Abstract
MBF and SBF transcription factors regulate a large family of coordinately expressed G1/S genes required for early cell-cycle functions including DNA replication and repair. SBF is inactivated upon S-phase entry by Clb/CDK whereas MBF targets are repressed by the co-repressor, Nrm1. Using genome-wide expression analysis of cells treated with methyl methane sulfonate (MMS), hydroxyurea (HU) or camptothecin (CPT), we show that genotoxic stress during S phase specifically induces MBF-regulated genes. This occurs via direct phosphorylation of Nrm1 by Rad53, the effector checkpoint kinase, which prevents its binding to MBF target promoters. We conclude that MBF-regulated genes are distinguished from SBF-regulated genes by their sensitivity to activation by the S-phase checkpoint, thereby, providing an effective mechanism for enhancing DNA replication and repair and promoting genome stability.
Keywords: cell cycle, DNA replication checkpoint, MBF, Nrm1, S. cerevisiae
Introduction
Periodic expression of families of genes during the cell division cycle is a critical mechanism imposing the orderly progression of cell-cycle events. The G1/S gene family encodes the components of the machinery required for cell-cycle initiation, DNA replication and repair (Breeden, 2003; Wittenberg and Reed, 2005). In the budding yeast Saccharomyces cerevisiae, expression of G1/S genes is regulated by the heterodimeric transcription factors SBF and MBF, each comprised of Swi6 and a sequence-specific DNA-binding protein, Swi4 or Mbp1, respectively. SBF primarily regulates genes involved in cell-cycle timing and morphogenesis, whereas MBF regulates the expression of many genes involved in DNA replication and repair (Koch et al, 1993; Bean et al, 2005; Wittenberg and Reed, 2005; de Bruin et al, 2006).
Many genes have DNA-specific recognition sequences for both SBF and MBF in their promoters (Iyer et al, 2001; Bean et al, 2005; Ferrezuelo et al, 2010), but neither the mechanism by which one transcription factor is favoured over the other nor the function of these redundant binding sites is known. Differences in the mechanisms for activation and repression of SBF and MBF may allow differential regulation of their target genes during the cell cycle. SBF is activated by relief of Whi5 repression (Costanzo et al, 2004; de Bruin et al, 2004), whereas MBF activation occurs independent of Whi5 (de Bruin et al, 2006). Furthermore, although both MBF- and SBF-regulated transcription are repressed by accumulation of B-type cyclin-associated CDK activity as cells enter S phase, MBF-regulated genes are repressed by a negative feedback loop wherein the co-repressor Nrm1, encoded by an MBF-target gene, binds to MBF as cells exit G1 phase (Amon et al, 1993; de Bruin et al, 2006).
Conservation of the G1/S transcriptional machinery belies its critical role in maintaining genomic stability and malignancy. The G1/S transcription pathway is altered in virtually all human cancers (Burkhart and Sage, 2008). The mammalian E2F transcription factor family (E2F1–8) (DeGregori and Johnson, 2006), functional homologues of SBF and MBF, regulate a large number of genes involved in cell-cycle progression, DNA replication and DNA repair. Mutations in Rb, the functional homologue of Whi5 in metazoans, lead to de-repression of those genes by liberating the active form of the E2F1 transcription factor, promoting cell-cycle progression even in the presence of DNA damage. Furthermore, the failure to inactivate the repressive forms of E2F promotes genomic instability by decreasing the levels of genes needed for DNA repair (Dominguez-Brauer et al, 2010).
To ensure genomic stability in the face of insults, including DNA replication stress and DNA damage, cells have developed checkpoint mechanisms that sense interference with cellular processes and transduce that information to the machinery regulating the cell cycle and other functions. When these checkpoints fail, cells are at risk of genome instability, transformation and malignancy (Hartwell and Weinert, 1989; Hanahan and Weinberg, 2000). In budding yeast, the intra-S-phase checkpoint, which senses DNA damage, and the DNA replication checkpoint, which detects DNA replication stress during S phase, exert their effects largely through the checkpoint effector protein kinase, Rad53, a functional homologue of human Chk1 and Chk2 (Sanchez et al, 1999). Among the many responses, the S-phase checkpoint induces the transcription of genes involved in DNA replication and repair, thereby promoting the resolution of replication stress to prevent genomic instability (Gasch et al, 2001). One of the pathways for this transcriptional response in budding yeast acts downstream of Rad53 via the Dun1 protein kinase to phosphorylate and inactivate the transcriptional repressor Crt1, thereby promoting expression of genes including those encoding ribonucleotide reductase (Zhou and Elledge, 1993; Huang et al, 1998). Although the importance of this transcriptional response has been well established in budding yeast, the same pathway is not conserved in fission yeast where DNA replication stress induces many genes involved in DNA replication and repair by activating the G1/S transcription factor MBF (de Bruin et al, 2008; Dutta et al, 2008).
Here, we describe a transcriptional regulatory pathway activated by the S-phase checkpoint in budding yeast that promotes the expression of MBF-target genes in cells responding to DNA damage and replication stress. Analysing this response using genome-wide RNA microarrays, we show that almost half of G1/S genes are induced in response to genotoxic stress during S phase, at least half of which is dependent upon Rad53. Nrm1, presumably acting via MBF, regulates 80% of those Rad53-dependent genes. We find that this response blocks repression by Nrm1, which is directly phosphorylated by Rad53. We conclude that an important hallmark of MBF-regulated genes that distinguishes them from SBF-regulated genes is their capacity to be activated by the S-phase checkpoint.
Results
DNA replication checkpoint induces expression of MBF-regulated genes
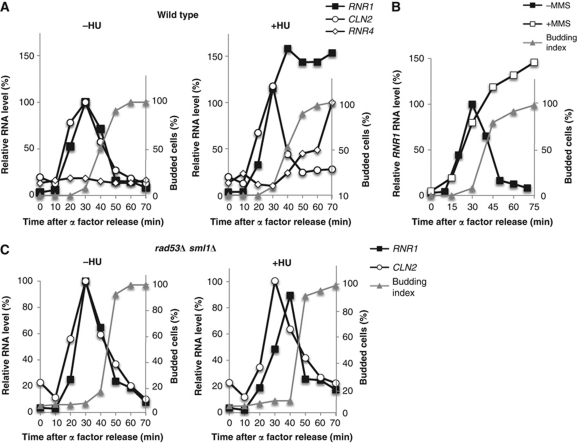
A large number of genes are induced in response to replication stress or DNA damage (Gasch et al, 2001). To understand the effect of replication stress on G1/S genes regulated by SBF and MBF, we analysed CLN2 and RNR1, respectively, in cells treated with hydroxyurea (HU). In an unperturbed cell cycle, the expression of both genes peaks during G1 phase and is repressed as cells enter S phase. However, in the presence of HU, the expression of the MBF target, RNR1, is induced and maintained (Figure 1A) whereas the SBF target, CLN2, behaves similarly in treated and untreated cells (Figure 1A). RNR4, which is regulated by Crt1, is repressed in untreated cycling cells but strongly induced by HU treatment (Figure 1A) (Zhou and Elledge, 1993).
