhanced cleavage at G143. In addition, C135, a hypersensitive site also appearing when 6S-1 RNA was incubated in RNase T1 buffer without enzyme, was partially protected in the pRNA:6S-1 RNA hybrid (Figure 3D, lanes 4 and 5), and the enhancement of Pb2+-induced hydrolysis at G141 was seen (lanes 8 and 9) as with 3′-endlabelled wt 6S-1 RNA (Figure 3B, lanes 6 and 7). Probing with the double strand-specific RNase V1 mainly revealed cleavage at nt 137, which was somewhat increased in the presence of pRNA (Figure 3C and D, lanes 6 and 7). An ambiguity was seen at G141, at which we observed a slight pRNA-mediated protection from RNase T1 in panel B (lanes 4 and 5), but partial enhancement of T1 cleavage in panel D (lanes 4 and 5). Similar probing results were obtained for hybrid structures composed of 6S-1 RNA and the all-LNA 8-mer instead of the RNA 14-mer (Supplementary Figure S2). Folding programs (RNAfold (Lorenz et al, 2011), mfold (Zuker, 2003)) predict hairpin formation in the 3′-part of the central bulge already in free 6S-1 RNA (Figure 3E, top; nt 135–149). Our probing results are consistent with the presence of the hairpin, although it appears to be in equilibrium with open conformations, indicated, for example, by susceptibility of stem nucleotides G147–149 to cleavage by the ssRNA-specific RNase T1 as well as the dsRNA-specific RNase V1 (Figure 3B and C). However, the observed pRNA-dependent changes in cleavage susceptibility (Figure 3) suggest that the hairpin is more structured upon pRNA association and/or is presented differently in free 6S-1 RNA versus the 6S-1:pRNA hybrid structure. To further explore the mechanistic role of this hairpin, we disrupted its stem by three mutations (C136U/G145U/C146A; Figure 4D). In an experiment equivalent to the one in Figure 2A (lanes 1–7), we could show that this mutant 6S-1 RNA is able to induce the mobility shift of 6S-1 RNA and to reduce complex formation with σA-RNAP (Figure 4A). Since we noticed, relative to wt 6S-1 RNA (Figure 2A), an increased smear of radioactivity below and above the mutant 6S-1 RNA:pRNA hybrid band in standard native PAA gels run in 1 × TBE buffer (Figure 4A, cf. lane 1 and lanes 2–7), we considered the possibility that this may arise from increased conformational flexibility of the mutant 6S-1 RNA. Consistent with this idea, sharper bands and less smear was seen when native gels were run in 0.5 × TBE, 160 mM KOAc, 5 mM Mg(OAc)2 and 1 mM DTT (Figure 4A, right panel), which can be attributed to RNA stabilization by the higher salt concentration. When we tested the capacity of the U136/U145U/A146 mutant 6S-1 RNA to direct pRNA transcription, reduced synthesis of pRNAs up to ∼14 nt in length and apparently increased synthesis of longer transcripts was observed (Figure 4B). The affinity of the mutant 6S-1 for σA-RNAP was two- to three-fold lower than that of the wt 6S-1 RNA, as inferred from native PAGE analyses of samples in which trace amounts of 5′-endlabelled 6S RNA were preincubated with increasing amounts of σA-RNAP in the absence of NTPs (Figure 4C). We conclude from these findings that the hairpin structure is neither absolutely essential for pRNA synthesis, nor for the conformational rearrangement or 6S-1 RNA release from RNAP, but nonetheless contributes to the overall efficiency of the process.
Figure 3.
