Abstract
Protease research has undergone a major expansion in the last decade, largely due to the extremely rapid development of new technologies, such as quantitative proteomics and in-vivo imaging, as well as an extensive use of in-vivo models. These have led to identification of physiological substrates and resulted in a paradigm shift from the concept of proteases as protein-degrading enzymes to proteases as key signalling molecules. However, we are still at the beginning of an understanding of protease signalling pathways. We have only identified a minor subset of true physiological substrates for a limited number of proteases, and their physiological regulation is still not well understood. Similarly, links with other signalling systems are not well established. Herein, we will highlight current challenges in protease research.
Keywords: cathepsin, inhibitor, protease, regulation, signal transduction
Introduction to proteases
Proteases control a great variety of physiological processes that are critical for life, including the immune response, cell cycle, cell death, wound healing, food digestion, and protein and organelle recycling. Their action is strictly controlled and imbalances in their activities have been found to be critical in a number of pathologies, such as cardiovascular diseases, inflammation, cancer, and neurodegenerative diseases, thereby suggesting proteases as suitable and valuable drug targets (Turk, 2006; Drag and Salvesen, 2010). In humans, there are almost 600 proteases and about 80 more can be found in the mouse, representing ∼2% of the genome. Proteases differ remarkably in their properties. Some are as small as 20–30 kDa and composed of essentially the catalytic domain(s) alone, such as the cysteine cathepsins, whereas some can be extremely large, multidomain protein complexes, such as the proteasome and tripeptidyl peptidase I. They vary significantly in their specificities from being highly selective, such as the proteases of the blood coagulation cascade which activate the subsequent protein in the cascade, or the caspases and granzyme B which cleave their substrates exclusively after Asp residues, to the highly non-specific, such as those involved in protein turnover and which include the cathepsins and the proteasome, as well as most exopeptidases. The different selectivitiy of the proteases has been widely exploited in different analytical applications from amino-acid sequencing and mass spectrometry analysis, where proteases like trypsin, which cleaves all unfolded protein substrates after every Arg and Lys residues only, to biotechnological applications such as removal of the tags from the recombinant proteins where proteases like thrombin or enterokinase have been frequently used.
Based on their different catalytic mechanisms, there are five major classes of proteases known in mammals including serine, cysteine, metallo, aspartic, and threonine proteases. Whereas cysteine, serine, and metalloproteases are widespread with ∼150–200 members identified in humans, threonine and aspartic proteases are much scarcer with <30 members of each class being identified (Lopez-Otin and Overall, 2002; Overall and Blobel, 2007; Lopez-Otin and Bond, 2008). However, not all proteases catalyse the cleavage of α-peptide bonds between the naturally occurring amino acids. For example, deubiquitinating enzymes (DUBs), a large group comprising almost 100 members in humans mostly belonging to cysteine proteases, hydrolyse the isopeptide bond(s) between ubiquitin or ubiquitin-like proteins and pro-proteins or target proteins (Reyes-Turcu et al, 2009). In addition to different catalytic mechanisms, proteases can cleave their substrates either at the termini, such as carboxypeptidases and aminopeptidases, or within the polypeptide chain, such as endopeptidases. Although exopeptidases and endopeptidases usually differ significantly, lysosomal cysteine cathepsins are an example where nature has been very economical. With just a handful of small changes, such as insertions of small structural elements including the loops and the remaining parts of propeptide regions onto an endopeptidase scaffold, endopeptidases have been converted into exopeptidases. Moreover, cathepsin B can act as both an endopeptidase or an exopeptidase, based on the opening or closing of the ‘occluding loop’, which depends on the pH of the local millieu (Turk et al, 2000, 2001c). Another important factor that affects protease activity and substrate selection is their localization. Approximately half of all proteases are extracellular, whereas the remainder proteases are intracellular, with a minor proportion being intramembrane, localized in plasma and organelle membranes. Interestingly, cysteine proteases that usually require reducing conditions for their activity are predominantly found intracellularly, whereas majority of the metalloproteases are extracellular. Collectively, all these different properties and abilities of proteases explain why they provide an excellent way of regulating cellular processes throughout the body (Lopez-Otin and Overall, 2002; Overall and Blobel, 2007; Lopez-Otin and Bond, 2008).