Figure 1.
MBF-dependent transcription is induced in response to DNA replication stress. (A) MBF-regulated transcription is induced in response to HU. Wild-type cells were grown in rich medium, synchronized by α-factor and subsequently released into media without (left panel) or with (right panel) 0.2 M HU. CLN2 (SBF-regulated) and RNR1 (MBF-regulated) RNA from arrested cells (0 min) and cells released from the arrest for the indicated interval was quantitated by RT–qPCR and shown as percentage of the maximal RNA level in untreated cells. (B) MBF-regulated transcription is induced in response to MMS. Wild-type cells were grown as in (A) and released into medium with or without 0.033% MMS. RNR1 RNA was assessed by RT–qPCR and displayed as in (A). (C) Rad53 is required for induction of MBF-regulated transcription in response to DNA replication stress. rad53Δ sml1Δ cells were grown and synchronized as in (A) and subsequently released into media without (left panel) or with (right panel) 0.2 M HU. CLN2 and RNR1 RNA levels were quantitated and displayed as in (A). Budding index is shown as a marker of cell cycle progression.
This induction is dependent on a functional MBF as mbp1Δ mutants were unaffected by the presence of HU. Because MBF acts, in part, as a transcriptional repressor, MBF-dependent transcription is constitutively activated in this mutant, regardless of treatment with HU (Supplementary Figure S1) (de Bruin et al, 2006). Moreover, the transcriptional response is not HU-specific, since a similar response is observed in cells experiencing DNA replication stress due to DNA damage induced by methyl methane sulphonate (MMS) (Figure 1B).
To establish whether the transcriptional response requires the checkpoint pathway invoked by DNA damage or replication stress, we evaluated the involvement of the Rad53 protein kinase, a central mediator of those pathways. RAD53 is an essential gene but is rendered non-essential by inactivation of the target, Sml1. When the experiment performed in Figure 1A was repeated using a rad53Δ sml1Δ strain, the expression of MBF-dependent genes was no longer induced or maintained in response to HU (Figure 1C). sml1Δ alone does not alter the expression of G1 genes. We conclude that the induction of RNR1, an MBF-target gene, in response to DNA replication stress during S phase is mediated via the DNA replication checkpoint.
DNA damage and replication stress induce a broad array of G1/S genes
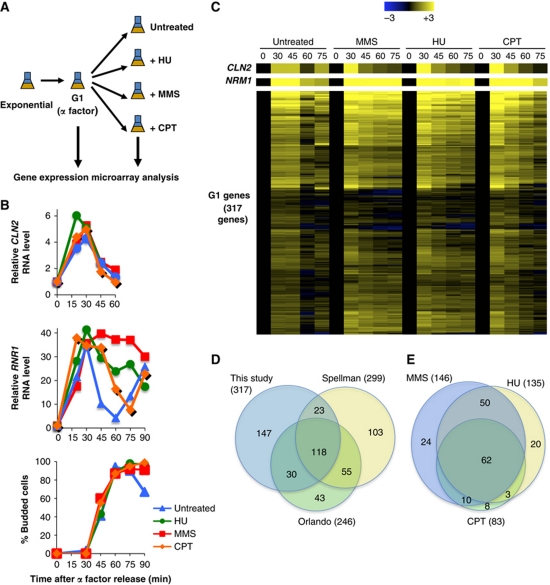
To determine whether the transcriptional response to DNA replication stress can be generalized to all MBF-regulated genes or is limited to a small subset of genes, we performed genome-wide expression analysis in cells responding to genotoxic agents that induce DNA replication stress or DNA damage during S phase. Wild-type cells synchronized in G1 phase by arrest with mating pheromone were released into the cell cycle and then treated with either 0.2 M HU, 0.03% MMS, 50 μM camptothecin (CPT) or left untreated (Figure 2A). Samples were taken at the peak of G1/S gene expression (30 min) and then at 15 min intervals until untreated cells completed a full cell cycle (Figure 2B). The well-established G1/S genes, CLN2 and RNR1, behaved as expected (Figure 2B). Treatment with either MMS or HU prevented the repression of RNR1 expression as cells exited G1 phase whereas CPT, which acts during S phase to induce DNA damage that is ultimately converted to double-strand breaks (DSBs) that are sensed and repaired during G2 phase, causes only a slight delay in repression of RNR1 expression. None of these treatments affected kinetics of passage through G1 phase as indicated by the appearance of budded cells or the repression of CLN2 gene expression. The RNA was analysed using Agilent yeast genome microarrays as described (see Materials and methods). The complete results of genome-wide expression analysis have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002) and are accessible through GEO Series accession number GSE33695 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33695).
Figure 2.
Genome-wide analysis of G1/S gene expression during the cell cycle in genotoxin-treated cells. (A) Schematic summary of the experimental design. (B) Relative RNA levels of CLN2 and RNR1 from the samples taken throughout the time course (A) were assessed by RT–qPCR. Values are relative to the 0′ time point. Budding index of the cells is shown (bottom panel). (C) Heat map of the expression of G1/S genes in untreated cells and cells treated with genotoxins. The selection of 317 G1/S genes based upon the expression profile in untreated synchronized cells is described in Materials and methods. The transcript level for each gene (one row) is expressed as a log2-fold change relative to the 0-min time point based upon the colour scale (top). CLN2 (SBF-regulated) and NRM1 (MBF-regulated) are provided as a reference (top). (D) Venn diagram showing the overlap between the G1/S genes identified in this study and those identified by Spellman et al (1998) and Orlando et al (2008; based upon subclusters (A–D) in Supplementary Figures S15 and S17). (E) Venn diagram representing the overlap between the G1/S genes induced in response to HU, MMS and CPT (see Materials and methods).