pRNA induces structural changes in 6S-1 RNA. (A) Structure probing using 5′-endlabelled 6S-1 RNA. Lane 1: 5′-endlabelled 6S-1 RNA directly loaded onto the gel; lane 2: alkaline hydrolysis ladder of 6S-1 RNA; lane 3: limited RNase T1 digest under denaturing conditions; lanes 4 and 5: free 6S-1 RNA subjected to the annealing procedure (lane 5) or not (lane 4) before lead probing; lane 6: 10 pmol 5′-endlabelled 6S-1 RNA and 100 pmol pRNA 14-mer were subjected to the annealing procedure in 6 μl 1 × TE buffer before lead probing. (B) Structure probing using 3′-endlabelled 6S-1 RNA. Lanes 1–3, 6 and 7: as lanes 1–3, 5 and 6, respectively, in (A); lanes 4 and 5: as lanes 6 and 7, but RNase T1 cleavage (under native conditions). Note that lane 1 was taken from a separate gel using the same batch of 3′-endlabelled 6S-1 RNA as in lanes 2–7. (C) Structure probing using 3′-endlabelled 6S-1 RNA. For lanes 1–5, see (B); lanes 6 and 7, cleavage by RNase V1. (D) Structure probing of the 5′-endlabelled circularly permuted 6S-1 RNA (6S-1 cp) variant (see Supplementary Figure S1). Lanes 1–7 correspond to lanes 1–7 in (C); lanes 8 and 9 correspond to lanes 6 and 7 in (B). For details of probing reactions, see Supplementary data. Asterisks in (B–D) mark the regions of band compression. (E) Model of the pRNA-induced structural rearrangement of the 6S-1 RNA core region (grey shaded in the 6S-1 RNA secondary structure at the top). The structural rearrangement can be induced by the RNA 14-mer (orange) but also by an all-LNA 8-mer (red; see Supplementary Figure S2) mimicking the 5′-terminal 8 nt of the 14-mer. The orientation of helical elements after the rearrangement is arbitrary, their exact orientation is unknown. RNAfold analysis of the 6S-1 RNA core structure (nt 15–81–(N)4–126–174) predicts that the central bulge collapse occurs when the first nine pRNA-encoding nucleotides are blocked (by constraint folding) for base-pairing with other 6S-1 RNA nucleotides. The three A–U base pairs (U51–53/A153–155) are predicted by RNAfold and mfold in dot plots of suboptimal structures with free energies close to that of the minimum free energy (MFE) structure.
Figure 4.
Functional analysis of the U136/U145/A146 mutant 6S-1 RNA unable to form a hairpin in the 3′-portion of the central bulge. (A) 5′-Endlabelled 6S-1 RNA was incubated with σA-RNAP in the presence of NTPs for different time periods as in Figure 2A. Samples were then either loaded on a 7.5% native PAA gel using 1 × TBE as electrophoresis buffer (left panel) or 0.5 × TBE, 160 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, pH 8.6 (right panel) as electrophoresis buffer. Lanes C1: samples incubated for 180 min at 37°C in the absence of NTPs; lanes C2: as lanes C1 but without RNAP. For further details, see legend to Figure 2. (B) In vitro transcription was conducted as in Figure 1A, comparing wt (wt 6S-1) and U136/U145/A146 mutant 6S-1 RNA (mut 6S-1) as template; C/G/UTP, NTP mixture lacking ATP. For further details, see legend to Figure 1A. (C) Binding of 5′-endlabelled wt 6S-1 RNA (open squares) and U136/U145/A146 mutant 6S-1 RNA (filled squares) to the σA-RNAP holoenzyme, based on experiments as shown in (A) (right), but in the absence of NTPs (see also Beckmann et al, 2011). Apparent Kd values, based on two independent experiments, were 0.24±0.06 μM for wt 6S-1 RNA and 0.56±0.17 μM for the mutant 6S RNA, as derived from non-linear regression analysis using the equation for a single ligand binding site. Errors, standard errors of the mean (s.e.m.); some error bars are small such that they are masked by the symbol. Similar binding curves were obtained by 7.5% native PAGE in 1 × TBE buffer. The experimental end point for complex formation is lower for the mutant versus wt RNA, suggesting that the fraction of 6S-1 mutant conformers that are unable to bind to RNAP is increased relative to wt 6S-1 RNA. (D) Model of the mutant 6S-1 RNA structure before and after the pRNA-induced structural rearrangement. The three base exchanges are highlighted.
Our probing results combined with in silico secondary structure predictions are consistent with the pRNA-induced structural rearrangement illustrated in Figure 3E (structure at the bottom right): Progressional pRNA transcription results in unwinding of the 6S-1 RNA closing stem adjacent to the central bulge (nt 28–34:151–157), which induces formation of a new base-pairing interaction between nt 42–53 and nt 153–164, resulting in what we term a ‘central bulge collapse’. An all-LNA 8-mer, disrupting the terminal two base pairs of the closing stem, is sufficient to trigger this structural rearrangement, in line with its capacity to induce the gel mobility shift of 6S-1 RNA, to reduce RNAP affinity when annealed to 6S-1 RNA (Figure 2C), and to cause similar changes as the pRNA 14-mer in the RNase T1 probing pattern of 6S-1 RNA (cf. Figure 3C and Supplementary Figure S2). Consistent with our findings, RNAfold analysis of the 6S-1 RNA core structure (nt 15–81–(N)4–126–174) predicts that the central bulge collapse occurs when the first nine pRNA-encoding nucleotides are blocked (via constraint folding) for base-pairing with other 6S-1 RNA nucleotides.