Protease signalling
Although proteases were not traditionally regarded as signalling molecules, this view is dramatically changing. However, there are important differences between protease signalling and the other types of cellular signalling, for example, receptor signalling or kinase signalling. Protease signalling is irreversible and the signal is transmitted through the cleavage(s) of protein substrates resulting in their activation, inactivation, or modulation of function (Turk, 2006). Proteases can thus not only activate proteins such as cytokines, or inactivate them such as numerous repair proteins during apoptosis, but also expose cryptic sites, such as occurs with β-secretase during amyloid precursor protein processing, shed various transmembrane proteins such as occurs with metalloproteases and cysteine proteases, or convert receptor agonists into antagonists and vice versa such as chemokine conversions carried out by metalloproteases, dipeptidyl peptidase IV and some cathepsins. In addition to the catalytic domains, a great number of proteases contain numerous additional domains or modules that substantially increase the complexity of their functions (Lopez-Otin and Overall, 2002; Lopez-Otin and Bond, 2008). A number of proteases act in cascades that allow much better amplification of the signal and more stringent regulation. Typical examples are the blood coagulation cascade, apoptosis, as well as activation of a number of metalloproteases from ADAM and MMP families by furin-like proprotein convertases (Turk, 2006). As it is often very difficult to draw the borders between different protease signalling events, it was suggested to use the terms protease networks or protease webs (Doucet et al, 2008; Krüger, 2009).
However, regardless of the mechanism or the pathway, the key signalling event in protease signalling is the change of function of a physiological substrate (Turk, 2006). A proper understanding of the protease signalling pathways therefore requires identification of the physiological substrates of proteases or protease degradomes (Lopez-Otin and Overall, 2002). To qualify as a physiological substrate of a protease, it is neither sufficient for a protein to contain the recognition sequence for a protease nor for it to be cleaved in an in-vitro assay, as practiced in the early days of biochemical studies of proteases. The substrate has to colocalize with the active protease, that is, be present in the same cellular compartment or at the same extracellular location, and then subsequently to be processed. Moreover, a number of studies have demonstrated that a large number of cellular proteins reside in multiprotein complexes, which could further limit their accessibility to proteases (Gavin et al, 2002; Janin and Seraphin, 2003). However, it is unclear at the moment, how many proteins undergoing proteolytic processing are indeed present in such complex forms. There are quite a few examples known where a protein substrate is in a complex during the cleavage reaction, such as ICAD (inhibitor of caspase-activated DNase) that is in a complex with CAD (caspase-activated DNase). Following ICAD cleavage by caspases during apoptosis, CAD is released from the complex, thereby initiating DNA fragmentation in the nucleus (Enari et al, 1998). However, no detailed studies have been performed to specifically address this question. This also raises a question as to the number of proteases active when in complexes, and how many can act alone. Clearly, proteases like the proteasome, γ-secretase as well as several serine proteases involved in blood coagulation such as the prothrombinase complex (a complex between Factor Xa and Factor Va required for thrombin activation) require complex formation to be able to process their physiological substrates. In a similar manner to the substrates, no real systematic studies have been performed to address these questions.
Every single protein synthesized is degraded by the proteasome and/or lysosomal proteases during its recycling or degradation and is therefore by default a physiological substrate of these proteases; a general degradation mechanism that is not generally considered as part of protease signalling. Consequently, to prevent undesired proteolysis, proteases involved in protein recycling and degradation are physically separated from the majority of other proteins by being contained within lysosomes or in a self-compartment (proteasome).