To understand the regulation of G1/S genes by genotoxic stress, it was necessary to select the data regarding members of that family from the genomic analysis. However, because there is little overlap between the members of the G1/S gene family defined by earlier genome-wide studies (Spellman et al, 1998; Iyer et al, 2001; Simon et al, 2001; Orlando et al, 2008), we defined a list of genes based upon our own analysis that were maximally induced at either 30 or 45 min after the release and repressed in untreated wild-type cells at 60 min. A group of 317 genes that conform to those parameters, including the well-established MBF- and SBF-target genes (Figure 2C and Supplementary Dataset 1), were compared with those identified in two other genome-wide expression analyses (Spellman et al, 1998; Orlando et al, 2008). Approximately half of the G1/S genes from our study overlap with those defined in each of the other studies (Figure 2D, left diagram) and about one third of our genes are found in all three lists. Focusing on the unique G1/S genes from each study, those from our study exhibit a greater enrichment in genes with Mbp1-, Swi4- and Swi6-binding motifs in their promoters, as well as a greater enrichment of genes falling into the cell cycle, DNA metabolic process and stress response GO slim categories (Supplementary Table S1). Along with the relatively poor overlap between studies, those factors highlight the need for simultaneous analysis of untreated, genotoxin-treated and mutant cells in the same study to ensure confidence in the conclusions regarding the transcriptional responses.
To identify G1 genes induced in response to treatments with genotoxic agents, we selected those that significantly increase in expression at 60 min compared with the same time point in untreated cells. Approximately half of the G1-specific genes are induced in response to MMS or HU (46 or 43%, respectively; Figure 2C; Supplementary Dataset 1). In general, MMS generated a higher level of induction of most of the affected genes than HU. Nevertheless, there is >75% overlap in the genes induced by these two treatments (Figure 2E).
Interestingly, the transcriptional response to CPT is strongly curtailed relative to MMS or HU (Figure 2C). However, 90% of the 83 G1/S genes induced in response to CPT are also induced by MMS or HU (Supplementary Dataset 1; Figure 2E). The significant overlap between these three treatments suggests that there is a common cluster of G1 genes induced in response to DNA damage and replication stress, independent of the genotoxic agent. The variation in the breadth of the response may be a consequence of differences in the mechanisms by which these drugs lead to DNA damage and, therefore, in the timing of checkpoint activation. MMS and HU both activate the checkpoint during S phase whereas the DSBs generated by CPT are primarily sensed during G2 phase, allowing cells to progress through S phase without fully activating the checkpoint (Redon et al, 2003).
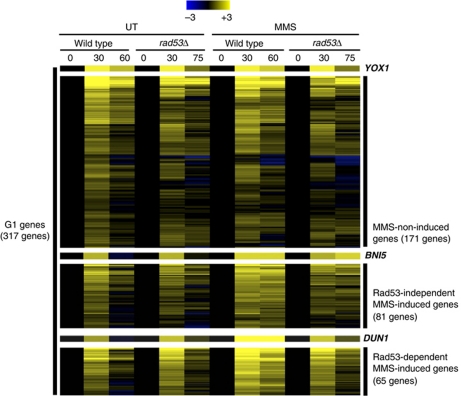
To determine whether this broad induction of the G1/S genes is dependent upon activation of the checkpoint, we repeated the genome-wide expression array using a rad53Δ mutant. Samples from the peak of G1/S gene expression (30 min) and the 75-min time point were analysed. Genes induced by MMS or HU in wild-type cells but significantly diminished in expression at 75 min relative to that at 30 min in the rad53 background were considered to be Rad53 dependent. In all, 65% of the HU-induced genes and ∼45% of the MMS-induced genes fit that criterion (Figure 3; Supplementary Figure S2 and Supplementary Dataset 1). Another group of genes maintained their induction after the addition of HU or MMS in the Rad53-deficient cells (Supplementary Figure S2). Some of these genes may appear Rad53 independent because they were insufficiently affected in the rad53Δ mutant to meet our criteria, others may be Rad53 independent but still dependent upon the checkpoint via Mec1 and yet others may be induced as a consequence of checkpoint-independent effects of the genotoxic compounds. The genes induced by MMS and HU that were classified as Rad53-dependent overlap 70% (Figure 4B, left diagram), whereas less than half of those classified as Rad53-independent overlap (Figure 4B, right diagram), consistent with coordinate regulation of a large portion of the genes through a common pathway.
Figure 3.
Analysis of the G1/S genes induced in response to replication stress in a checkpoint-dependent manner. Heat map of the expression of G1/S genes classified based upon MMS induction and Rad53 dependence (see Materials and methods). Transcripts levels are expressed as a log2-fold change relative to the 0-min time point based upon the colour scale shown (top). Values are shown for untreated and MMS-treated wild-type and rad53Δ cells at the time point where expression is maximal (30 min) and maximally repressed (60 and 75 min, respectively).
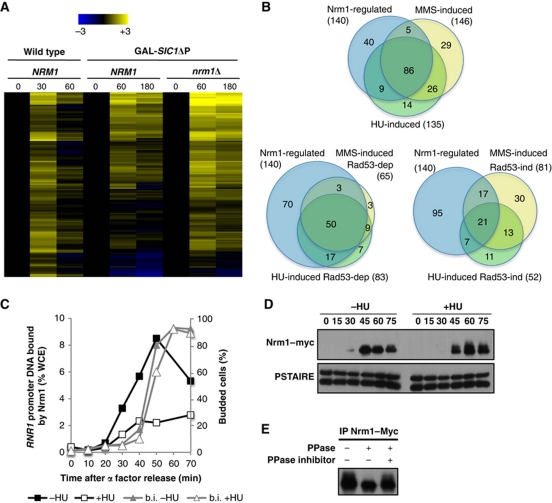
Figure 4.