Specific structural changes in 6S RNA are conserved among bacteria
Formation of a sufficiently stable pRNA:6S RNA hybrid helix of ⩾8 bp is the key event in the conformational switch of 6S RNA structure, which in the case of 6S-1 RNA involves a central bulge collapse as inferred from the probing data in Figure 3. An in silico screen of all known 6S RNA sequences available from Rfam (Gardner et al, 2009) was conducted to investigate whether the conformational rearrangement observed for B. subtilis 6S-1 RNA is a general feature of bacterial 6S RNAs. Indeed, 161 out of 162 sequences had the potential to form a reasonably stable hairpin in the 3′-part of the central bulge, many particularly after pRNA-induced partial unwinding of 6S RNA structure (Supplementary Figure S3). For 147 of the 162 sequences, a central bulge collapse (see Figure 3E) was found to be possible and likely for the majority of 6S RNAs when the anticipated pRNA-coding region (starting in the 5′-part of the central bulge) was blocked (via constraint folding) for intramolecular base-pairing (Supplementary Figure S3A). Interestingly, while in one group of 6S RNAs, the hairpin is predicted to be formed already in the free RNA (e.g., in B. subtilis 6S-1 RNA), stable formation of a hairpin is predicted in many γ-proteobacterial 6S RNAs (e.g., in E. coli 6S RNA) only after pRNA-induced disruption of 6S RNA structure. Intriguingly, in the latter cases, the hairpin is particularly stable and a central bulge collapse is not predicted to occur (Supplementary Figure S3B). In a third group of 6S RNAs, such as the one from Aquifex aeolicus, both mechanistic components, pRNA-induced hairpin formation and central bulge collapse, seem to be implemented simultaneously (Supplementary Figure S3B).
RNAP release and 6S-1 RNA degradation are coupled
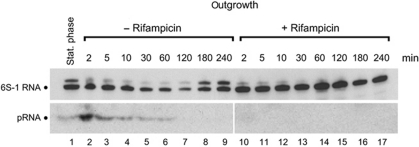
As outgrowth from stationary phase has been shown to be the time point of massive pRNA production in B. subtilis (Beckmann et al, 2011), we probed the levels of pRNA and 6S-1 RNA at different time points after induction of outgrowth. We induced outgrowth of extended stationary cells by addition of fresh LB medium either without (Figure 5, lanes 2–9) or supplemented with rifampicin (lanes 10–17) to inhibit pRNA transcription. As expected, rifampicin treatment blocked pRNA synthesis (Figure 5, bottom panel, lanes 10–17) and de novo 6S-1 RNA synthesis (top panel), the latter inferred from the disappearance of the 6S-1 RNA precursor signal. Of note, we observed a stabilization of mature 6S-1 RNA levels in the presence of rifampicin, whereas 6S-1 RNA levels decreased in the course of outgrowth in the absence of the antibiotic (Figure 5, cf. lanes 2–7 and 10–15). In the absence of rifampicin, 6S-1 RNA levels rose again when cells entered a new stationary phase (lanes 8 and 9). Thus, pRNA synthesis and 6S-1 RNA degradation are coupled processes (see Discussion).
Figure 5.
Effect of rifampicin on pRNA synthesis and 6S-1 RNA stability. Analysis of 6S-1 RNA steady-state levels following outgrowth from stationary phase by northern blots of cellular RNA extracted at different time points after induction of outgrowth, either in the absence of rifampicin (lanes 2–9) or in the presence of rifampicin (100 μg/ml; lanes 10–17). Lane 1: RNA extracted from stationary cells immediately before 1:5 dilution in fresh LB medium. An antisense 6S-1 RNA, internally labelled with digoxigenin-UTP (top panel), or a 5′-digoxigenin-labelled LNA/DNA mixmer complementary to the pRNA 14-mer (bottom panel) were used as northern probes; for details see Beckmann et al (2010).
Discussion
Using differential high-throughput RNA sequencing (dRNA-seq; Sharma et al, 2010), we recently identified pRNAs transcribed in vivo from B. subtilis 6S-1 RNA as template (Beckmann et al, 2011). 6S-1 pRNA levels were found to be low during exponential, to increase in stationary, and to burst during outgrowth from stationary phase. In vitro transcription indicated that the major pRNA species are ⩽5, 8/9 and 14 nt in length (Beckmann et al, 2011) (Figure 1A). dRNA-seq further revealed an increase in the relative abundance of ∼14-mers compared with 8–12-mers in outgrowing versus stationary cells, suggesting that the length distribution of pRNAs varies upon the physiological state and may have functional consequences (Beckmann et al, 2011). Here, we have shown that a pRNA 8-mer is unable to stably associate with 6S-1 RNA and to induce the mobility shift of 6S-1 RNA that decreases the affinity for σA-RNAP. In contrast, an all-LNA 8-mer has this capacity, clearly demonstrating that the high rate constant for dissociation (koff) of an 8-bp RNA/RNA duplex is the key feature here, rather than duplex length per se. A comparison of pRNA 12-, 13- and 14-mers (Figure 2B) further revealed that only the naturally abundant 6S-1 RNA:pRNA 14-mer complexes have a sufficiently low koff to resist any dissociation during native PAGE (Figure 2B). Of note, native PAGE does not mimic equilibrium conditions, and complexes undergoing repeated dissociation and reassociation cycles in gel ‘cages’ are at risk to dissociate irreversibly when they fall below a certain thermodynamic stability. Nonetheless, the finding that pRNA 12- and 13-mers were able to reduce the fraction of 6S-1 RNA bound to RNAP suggests that the life-time of 6S-1 RNA:pRNA 12/13-mer complexes is sufficiently long under equilibrium conditions to rearrange 6S-1 RNA as a prerequisite for destabilizing σA-RNAP:6S-1 RNA complexes.