Identification of physiological protease substrates
The identification of physiological protease substrates is currently one of the major challenges in protease research. Initial studies essentially used a bottom-up approach, that is, identification of the protease responsible for the processing of an orphan substrate, thereby simultaneously validating the results. The first such studies, performed over half a century ago, led to the discoveries of the renin-angiotensinogen system (Page and Helmer, 1940) and angiotensin-converting enzyme (ACE; Skeggs et al, 1956). This approach was also successfully applied to identification of the proteases in the blood coagulation cascade (Davie and Ratnoff, 1964), furin as the processing enzyme of many prohormones in mammals, caspase-1 as the interleukin-1β processing enzyme (Thornberry et al, 1992), dipeptidyl peptidase IV as the processing enzyme of insulin-related hormones (Demuth et al, 2005) and intramembrane-cleaving proteases (Weihofen and Martoglio, 2003; Wolfe, 2009). This approach is still in use, and has recently led to the identification of cathepsin L/V as the histone H3-processing enzyme (Duncan et al, 2008). The usefulness of this approach is further demonstrated by the fact that a number of proteases identified in this way have also been validated as drug targets. Moreover, ACE inhibitors are still the most commonly used protease-targeting drugs (Turk, 2006; Drag and Salvesen, 2010).
The applicability of this approach is, however, limited as it is very labour intensive. The majority of proteases process more than one substrate, resulting in a functional redundancy that may mask the validation process. Therefore, additional approaches have been developed over the years, such as combinatorial fluorescent substrate libraries, positional scanning libraries based on covalent inhibitors, and phage display peptidic libraries (Matthews and Wells, 1993; Thornberry et al, 1997; Turk et al, 2001a). These approaches generated vast amount of data from which information could only be extracted with the simultaneous development of bioinformatic tools. Using these approaches, substantial success has been achieved in determining substrate specificities of several proteases, such as caspases (Thornberry et al, 1997). This latter seminal work identified the DXXD↓X amino-acid sequence as the consensus cleavage sequence for caspases-3 and -7 with an Asp residue in the P1 position being absolutely required. This information was then extensively used in a number of subsequent studies on identification of natural caspase-3 substrates. This approach proved highly successful since mutation of a selected Asp residue in the target protein was usually found sufficient not only to validate the protein as a caspase target, but also the selected cleavage site. Such studies were therefore of great help in the identification of substrate specificities of proteases and important for the development of in-vitro assays to follow protease activities in vitro or in complex samples and as a result they have been heavily employed in academic and industrial laboratories (Figure 1; Nicholson, 2000).
Figure 1.
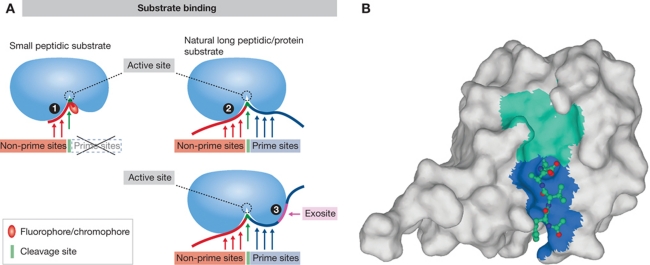
Binding of a substrate to a protease. (A) Schematic representation of substrate binding. The basic differences between binding of a small peptidic or peptidomimetic substrate and a larger peptide or protein substrate are shown. Small substrate binds tightly to the non-prime binding sites whereas the usually bulky fluorophore/chromophore (leaving group) binds to the S1′ site while the other prime sites remain empty (1). Larger peptidic substrates bind to both prime site and non-prime site binding sites, while the interaction with some individual binding sites can be looser (2). Exosites on protease surface serve as additional binding elements for large substrates to strengthen the interaction with the protease and allow recognition (3). They can be also used for discrimination between the different substrates. (B) Small substrate-mimicking inhibitors that helped tremendously in elucidating the substrate binding mechanism(s) often bind the same way as the small substrates (some more differences with metalloproteases), except that in covalent inhibitors the leaving group is replaced by a warhead (see A for a schematic representation). Crystal structure of caspase-1 in complex with the inhibitor YVAD-CHO (Wilson et al, 1994; 1ICE) revealed the insight into the mechanism of substrate binding to the active site of caspases as an example of such small molecule (substrate or inhibitor) binding. The surface of caspase 1 is shown in light grey. In the active site cleft, the non-primed and primed parts are shown as cyan and blue surface. The structure of the inhibitor (shown as a ball-and-stick model) has revealed the S1, S2, and S3 substrate binding sites. The non-hydrogen atoms C, N, and O are shown in blue, dark blue, and red, respectively. Figure was prepared with MAIN (Turk, 1992) and rendered with POV-Ray.