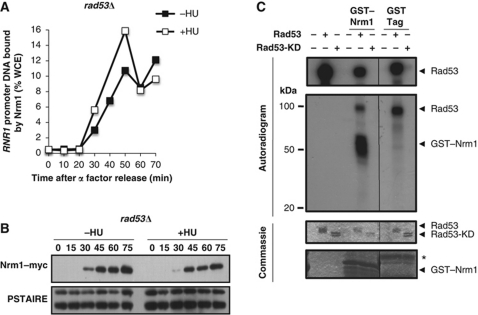
G1/S genes induced by genotoxic stress are primarily regulated by Nrm1. (A) Heat map showing expression of the 140 Nrm1-regulated G1/S genes. Transcript levels are shown for α-factor-synchronized wild-type cells (30 and 60 min) or wild-type and nrm1Δ cells expressing GAL-SIC1ΔP (60 and 180 min). Transcripts levels are expressed as a log2-fold change relative to the 0-min time point using the colour scale provided (top). (B) Venn diagrams showing the overlap between Nrm1-regulated G1/S genes and either HU- or MMS-induced G1/S genes (top), Rad53-dependent HU- or MMS-induced G1/S genes (left) and Rad53-independent HU- or MMS-induced genes (right). (C) HU abrogates Nrm1 binding to MBF-regulated promoters. Quantitation of Nrm1–myc binding to RNR1 promoter DNA by ChIP in wild-type cells synchronized by α-factor and released into medium with or without 0.2 M HU. Values are shown as a percentage of whole cell extract (WCE). (D) Nrm1 protein is phosphorylated in response to DNA replication stress. Immunoblot of Nrm1–myc from cells from the same time course as in (C). Anti-PSTAIRE is shown as a loading control. (E) Nrm1–Myc immunoprecipitated from cells treated with 0.2 M HU for 1 h was treated with λ phosphatase (PPase) in the presence or absence of phosphatase inhibitors (PPase inhibitor).
Most Rad53-dependent genes are regulated by Nrm1
The MBF-regulated gene RNR1 is de-repressed by DNA replication stress whereas the SBF-regulated gene CLN2 is unaffected. To determine whether the subset of G1/S genes induced by DNA replication stress are primarily those regulated by MBF, we identified the subset of the G1/S genes that are repressed by MBF-associated co-repressor Nrm1. We exploited the fact that, in cells lacking Clb/CDK activity, the repression of MBF-regulated genes as cells exit G1 phase depends upon Nrm1 (de Bruin et al, 2006), whereas SBF-regulated genes remain active because they are repressed in a Clb/CDK-dependent manner (Amon et al, 1993). Therefore, genome-wide expression analysis was performed in Nrm1-deficient or wild-type strains expressing a stabilized form of the Clb/CDK inhibitor Sic1 (SIC1ΔP) from the inducible GAL1 promoter. Cells were synchronized in G1 phase with mating pheromone, released into medium containing galactose to induce SIC1ΔP expression and RNA analysed from samples taken at 60 min (peak of induction for G1 genes) and 180 min.
We identified 140 genes that depend upon Nrm1 for repression based upon comparison of expression in wild-type and nrm1Δ mutants, including those genes generally considered to be MBF targets (see Materials and methods; Figure 4A; Supplementary Dataset 1). Unexpectedly, some genes previously considered to be SBF targets were also among the Nrm1-dependent genes (discussed below). The majority of HU and MMS-induced genes overlap with the Nrm1-dependent genes and that overlap increases when only the Rad53-dependent genes are considered (Figure 4B). In all, 80% of the Rad53-dependent genes induced in response to either HU or MMS are Nrm1-dependent genes, whereas only half of Rad53-independent genes induced by those genotoxins are dependent upon Nrm1. This shows that genes activated by the Rad53-dependent checkpoint pathway are primarily those regulated by MBF.
Nrm1 dissociates from MBF targets upon activation by DNA replication stress
MBF-regulated transcription in budding and fission yeast is repressed by the transcriptional co-repressor Nrm1 (Figure 4) (de Bruin et al, 2006). In fission yeast, Nrm1 is the target of the DNA replication checkpoint (de Bruin et al, 2008). We, therefore, evaluated whether Nrm1 binding to MBF-regulated genes in budding yeast is also regulated by replication stress using chromatin immunoprecipitation (ChIP). Nrm1 associates with the RNR1 promoter as cells progress into S phase during an unperturbed cell cycle whereas binding is abrogated in the same cells when replication is blocked with HU (Figure 4C). Moreover, a low mobility species of Nrm1 sensitive to treatment with protein phosphatase accumulates in the presence of HU (Figure 4D and E), indicating that it is phosphorylated. These data show that the interaction of Nrm1 with MBF is abrogated, perhaps as a consequence of phosphorylation, in response to DNA replication stress, leading to the maintenance of the MBF-regulated gene expression during S phase.
Induction of MBF-regulated genes is dependent upon Rad53
When activated in response to DNA replication stress or damage, Rad53 phosphorylates and activates Dun1, which then phosphorylates the transcriptional repressor Crt1 preventing it from binding to target promoters (Huang et al, 1998). However, our analysis of Dun1 showed that it is not required for transcriptional induction of RNR1, the phosphorylation of Nrm1 or the dissociation of Nrm1 from the RNR1 promoter (Supplementary Figure S4). This shows that Dun1 is dispensable for Nrm1 regulation by DNA replication stress during S phase.
Next, we determined whether Rad53 is required for Nrm1 phosphorylation and dissociation from promoters in response to DNA replication stress as suggested by the defect in induction of MBF-target genes in a rad53Δ mutant (Figure 1C). Treatment of rad53Δ mutants with HU leads neither to reduction in Nrm1 mobility (Figure 5B) nor to dissociation of Nrm1 from the RNR1 promoter (Figure 5A). Together, these results demonstrate that Rad53 is required for both responses, consistent with the requirement for transcriptional activation of MBF targets.
Figure 5.
Rad53, but not Dun1, is required for the Nrm1 inactivation in response to DNA replication stress. (A) Rad53 is required for inactivation of Nrm1 binding to MBF-regulated promoters in response to DNA replication stress. Nrm1–myc binding to RNR1 promoter DNA was determined by ChIP in samples from the same time course as in Figure 1C. Values are shown as percentage of WCE. (B) Rad53 is required for Nrm1 phosphorylation in response to DNA replication stress. Nrm1–myc was detected by immunoblot using cells from the same time course as in Figure 1C. Anti-PSTAIRE is shown as a loading control. (C) Rad53 directly phosphorylates Nrm1 in vitro. Phosphorylated products from an in vitro kinase assay performed using γ-32P-ATP and bacterially expressed recombinant Rad53 protein kinase or mutationally inactivated recombinant Rad53 kinase (Rad53-KD) and either recombinant GST–Nrm1 or GST-tag alone (second panel). Rad53 protein kinase activity is indicated by its autophosphorylation (top panel). Protein levels on the same gel are shown by Commassie Blue staining (lower two panels).