pRNA transcription from 6S RNA is reminiscent of abortive transcription initiation at DNA promoters, where RNAP can go through iterative cycles of abortive transcription until the enzyme surpasses the transition to a ternary elongation complex (TEC) that processively synthesizes longer transcripts. Abortive transcription is strongly promoter dependent and depends on (i) the equilibrium constant KB for the formation of the closed complex (RPc) from promoter DNA and σA-RNAP, (ii) on the forward and reverse rate constants k2 and k−2 for the isomerization of RPc to an open complex (RPo) and (iii) on kE for the transition from a non-productive RPo involved in catalysing abortive transcription to a productive elongation complex (TEC) (Hsu, 2009). The length of abortive transcripts is also strongly promoter dependent and varies between 2–16 nt (Hsu, 2009). For example, the T5N25anti promoter, displaying a high ratio of abortive to productive transcription, was shown to release mainly transcripts of 7–9 and 13–14 nt in length (Hsu, 2009), thus strongly resembling the pRNA transcript profile of 6S-1 RNA. Fluorescence resonance energy transfer and single-molecule DNA nanomanipulation experiments have revealed that initial abortive transcription proceeds through a DNA scrunching mechanism, where RNAP pulls downstream DNA into its active site and accommodates the accumulated DNA as flexible single-stranded bulges. Upon release of the abortive transcript, RNAP extrudes the accumulated DNA to restore the initial state (Kapanidis et al, 2006; Revyakin et al, 2006). We think it very likely that pRNA transcription essentially proceeds as abortive initiation at DNA promoters, with 6S RNA mimicking an RPo as proposed previously (Barrick et al, 2005). Apart from the abortive transcription-like profile of pRNAs, 6S RNAs are flexible in their central bulge regions, where RNA folding programs predict multiple structures and relatively weak base-pairing probabilities. This flexibility should favour 6S RNA scrunching. Compared with DNA promoters, the rate constant kE for the transition to a TEC appears to be very low for 6S RNAs, as inferred from the low number of transcripts exceeding 14 nt in the case of B. subtilis 6S-1 RNA (Beckmann et al, 2011).
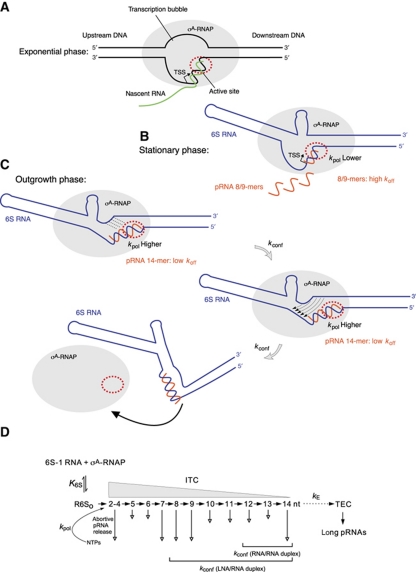
A mechanistic element that distinguishes transcription from 6S RNAs relative to DNA promoters is the structural/conformational rearrangement in 6S RNA as a consequence of the synthesis of longer pRNAs (e.g., 14-mers). This and the similarities to abortive transcription at DNA promoters lead us to propose a kinetic model (Figure 6), according to which 6S-1 RNA:pRNA hybrids can either decompose (determined by the koff of 6S-1 RNA:pRNA duplexes) or induce the structural rearrangement in 6S RNA (described by the rate constant kconf) that reduces the affinity for σA-RNAP. Also, the rate constant for nucleotide incorporation (kpol) will be critical at pRNA positions such as nt 8, where koff is high, but will be clearly uncritical at, for example, position 14 where koff is negligible. If the rate of pRNA polymerization and thus the average pRNA length distribution varies depending on the physiological state, as suggested by our dRNA-seq data (Beckmann et al, 2011), then our model predicts more ‘non-effective’ rounds, for example, of pRNA 8/9-mer synthesis during stationary phase compared with outgrowth conditions (Figure 6). In the experiment in Figure 2A, roughly 70% of 6S-1 RNA molecules were bound to RNAP (lane 1) and it seems that half of these molecules are rapidly (within 10 min; lane 2) converted to the gel-resolvable shifted conformer, indicating that the latter represent pRNA ∼14-mer:6S-1 RNA complexes. Under comparable conditions, 6S-1 RNA-templated in vitro transcription for 1 h resulted in a transcript pattern according to which pRNAs migrating as ⩽5-mers and 8/9-mers constitute the major fraction of length species (Figure 1A, lane 1). If the transcript pattern in Figure 1A had resulted from single transcription rounds, then the efficient formation of gel-resolvable pRNA ∼14-mer:6S-1 RNA complexes in Figure 2A (lanes 1–7) could not be explained. Thus, a straightforward interpretation of our results is that σA-RNAP molecules repeatedly traverse ‘non-effective’ rounds of pRNA ⩽9-mer synthesis before they stochastically succeed in synthesizing a longer pRNA that is able to trigger the ultimate release of RNAP. Figure 2A (lane 7) further indicates that a fraction of 6S-1 RNA remains bound to RNAP even after extended incubation. An explanation is that this represents a conformational subpopulation of RNAP:6S-1 RNA complexes that cannot synthesize pRNA 14-mers and possibly not even shorter pRNAs.