However, these studies were found to have a limited potential in identification of physiological protein substrates largely due to differences in the binding of small substrate molecules when compared with large protein substrates. Small peptidic substrates with a fluorophore attached only explore a small part of the non-prime substrate residues (i.e., substrate residues towards the N-terminus of the substrate from the scissile bond), whereas the prime residues (i.e., residues towards the C-terminus of the substrate from the scissile bond) remained largely unexplored (Figure 1). Another reason is that proteases may interact with their protein substrates through a considerably larger surface area as compared with the small peptidyl substrates (Figure 1). In a case study, this was demonstrated for the caspases. A recent proteomic study thus enabled discrimination between the substrate specificities of caspases-3 and -7 based on an undecapetide motif (P6-P5′; Demon et al, 2009). A further difficulty is imposed by those proteases that use exosites (i.e., those parts of the protease that are distal to the catalytic site that participate in substrate binding; Figure 1) for an efficient recognition of various physiological substrates, such as thrombin (Bar-Shavit et al, 1983) or various metalloproteases including ADAMTS-4 (ADAM with thrombospondin motif-4) and most MMPs (Overall and Blobel, 2007). The problem was at least partially addressed for MMPs with the approach called exosite scanning, which used MMP exosite domains as baits in the yeast-two hybrid screens to identify potential substrates (Overall et al, 2002).
More recently, advanced proteomics studies were found superior in this aspect. Here, research went in two directions: one devoted to identification of physiological substrates of proteases and the other devoted to determination of extended substrate specificities of proteases. Initial studies using DIGE and other forms of two-dimensional electrophoresis for sample separation resulted in identification of a few physiological substrates, such as demonstrated for caspases (Brockstedt et al, 1998; Gerner et al, 2000), granzymes A and B (Bredemeyer et al, 2004), MMP-14 (Hwang et al, 2004), and ADAMTS1 (Canals et al, 2006). On the other hand, non-gel-based methods such as combined fractional diagonal chromatography (COFRADIC; Gevaert et al, 2003), proteomic identification of protease cleavage sites (PICS; Schilling and Overall, 2008), terminal amine isotopic labelling of substrates (TAILS; Kleifeld et al, 2010), ICAT and iTRAQ variations (Tam et al, 2004; Dean and Overall, 2007; Enoksson et al, 2007), or genetically engineered enzyme use (Mahrus et al, 2008) were found to be superior, leading to identification of a great number of protease substrates, including those of caspases, granzyme B, and various MMPs (Van Damme et al, 2005, 2009; Mahrus et al, 2008; Morrison et al, 2009). These methods, in addition to identifying cleavage sites within protein substrates, enable determination of protease specificities, especially when used on the larger peptidic fragments that are generated from proteins by trypsin, endo GluC or some other protease cleavage, or on total cellular lysates. This offers a significant advantage compared with the combinatorial libraries as it can be used to define not only non-prime site specificity, but also extended prime site specificity. However, methods like PICS have to be performed with care. Generation of the larger initial peptides by trypsin or any other selective protease might be problematic as it can result in an apparently completely different specificity of proteases under investigation that readily accept the same P1 residues as the protease used in peptide generation. A typical example is cysteine cathepsins, where basic residues are well accepted in the P1 position, when combined with the use of trypsin for peptide generation. Recently, the approach has been expanded allowing the C-termini of the cleavage sites within the substrates to be identified, thereby enhancing the possibility of identifying the cleavage site (Schilling et al, 2010; Van Damme et al, 2010). Another approach that simultaneously allows identification of cleavage sites and identification of stable fragments is Protein Topography and Migration Analysis Platform (PROTOMAP). Due to the identification of stable fragments, this approach at the same time offers an advantage of initial validation of cleavage sites (Dix et al, 2008). In addition, there are various bioinformatic methods that have been found to be indispensable in extracting information from the database libraries and have helped in identifying additional protease substrates, such as demonstrated for granzymes (Barkan et al, 2010).