The loss of Nrm1 phosphorylation in the rad53Δ mutant suggested that the Rad53 protein kinase might directly phosphorylate Nrm1. To examine that possibility, we evaluated the capacity of bacterially expressed, enzymatically active Rad53 to phosphorylate GST–Nrm1 purified from bacteria (Figure 5C). The activity of Rad53 was assessed as the capacity of Rad53 to autophosphorylate itself (top panel, Figure 5C). Rad53 phosphorylated GST–Nrm1, but not GST alone, in vitro in the absence of other yeast proteins. In contrast, an enzymatically inactive mutant of Rad53 fails to phosphorylate GST–Nrm1. These results are consistent with our in vivo analyses (Figures 4D and 5B). Thus, we conclude that the S-phase checkpoint regulates MBF-dependent transcription via the direct Rad53-dependent phosphorylation of Nrm1.
Elimination of Rad53-dependent phosphorylation sites in Nrm1 abrogates induction of transcription by DNA replication stress
Rad53-dependent phosphorylation sites in Nrm1 were identified by two independent methods. First, a single serine residue, S145, was found to be phosphorylated by mass spectrometry in wild-type, but not rad53Δ mutant, cells responding to HU (see Materials and methods). Next, residues that were directly phosphorylated by Rad53 were identified by mass spectrometry using GST–Nrm1 phosphorylated in vitro in a Rad53 kinase assay performed with unlabelled ATP, as described above (see Materials and methods). That approach identified three phosphorylated residues with high confidence, T42, S47 and S240.
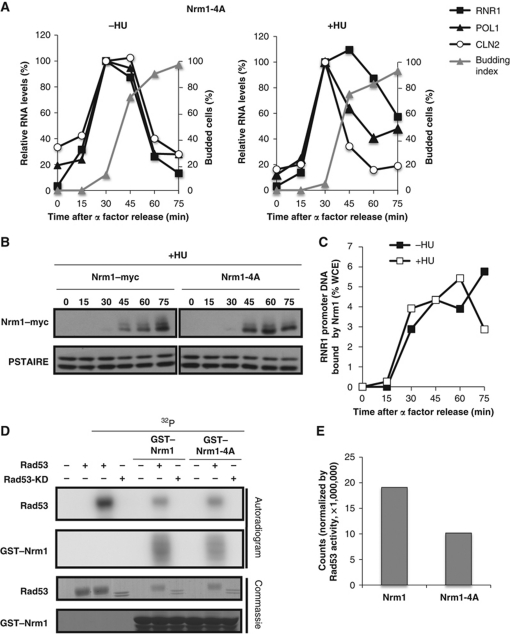
To assess the effect of those phosphorylated residues on regulation of Nrm1 binding and transcriptional repression, we generated a non-phosphorylatable mutant by mutating the four S/T residues to alanine (Nrm1-4A) and integrated this resulting mutant into nrm1Δ cells at the URA3 locus. We analysed the expression of CLN2, RNR1 and POL1, in cells expressing Nrm1-4A in the presence and absence of HU. The expression pattern of these genes in unperturbed cells is indistinguishable from untreated wild-type cells (Figures 1A and 6A). However, partial abrogation of the induction and maintenance of expression of MBF-target genes, RNR1 and POL1 is observed in NRM1-4A mutant following DNA replication stress (Figure 6A). That defect correlates with the decreased extent of phosphorylation of the Nrm1-4A mutant as compared with the wild-type protein based upon the difference in electrophoretic mobility (Figure 6B). Our data suggest that expression of MBF-regulated genes during S phase in response to replication stress occurs as a consequence of Nrm1 dissociation from MBF and the promoters induced by Rad53-dependent phosphorylation. Because MBF-regulated transcription fails to be appropriately maintained in the NRM1-4A mutant in cells responding to DNA replication stress, we used ChIP to evaluate whether replication stress failed to block association of the Nrm1-4A mutant protein with promoters. As expected, Nrm1-4A binds to promoters as cells progress from G1 into S phase even in the presence of HU (Figure 6C). Consistent with the reduced mobility of Nrm1-4A observed in HU-treated cells, phosphorylation of the mutant protein by Rad53 in vitro is reduced by nearly 50% relative to the wild-type protein (Figure 6D and E). The sites responsible for the remaining Rad53-dependent phosphorylation observed in vivo and in vitro have not been identified. Nevertheless, these results support a role for direct phosphorylation of Nrm1 by Rad53 in the abrogation of Nrm1 binding to MBF promoters and, thereby, in transcriptional induction of MBF-regulated genes in response to replication stress.
Figure 6.
Mutation of Rad53-dependent phosphorylation sites in Nrm1 impairs its function in response to DNA replication stress. (A) MBF-regulated transcription is not induced in response to HU in the Nrm1-4A mutant. Nrm1-4A mutant cells were grown in rich medium, synchronized by α-factor and subsequently released into media without (left panel) or with (right panel) 0.2 M HU. CLN2 (SBF-regulated) and RNR1 and POL1 (MBF-regulated) RNA from arrested cells (0 min) and cells released from the arrest for the indicated interval was quantitated by RT–qPCR and shown as percentage of the maximal RNA level in untreated cells. (B) Nrm1-4A is less phosphorylated in vivo in response to replication stress. Nrm1–myc and Nrm1-4A–myc were detected by immunoblot using cells from the same time course as in (A). Anti-PSTAIRE is shown as a loading control. (C) Nrm1-4A binds to MBF-dependent promoters despite the presence of HU. Nrm1-4A–myc binding to RNR1 promoter DNA was determined by ChIP in samples from the same time course as in (A). Values are shown as percentage of WCE. (D) Nrm1-4A is less phosphorylated than Nrm1 wild-type in vitro by Rad53. An in vitro kinase assay was performed using γ-32P-ATP and bacterially expressed recombinant Rad53 protein kinase or Rad53-KD and either recombinant GST–Nrm1 or GST–Nrm1-4A (second panel). Rad53 protein kinase activity is indicated by its autophosphorylation (top panel). Protein levels on the same gel are shown by Commassie Blue staining (lower two panels). (E) Quantification of Nrm1 and Nrm1-4A phosphorylation by Rad53 in the in vitro kinase assay shown in (D). Counts are normalized by Rad53 activity in each sample.