Figure 6.
Model of the pRNA length-controlled structural rearrangement of 6S-1 RNA and its release from σA-RNAP. (A) In exponential phase, where cellular 6S-1 RNA concentrations are low, σA-RNAP is primarily engaged in transcription at DNA promoters. (B) During late exponential and stationary phase, 6S-1 RNA levels raise and a substantial fraction of σA-RNAP becomes trapped in complexes with 6S-1 RNA. Here, abortive transcription produces primarily short pRNAs of <10 nt due to a lower kpol for nucleotide addition than under outgrowth conditions (at present, we cannot define at which nucleotide position(s) kpol is critically reduced and to which extent). As a consequence, the high rate constant koff for an RNA/RNA duplex of <10 bp causes the vast majority of pRNAs to dissociate, prompting σA-RNAP to reinitiate transcription. (C) During outgrowth, when nutrients including NTPs are resupplied, the fraction of longer pRNAs (⩾14 nt) increases (Beckmann et al, 2011), because kpol for nucleotide addition increases. When the pRNA transcript reaches a length of 12–14 nt, the hybrid duplex is stable enough to trigger the structural rearrangement (described by the rate constant kconf) that results in dissociation of 6S-1: pRNA hybrids from the enzyme (bottom panel). In the top and middle sketches, the hairpin in the 3′-portion of the central bulge is shown with a curved stem to indicate that it is assumed to be in equilibrium with an open structure (see Figure 3). An inflection of 6S-1 RNA is shown because formation of the hairpin constricts this side of the central bulge. In the top panel, stippled lines depict the progressive disruption of the closing stem, and thin arrows in the middle panel indicate the central bulge collapse. The bottom sketch illustrates the substantial extent of the final structural rearrangement, although the exact relative orientation of the helical elements is unknown. (D) Kinetic scheme adapted from abortive transcription initiation at DNA promoters (Hsu, 2009). Since 6S RNA is thought to mimic an open RNA polymerase–promoter complex (RPo), the equilibrium constant K6S defines formation of the enzyme complex with 6S RNA R6So for simplification, the competition between 6S RNA and DNA promoters as well as other 6S RNAs (such as 6S-2 RNA in B. subtilis) for binding to σA-RNAP is not considered here. Upon addition of NTP substrates, R6So complexes act as initial transcribing complexes (ITCs) releasing abortive transcripts of various length (here 2–14 nt) to different extents (indicated by the length of the vertical arrow). Only pRNAs of ⩾12 nt or isosequential pLNAs ⩾8 nt are capable of inducing the structural rearrangement described by the rate constant kconf. The rate constant for the transition from an ITC to a productive elongation complex (TEC), kE, is thought to be very low if occurring at all with 6S RNAs as transcription templates.
Cotranscriptional formation of a stable duplex between pRNA transcript and 6S-1 RNA introduces rigidity into the 5′-portion of the central bulge and likely constricts this region. Together with the pRNA-induced central bulge collapse, this will likely restrict the conformational flexibility of the RNA's core structure and change the orientation of the helical arms flanking the core structure on both sides (Figure 6C). Since the scrunching mechanism underlying abortive transcription requires template flexibility, we propose that this flexibility is abrogated by the conformational rearrangement in 6S RNA. As a consequence, the 6S RNA:pRNA hybrid is irreversibly extruded by RNAP during scrunching and cannot rebind to the active site owing to its reduced flexibility and disruption of contacts to σA-RNAP. It remains to be clarified what the spatial orientation of the helical elements in the core of 6S RNA:pRNA hybrids really looks like. The fact that 6S RNA:pRNA hybrids migrate substantially slower in native PAA gels than free 6S RNAs would be consistent with a less flexible and bulkier structure of the hybrid RNA.
The fate of 6S-1 RNA:pRNA hybrids after dissociation from RNAP is unknown so far. Our results demonstrate that pRNA synthesis is a prerequisite for the reduction of 6S-1 RNA levels during extended exponential growth phase (Figure 5). We interpret these findings as follows: pRNA synthesis triggers the intracellular 6S-1 RNA release from RNAP; as a consequence, 6S-1 RNA becomes susceptible to degradation. In the presence of rifampicin and thus the absence of pRNA synthesis, 6S-1 RNA remains bound to RNAP, which protects the RNA from nucleolytic attack. An alternative explanation could be that rifampicin blocks expression of an RNase involved in 6S-1 RNA decay. We thus tested 6S-1 RNA degradation in extracts from outgrowing cells treated with and without rifampicin. No differences in the decay rate were observed (Supplementary Figure S4), arguing against this alternative explanation. We also tested if pre-annealed 6S-1 RNA:pRNA 14-mer hybrids may accelerate 6S-1 RNA decay. Again, no significant differences were found when we compared the decay rates for radiolabelled 6S-1 RNA in its free form or complexed with the pRNA 14-mer (Supplementary Figure S4), making a direct effect of pRNA on 6S-1 RNA degradation unlikely. RNases involved in degradation of 6S RNAs have not yet been identified in B. subtilis nor in any other bacterium.
Finally, we have provided evidence that a pRNA-induced structural rearrangement, as revealed for B. subtilis 6S-1 RNA, is a key mechanistic component shared among all bacteria that possess a 6S RNA. This observation is remarkable, taking into account that 6S RNAs are very weakly conserved on the sequence level (Barrick et al, 2005; Trotochaud and Wassarman, 2005). Interestingly, a C132A mutant of E. coli 6S RNA was recently observed to be decreased in the rate of 6S RNA:pRNA release from RNAP (Shephard et al, 2010). This mutation would disrupt the base pair at the base of the extended hairpin predicted to form in the rearranged structure of E. coli 6S RNA (Supplementary Figure S3B), and would favour a new interaction between nt 52–54 and 152–154 (predicted by RNAfold for the E. coli 6S RNA core, nt 21–78–(N)4–113–165). Future studies will have to explore if the C132A mutation indeed affects the kinetics of the 6S RNA rearrangement via impairing formation of the extended hairpin and/or by causing folding alterations.
Two types of 6S RNA-templated pRNA transcripts were identified by deep sequencing (dRNA-seq) in Helicobacter pylori: 12–13 nt long canonical pRNAs initiated in the 5′-part of the central bulge, and a second type of pRNA, pRNA*, initiated ∼70 nt downstream of the canonical pRNA initiation site and ∼17 nt in length (Sharma et al, 2010). Such a second pRNA transcript complementary to 6S-1 RNA was not detected by dRNA-seq in B. subtilis (Beckmann et al, 2011). Intriguingly, when either the pRNA- or pRNA*-encoding sequence of H. pylori 6S RNA is blocked for intramolecular base-pairing in RNAfold analyses, the probability of extended hairpin formation in the 3′-part of the central bulge is increased in both cases (Supplementary Figure S5). The hairpin is very similar (9–10 bp) to that predicted for E. coli 6S RNA (Supplementary Figure S3B). Although H. pylori 6S RNA is predicted to refold more substantially (entire rearrangement of the apical helical arm) as a consequence of pRNA* synthesis (relative to pRNA synthesis), stable formation of this hairpin in the 3′-part of the central bulge seems to be a common feature induced by pRNA or pRNA* duplex formation with 6S RNA. Thus, it is tempting to hypothesize that both pRNA and pRNA* use this common mechanistic feature to rearrange H. pylori 6S RNA for release from RNAP. This raises several questions to be analysed in future studies, for example, if H. pylori RNAP binds 6S RNA randomly in the two reverse orientations, or if degradation of 6S RNA:pRNA and 6S RNA:pRNA* complexes differs.
The pRNA-induced conformational rearrangement of 6S RNA also shares features with larger regulatory riboswitches; for example, those involved in transcriptional attenuation, such as the M-box Mg2+ sensor, the FMN and the lysine riboswitches (for review, see Serganov, 2009). These riboswitches change their conformation upon ligand binding, such that so-called antiterminator sequences engage in an alternative base-pairing interaction that is mutually exclusive with formation of a Rho-independent transcription terminator. Another mechanistic resemblance becomes evident when comparing 6S RNA with the guanine-sensing riboswitch aptamer domain (GSRapt) of the B. subtilis xpt-pbuX operon: here, NMR analyses showed the free riboswitch to exist as a broad ensemble of flexible structures, where its three helices protruding from the central multiloop adopt many different relative orientations; however, upon ligand binding to the core region, the conformation becomes more restrained with decreased fluctuation of interhelical angles (Buck et al, 2007). This is in line with our bioinformatic evidence that the structure of 6S RNA:pRNA hybrids is rigidified relative to free 6S RNA. Unique features of 6S RNA relative to other riboswitches are its function as a global regulator of transcription and that the RNA directs the synthesis of its own ligand (pRNA) in cis.