Discussion
Cells exposed to genotoxic stress during S phase maintain genomic stability by activation of the S-phase checkpoint pathway. One well-characterized transcriptional response to checkpoint activation is mediated by the Dun1 protein kinase acting downstream of Rad53 to phosphorylate and inactivate the transcriptional repressor Crt1 (Zhou and Elledge, 1993; Huang et al, 1998). Here, we report another mechanism for checkpoint activation of genes with important S-phase functions that acts independent of Dun1. We show that Rad53 directly phosphorylates the Nrm1 transcriptional repressor and inhibits its binding to MBF-regulated promoters, thereby, maintaining the expression of MBF-target genes in cells responding to DNA replication stress. MBF-target genes play roles in DNA replication and repair as well as other cell-cycle functions. This includes the S-phase cyclin Clb6, previously shown to be induced via a Dun1-independent mechanism by replication stress (Palou et al, 2010).
We have identified four residues in Nrm1 that are phosphorylated in a Rad53-dependent manner. When mutated, Rad53-dependent phosphorylation of Nrm1 is reduced and its binding to promoters is no longer blocked by replication stress (Figure 6). Consequently, MBF targets fail to be expressed during S phase. The incomplete effect of the Nrm1 phosphorylation site mutant on transcriptional induction can be attributed to the fact that Rad53 can still partially phosphorylate Nrm1 and, therefore, retains the ability to partially induce transcription. Although we presume that identification of the remaining Rad53 phosphorylation sites in Nrm1 will allow us to construct a mutant that completely abrogates both phosphorylation and checkpoint-dependent transcriptional induction, it remains possible that the regulation reported here acts in conjunction with the previously reported phosphorylation of Swi6 by Rad53 (Sidorova and Breeden, 2003).
The conservation of this regulatory pathway suggests that the induction of G1/S genes in response to DNA damage and replication stress is beneficial, perhaps due to the enhancement of genome integrity. We find that almost half of the G1/S transcripts induced by genotoxic stress are involved in DNA replication and repair, cell cycle and the general stress response (Supplementary Figure S3). Consistent with that notion, cells become hypersensitive to replication stress when an indestructible form of Nrm1 is overexpressed (de Bruin et al, 2006). However, the relative importance of the components of the checkpoint response is unclear. In S. cerevisiae, stabilization of replication forks appears to be the primary mechanism for the maintenance of cell viability upon exposure to DNA replication stress (Lopes et al, 2001; Tercero and Diffley, 2001). If that mechanism remains intact, disrupting other checkpoint-induced responses, for example, late origin firing, does not affect cell viability following replication stress (Zegerman and Diffley, 2010). Although the importance of activation of G1/S transcription for resistance to replication stress has been clearly demonstrated in Schizosaccharomyces pombe, its effect is modest compared with that of replication fork stabilization (de Bruin et al, 2008; Dutta and Rhind, 2009). Thus, it seems likely that the induction of G1/S genes by the checkpoint is one of a constellation of responses facilitating genomic stability during genotoxic stress.
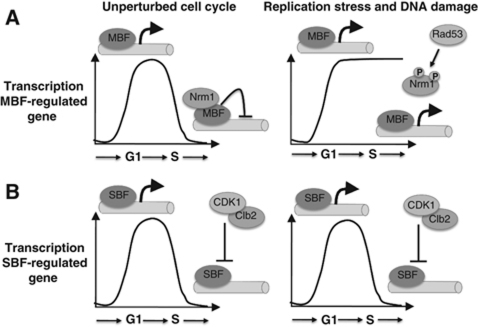
The involvement of two transcription factors, SBF and MBF, that promote a very similar, if not identical, pattern of transcription during the cell cycle has been enigmatic. Our findings suggest that the diversification of the SBF and MBF transcription factors provides an additional level of regulation relative to an organism with a single G1/S transcription factor, like the fission yeast in which G1/S transcription relies only upon MBF. The regulation of distinct set of genes by the two transcription factors presents the potential for two distinct patterns of gene regulation (Figure 7). For example, SBF, but not MBF, is subject to repression during early G1 phase by Whi5 (Costanzo et al, 2004; de Bruin et al, 2004). Another distinction between SBF- and MBF-regulated genes is in their mechanisms of repression. Whereas SBF targets appear to be repressed via a mechanism that is dependent solely upon Clb/CDK, MBF targets are additionally repressed via the co-repressor Nrm1 (Amon et al, 1993; de Bruin et al, 2006). We show here that the capacity of Nrm1-repressed genes to be activated by the DNA replication checkpoint pathway distinguishes them from SBF targets that are inactivated under the same conditions. Thus, budding yeast exhibit regulatory complexity of G1/S gene expression that is not available in fission yeast. The significance of that distinction may stem from differences between the two yeasts in terms of the importance of G1 phase for regulation of commitment to a new cell cycle. Whereas the primary checkpoint for cell cycle commitment in budding yeast occurs during G1 phase, that checkpoint is largely imposed during G2 phase in fission yeast. Most human cells resemble budding yeast, having a prominent G1-phase checkpoint, which may, in part, explain the involvement of a multiplicity of E2F transcriptional regulators and pocket proteins in that organism.
Figure 7.
Model for the regulation of G1/S genes by genotoxic stress. (A) The transcription of MBF-regulated genes is inactivated as cells progress through S phase by Nrm1 binding to their promoters. In response to genotoxic stress, Rad53 is activated and phosphorylates Nrm1, impeding its binding and allowing the expression of MBF genes to be maintained. (B) SBF-regulated genes are inactivated by B-type cyclin/CDK activity in S phase. In response to genotoxic stress, cells continue to have B-type cyclin/CDK activity and, thus, SBF-regulated genes are inactivated in a regular manner.
We used genome-wide microarrays to compare the effect of genotoxic agents on the abundance of RNA transcripts during the cell cycle in synchronized populations of cells. Although similar studies of gene expression in synchronous populations of cells traversing the cell cycle have been performed previously (Spellman et al, 1998; Orlando et al, 2008; and others), the effect of genotoxic stress has only been evaluated using asynchronous populations (Gasch et al, 2001). Using synchronous population of cells has enabled us to visualize effects that occur transiently during the cell cycle as well as revealing responses that are masked as a consequence of averaging abundance over an asynchronous population.
The size of the G1/S gene family (317 genes) described in this study is comparable to previous studies. Approximately one-third of the genes identified in those studies overlap including all of the commonly recognized G1/S genes (Figure 2 and Supplementary Dataset 1). The genes unique to this study are more highly enriched for Mbp1- and Swi4-binding sites and are assigned to the cell cycle, DNA metabolic processes and stress response GO slim categories (Supplementary Table S1), suggesting that we have captured a relatively larger portion of the G1/S genes. That, together with the disparity between the various studies, reinforces the need for a comprehensive study comparing the effect of multiple genotoxic agents on synchronized populations of both checkpoint-proficient and checkpoint-deficient cells.