The unique mechanism of 6S RNA release from RNAP via pRNA synthesis raises the question if pRNAs, which accumulate to considerable levels in outgrowing cells, might play additional roles in the regulation of gene expression (Wurm et al, 2010). Although the question still awaits a final answer, our results clearly show that a central pRNA function is exerted in cis through inducing a switch in the structure of 6S RNA in order to lift the block of RNAP.
Materials and methods
For northern blot analysis, structure probing, construction of the U136/U145U/A146 mutant 6S-1 RNA and bioinformatics see Supplementary data.
6S-1 RNA:pRNA annealing, gel shift assays and in vitro transcription
6S-1 RNA:pRNA annealing. In vitro transcribed 190 nt (mature) 6S-1 RNA was incubated with pRNA oligonucleotides (Noxxon; the LNA 8-mer, 5′-GTTCGGTC, was from RiboTask) and trace amounts of either 5′-32P-labelled 6S-1 RNA or pRNA in a volume of 4–6 μl 1 × TE buffer for 5 min at 95°C. For concentrations, see legends of Figures 1–3. Annealing of the denatured RNAs was then accomplished by stepwise cooling (5 min 80°C, 5 min 70°C, 5 min 50°C and 5 min 37°C) in a thermocycler (Biometra). The same annealing procedure was applied to samples were only one of the two RNAs (6S-1 RNA or pRNA) was present. Samples were mixed with 5 μl of 2 × native RNA loading buffer (2 mM MgCl2, 0.025% bromophenol blue (w/v), 0.025% xylene cyanol blue (w/v), 10% glycerol) and subsequently separated on a 10% native PAA gel (1 × TBE).
6S-1 RNA:RNAP gel shifts without transcription. To 6μl pre-annealed 6S-1 RNA (without or with pRNA) we added 2 μl of 5 × activity buffer (200 mM Tris–HCl pH 8.0, 25 mM MgCl2; 800 mM KCl, 5 mM DTT), 1 μl of heparin solution (400 ng/μl) and 1.06 μl σA-RNAP holoenzyme (8 μg/μl; prepared as described in Sogo et al, 1979) and samples were incubated for 30 min at 37°C to promote 6S-1 RNA:RNAP complex formation (f.c. σA-RNAP: 2 μM; f.c. 6S-1 RNA: 1 or 10 μM); samples were mixed with an equal volume of 2 × native RNA loading buffer (see above) and loaded onto a 7.5% native PAA gel (1 × TBE, or 0.5 × TBE, 160 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, pH 8.6).
6S-1 RNA:RNAP gel shifts with transcription. To 4μl pre-annealed 6S-1 RNA (6.25 μM) we added 2 μl of 5 × activity buffer, 1 μl of heparin solution (400 ng/μl) and 1.06 μl σA-RNAP holoenzyme (8 μg/μl) and samples were incubated for 30 min at 37°C. Transcription was then started by adding 2 μl NTP mix (1 mM each ATP, CTP, GTP and UTP; or an NTP mix lacking ATP; or 2 μl ddH2O instead); the f.c. of σA-RNAP was 2 μM, and that of 6S-1 RNA 2.5 μM.
Transcription of 32P-labelled pRNAs using 6S-1 RNA as template. 6S-1 RNA (f.c. 2.5–3.2 μM) was mixed with 2 μl 5 × activity buffer, 2 μl NTP mix B (1 mM each ATP, CTP, GTP and 0.25 mM UTP), 0.5 μl α-32P-UTP (20 000 Cerenkov c.p.m.) and 1.06 μl σA-RNAP holoenzyme (8 μg/μl) in a final volume of 10 μl, followed by incubation for 1 h at 37°C. Samples were mixed with an equal volume of 2 × denaturing RNA loading buffer (0.02% (w/v) bromophenol blue, 0.02% (w/v) xylene cyanol blue, 2.6 M urea, 66% (v/v) deionized formamide, 2 × TBE, pH 8.0) and loaded onto 20% denaturing PAA gels (1 × TBE).
Structure probing. In vitro transcribed 6S-1 RNA was 5′-endlabelled using T4 PNK (Fermentas) and (γ-32P-ATP) or 3′-endlabelled using T4 RNA ligase (Fermentas) and (5′-32P)pCp. After gel purification, 10 μM unlabelled 6S-1 RNA, trace amounts of radiolabelled 6S-1 RNA (usually 20 000 c.p.m. per reaction) or pre-annealed 6S-1 RNA:pRNA hybrids (see above) were subjected to partial enzymatic digestion using RNase T1, RNase V1 or partial chemical cleavage using NaHCO3 or Pb2+-ions. For further details see Supplementary data.
Supplementary Material
Acknowledgments
We thank D Helmecke for technical support. Funding from the Spanish Ministry of Science and Innovation to MS (Grant BFU2008-00215), and from the Deutsche Forschungsgemeinschaft to MM (GRK 1384) and to RKH (GRK 1384, SPP 1258, HA-1672/16–1) is acknowledged.
Author contributions: BMB performed 50% of the experiments and contributed to experimental design and manuscript writing; PGH performed 50% of the experiments and contributed to experimental design; MM performed the bioinformatic analysis (Supplementary Figure S3); DKW was involved in supervision of BMB and contributed to project design; MS purified the B. subtilis σA-RNAP holoenzyme; RKH was the principal investigator and contributed to experimental design, bioinformatics (Supplementary Figure S5) and did 80% of manuscript writing.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ando Y, Asari S, Suzuma S, Yamane K, Nakamura K (2002) Expression of a small RNA, BS203 RNA, from the yocI-yocJ intergenic region of Bacillus subtilis genome. FEMS Microbiol Lett 207: 29–33 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR (2005) 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11: 774–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann BM, Burenina OY, Hoch PG, Sharma CM, Kubareva EA, Hartmann RK (2011) In vivo and in vitro analysis of 6S RNA-templated short transcripts in Bacillus subtilis. RNA Biol 8: 839–849 [DOI] [PubMed] [Google Scholar]
- Beckmann BM, Grünweller A, Weber MHW, Hartmann RK (2010) Northern blot detection of endogenous small RNAs (∼14 nt) in bacterial total RNA extracts. Nucleic Acids Res 38: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee GG (1971) Sequence of 6S RNA of E. coli. Nat New Biol 229: 147–149 [DOI] [PubMed] [Google Scholar]
- Buck J, Fürtig B, Noeske J, Wöhnert J, Schwalbe H (2007) Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc Natl Acad Sci USA 104: 15699–15704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, GriffithsJones S, Eddy SR, Bateman A (2009) Rfam: updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildehaus N, Neusser T, Wurm R, Wagner R (2007) Studies on the function of the riboregulator 6S RNA from E coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA directed de novo transcripts. Nucleic Acids Res 35: 1885–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünweller A, Hartmann RK (2007) Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs 21: 235–243 [DOI] [PubMed] [Google Scholar]
- Hsu LM (2009) Monitoring abortive initiation. Methods 47: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH (2006) Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314: 1144–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Hoener ZU, Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR (2006) Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314: 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A (2009) The long and the short of riboswitches. Curr Opin Struct Biol 19: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, Stadler PF, Vogel J (2010) The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Shephard L, Dobson N, Unrau PJ (2010) Binding and release of the 6S transcriptional control RNA. RNA 16: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Inciarte MR, Corral J, Viñuela E, Salas M (1979) RNA polymerase binding sites and transcription map of the DNA of Bacillus subtilis phage Φ29. J Mol Biol 127: 411–436 [DOI] [PubMed] [Google Scholar]
- Suzuma S, Asari S, Bunai K, Yoshino K, Ando Y, Kakeshita H, Fujita M, Nakamura K, Yamane K (2002) Identification and characterization of novel small RNAs in the aspS-yrvM intergenic region of the Bacillus subtilis genome. Microbiology 148: 2591–2598 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM (2005) A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol 12: 313–319 [DOI] [PubMed] [Google Scholar]
- Vester B, Wengel J (2004) LNA (locked nucleic acid): high affinity targeting of complementary RNA and DNA. Biochemistry 43: 13233–13241 [DOI] [PubMed] [Google Scholar]
- Wassarman KM (2007) 6S RNA: a regulator of transcription. Mol Microbiol 65: 1425–1431 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Saecker RM (2006) Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 314: 1601–1603 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Storz G (2000) 6S RNA regulates E. coli: RNA polymerase activity. Cell 101: 613–623 [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willkomm DK, Hartmann RK (2005) 6S RNA an ancient regulator of bacterial RNA polymerase rediscovered. Biol Chem 386: 1273–1277 [DOI] [PubMed] [Google Scholar]
- Wurm R, Neusser T, Wagner R (2010) 6S RNA-dependent inhibition of RNA polymerase is released by RNA-dependent synthesis of small de novo products. Biol Chem 391: 187–196 [DOI] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.