Genome-wide expression analysis of synchronized populations of MMS and HU-treated cells revealed an effect on the timing of expression in approximately half of the genes of the G1/S family. The transcriptional response to MMS and HU is remarkably similar, consistent with the fact that both agents induce fork stalling, HU by depleting nucleotide pools (Lopes et al, 2001) and MMS via steric hindrance caused by DNA damage (Tercero and Diffley, 2001), resulting in the uncoupling of helicase and polymerase activities, thereby, activating the intra-S-phase checkpoint (Zou and Elledge, 2003; Byun et al, 2005). The fact that HU and MMS induce a very similar cluster of G1/S genes suggests that the S-phase checkpoint, regardless of its mechanism of activation, has a nearly identical transcriptional signature. This is consistent with our finding that 71% of the Nrm1/MBF-regulated genes were induced by either HU or MMS. We conclude that, among the G1/S genes, the transcriptional signature for genotoxins inducing replication stress is largely associated with regulation by MBF.
To our knowledge, the group of genes identified in this study represents the largest single group co-regulated by DNA damage and replication stress and is likely to represent an important component of the mechanism to avoid genomic instability. MMS and ionizing radiation were previously shown to act via Mec1, budding yeast ATR homologue, to induce a cluster of only nine genes representing a DNA damage signature (Gasch et al, 2001). We have identified 65 G1/S genes induced in response to MMS via a mechanism depending upon Rad53, only two of which, DUN1 and ALG14, were identified in that study (Gasch et al, 2001). The other members of the DNA damage signature were among the genes induced by genotoxins but were not members of the G1/S gene family. The G1/S genes identified here were likely missed in that study because the effect on expression occurs during a specific phase of the cell cycle and would be averaged over all the other phases represented in the untreated asynchronous population. Furthermore, that study may have eliminated some genes based upon their induction by other environmental stress conditions (Gasch et al, 2000), some of which also cause DNA replication stress and DNA damage.
Unexpectedly, our analysis revealed a small number of genes, previously considered to be specifically regulated by SBF, that, like MBF-regulated genes, are induced by DNA replication stress. No mechanism related to that describe here for MBF has been described for SBF-regulated genes. Genes binding SBF and MBF transcription factors have been previously described (Iyer et al, 2001; Bean et al, 2005; de Bruin et al, 2006; Ferrezuelo et al, 2010) but the relevance of that binding in terms of regulation of gene expression was not known. However, it is now clear that SBF binds to those genes during G1 phase but is replaced by MBF as cells pass from G1 into S phase (Bastos de Oliveira et al, 2012) and, although their expression is indistinguishable from other G1/S genes in untreated cells, their association with MBF renders them inducible by genotoxic stress during S phase.
In contrast to the robust induction of MBF targets observed in response to MMS and HU, CPT, an agent that only weakly impedes DNA replication, induces only a quarter of G1/S genes. Nevertheless, those G1/S genes that are induced overlap substantially with genes induced by MMS and HU. CPT induces DNA damage by stabilizing topoisomerase I–DNA complexes, which collide with replication forks to generate DSBs (Kaufmann, 1998). In S. cerevisiae, those breaks are sensed and repaired during G2/M and, consequently, do not robustly activate the intra-S-phase checkpoint, as indicated by the partial phosphorylation of Rad53 (Redon et al, 2003). That weak activation likely accounts for the induction of a smaller number of G1/S genes but whether it also reflects a difference in the nature of the signal remains to be established. Nevertheless, the extensive overlap between genes induced by CPT, MMS and HU is, again, consistent with a common G1/S transcriptional signature for the DNA replication checkpoint.
The regulation of G1/S transcription by the DNA replication checkpoint is conserved in the fission yeast, S. pombe, and in humans. In fission yeast, DNA replication stress induces the phosphorylation of Nrm1 by Cds1, a Rad53 orthologue (de Bruin et al, 2008). In humans, Chk1, an orthologue of Rad53 and Cds1, promotes E2F-dependent transcription of G1/S genes by preventing a repressive E2F from binding to target promoters (C Bertoli, S Klier, C Wittenberg and R de Bruin, unpublished results). The E2F-regulated gene family, like the MBF-regulated gene family in yeast, is rich in DNA replication and repair factors, suggesting that this response to genotoxic stress has been conserved throughout the eukaryotic kingdom to facilitate the maintenance of genome integrity.
Materials and methods
Strains
The table of yeast strains used is presented as Supplementary data. The PCR method of Longtine et al (1998) was used to disrupt SML1, RAD53, DUN1 and NRM1, to incorporate 13xmyc tag at the carboxy terminus of NRM1 and to introduce GAL1 promoter in the SIC1 locus to generate GAL-SIC1ΔP. Nrm1-4A mutant (T42A, S47A, S145A and S240A) was constructed via several rounds of site-directed mutagenesis with Quickchange XL site-directed mutagenesis kit (Agilent Technologies), followed by DNA sequencing to confirm the generation of mutations.
Cell synchronization
Mating pheromone arrest synchrony experiments were carried out as described (de Bruin et al, 2006). Genotoxic agents were used as follows: 0.2 M HU, 0.033% MMS or 50 μM CPT. In the experiments involving GAL-SIC1ΔP, cells were released from mating pheromone arrest (YEP-raffinose) into YEP-galactose medium.
Real-time PCR and RT–PCR
Total RNA was isolated using the RNeasy Kit (Qiagen). The iQ SYBR Green Supermix (Bio-Rad) was used for quantitative PCR on ChIP samples and the iScript OneStep RTPCR Kit with SYBR Green (Bio-Rad) was used for RT–PCR. Reactions were run on the Chromo-4 qPCR I system (MJ Research) using standard PCR and RT–PCR conditions. Data were analysed by using MJ Opticon Monitor Analysis Software 3.0.
Microarray expression profiling
Total RNA was isolated using the RNeasy Kit (Qiagen). Samples were processed as previously described (Kuo et al, 2010). Arrays were scanned using a GenePix 4000A microarray scanner and quantied with the GenePix 6.0 software package.
Microarray expression analysis
Prior to further analysis, the data from each array were subjected to background correction and LOESS normalization with the intensity values of within-array technical replicates (identical probes on the same array) averaged (Smyth, 2005). Quantile normalization was then applied to the entire data set (Smyth, 2005). Expression values were extracted using an empirical Bayes linear model with the R package LIMMA (Smyth, 2005). Genes were considered differentially expressed for P<0.05 after FDR multiple-test correction (Benjamini and Hochberg, 1995).
ChIP analysis
ChIP was performed as described (de Bruin et al, 2008).
Phosphatase assay
Nrm1-13xmyc was immunoprecipitated from cells treated with 0.2 M. HU for 1 h, using denaturing conditions and incubated at 30°C for 1 h with or without λ phosphatase, and with or without 2 mM NaF. Samples were resolved by 7.5% SDS–PAGE.
In vitro kinase assay
GST–Rad53 and kinase dead GST–Rad53 (Rad53-KD) were expressed in Escherichia coli BL21(DE3)RIL, purified by using glutathione-Sepharose beads and released from beads by overnight incubation with PreScission protease. GST–Nrm1 or GST alone was also expressed in E. coli BL21(DE3)RIL and purified by using glutathione-Sepharose beads. Kinase assay was performed at 30°C for 30 min in the presence of 100 μM ATP, and 5 μCi of [γ-32P]-ATP in 30 μl of kinase reaction buffer (50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 1 mM dithiothreitol and 2 mM NaF). Reactions were resolved by 10% SDS–PAGE, stained with Coomassie blue and subjected to autoradiography.
Identification of phosphorylated sites
For the identification of the Nrm1 phosphopeptide containing phosphorylation on S145, cells were treated with 100 mM HU for 2 h and protein extract was prepared for phosphoproteomic analysis as previously described (Albuquerque et al, 2008). In short, the protein extract was trypsinized and phosphopeptides were enriched by preparative IMAC purification and subsequently fractionated by hydrophilic interaction chromatography. Fractions were then analysed by LC-MS/MS using an Orbitrap XL mass spectrometer.
For the identification of the in vitro phosphorylation sites of Nrm1, T42, S47 and S240, a cold in vitro kinase assay was performed as described above. The sample was denatured and then reduced and alkylated prior to overnight digestion with trypsin. The protein digest was loaded onto a MudPIT column that was placed in-line with an 1100 quaternary HPLC pump (Agilent Technologies) and the eluted peptides were electrosprayed directly into an LTQ Orbitrap XL mass spectrometer (Thermo Scientific) using a 10-step MudPIT method. MS/MS spectra were extracted using RawXtract (version 1.9.9) and searched with the Sequest algorithm. Sequest search results were assembled and filtered using the DTASelect (version 2.0) algorithm.
Supplementary Material
Acknowledgments
We thank the members of the TSRI Cell Cycle Group for helpful discussions and comments. We thank David G Quintana for Rad53 and Rad53-KD plasmids. This work was supported by USPHS Grants GM084279 and GM085764 to TI, P41 RR011823 to JY and R01 GM059441 to CW.
Author contributions: AT, DK, RAMdB, TI and CW participated in designing the experiments; AT, TIK and MG performed most of the experiments; DK performed microarray analysis; AT and DK analysed the data; AA, JRY and MBS identified phosphorylation sites; and AT and CW wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7: 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A, Tyers M, Futcher B, Nasmyth K (1993) Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74: 993–1007 [DOI] [PubMed] [Google Scholar]
- Bastos de Oliveira FM, Harris MR, Brazauskas P, de Bruin RAM, Smolka MB (2012) Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J 31: 1798–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR (2005) High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics 171: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B 57: 289–300 [Google Scholar]
- Breeden LL (2003) Periodic transcription: a cycle within a cycle. Curr Biol 13: R31–R38 [DOI] [PubMed] [Google Scholar]
- Burkhart DL, Sage J (2008) Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004) CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913 [DOI] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, Aslanian A, Wohlschlegel J, Chahwan C, Yates JR 3rd, Russell P, Wittenberg C (2008) DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc Natl Acad Sci USA 105: 11230–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J 3rd, Russell P, Wittenberg C (2006) Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell 23: 483–496 [DOI] [PubMed] [Google Scholar]
- de Bruin RA, McDonald WH, Kalashnikova TI, Yates J 3rd, Wittenberg C (2004) Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117: 887–898 [DOI] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG (2006) Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med 6: 739–748 [DOI] [PubMed] [Google Scholar]
- Dominguez-Brauer C, Brauer PM, Chen YJ, Pimkina J, Raychaudhuri P (2010) Tumor suppression by ARF: gatekeeper and caretaker. Cell Cycle 9: 86–89 [DOI] [PubMed] [Google Scholar]
- Dutta C, Patel PK, Rosebrock A, Oliva A, Leatherwood J, Rhind N (2008) The DNA replication checkpoint directly regulates MBF-dependent G1/S transcription. Mol Cell Biol 28: 5977–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta C, Rhind N (2009) The role of specific checkpoint-induced S-phase transcripts in resistance to replicative stress. PLoS One 4: e6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, Futcher B, Aldea M (2010) The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol 11: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO (2001) Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12: 2987–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA (1989) Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634 [DOI] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605 [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO (2001) Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533–538 [DOI] [PubMed] [Google Scholar]
- Kaufmann SH (1998) Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim Biophys Acta 1400: 195–211 [DOI] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K (1993) A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261: 1551–1557 [DOI] [PubMed] [Google Scholar]
- Kuo D, Tan K, Zinman G, Ravasi T, Bar-Joseph Z, Ideker T (2010) Evolutionary divergence in the fungal response to fluconazole revealed by soft clustering. Genome Biol 11: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JE, Iversen ES, Hartemink AJ, Haase SB (2008) Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453: 944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palou G, Palou R, Guerra-Moreno A, Duch A, Travesa A, Quintana DG (2010) Cyclin regulation by the S phase checkpoint. J Biol Chem 285: 26431–26440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM (2003) Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep 4: 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ (1999) Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286: 1166–1171 [DOI] [PubMed] [Google Scholar]
- Sidorova JM, Breeden LL (2003) Rad53 checkpoint kinase phosphorylation site preference identified in the Swi6 protein of Saccharomyces cerevisiae. Mol Cell Biol 23: 3405–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, Young RA (2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697–708 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2005) Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor, Gentleman RVC, Dudoit S, Irizarry R, Huber W (eds) pp 397–420. New York: Springer [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI (2005) Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24: 2746–2755 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2010) Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ (1993) DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75: 1119–1127 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.