Unfortunately, none of the methods mentioned above is ideal and each suffers from certain limitations. A recent analysis of several cleavage site prediction programs, based on such experimental methods, has revealed that our understanding of protease processing is still relatively confined. In particular, a clear insight into the processing roles of non-specific proteases is still lacking (Verspurten et al, 2009). In addition to the non-specific proteases such as the cathepsins, investigating proteases with a requirement for an exosite (e.g., thrombin) is still a serious technological challenge. Even more challenging is the validation of these findings. In this aspect, mouse knockout/knock-in and transgenic models and the use of RNAi were found to be absolutely indispensable (Turk, 2006; Overall and Blobel, 2007). Alternatively, protease inhibitors can also be used in validation. However, they have to be used with extreme care as their specificity is often limited (Rozman-Pungerčar et al, 2003; Turk, 2006; Drag and Salvesen, 2010). All these combined efforts have generated a vast amount of data, which has greatly advanced our knowledge.
Challenges in protease signalling
The primary cleavage of a protein substrate is largely responsible for the change of its function (Turk, 2006). However, defining this primary cleavage event could not be satisfactorily resolved by the above-mentioned approaches due to the predominant use of long-term incubation times. More specifically, only the surface-exposed regions of substrates are accessible to proteases, whereas internal regions can only be cleaved after substantial structure disassembly takes place as a consequence of protein degradation and/or unfolding. And these secondary cleavages resulting from the prolonged incubation were usually observed, masking the true picture.
Until recently, proteases were believed to initially process their substrates either at the termini or within the exposed loops, whereas cleavages within the secondary structure regions were considered unlikely. However, a recent study has demonstrated that cleavage can occur within the α helices far more frequently than expected, occurring almost as frequently as cleavages within the unstructured regions (Timmer et al, 2009). Using a very elegant approach, this group has overcome the major problem in such an experiment that of the sequential processing of a substrate. In order to reduce the number of cleavages they have used two highly specific proteases, GluC and caspase-3, which cleave specifically after Glu and Asp residues, respectively. In addition, they have designed a procedure by which only the single cleavage events were considered. The proteomic findings were verified in an in-vitro system using individual recombinant proteins. Since E. coli does not express proteases with Asp or Glu specificity, its proteome has been considered as unbiased. Interestingly, a kinetic comparison between the activity of mammalian caspase-3 on E. coli substrates and its natural mammalian substrates has revealed that the former were largely inferior to the natural substrates with up to a few orders of magnitude lower kcat/Km values. This suggests that proteases and their substrates have co-evolved and that this is crucial for successful signalling. Nevertheless, despite the high frequency of the cleavage sites found within helices, extended loops and unstructured interdomain linker regions still seem to be the preferable processing sites for the activation of proteins (Timmer et al, 2009). This is, however, true only for folded substrates. Two excellent examples of this are the activation of Bid in apoptosis and the inhibition mechanism of serpins. Bid has been found to be processed at different sites by a number of proteases (caspase-8, calpains, cathepsins B, L, S, H, D, granzyme B; Turk and Turk, 2009). The sites are all located within the same large loop (Chou et al, 1999). Similarly, serpins are initially cleaved by their target proteases within the reactive site loop (RSL) but remain trapped by covalent bond to the catalytic serine residue of the protease through the P1 residue. In the next step, the cleaved serpin undergoes a major conformational change in which the cleaved RSL inserts into the centre of the β-sheet, resulting in a huge protease movement to the opposite end of the serpin (∼70 Å) (Schulze et al, 1990; Huntington et al, 2000). Moreover, in the case of crossreactive serpins, which inhibit serine and/or cysteine proteases, the cleavage site varies within the loop depending on the protease (Schick et al, 1998; Al-Khunaizi et al, 2002).
Even though the problem of the initial cleavage can be at least partially resolved by measuring cleavage kinetics as recently demonstrated for granzyme B (Plasman et al, 2011), there is still another, even larger, problem remaining: that of differentiation between the signalling and the bystander events. There is no guarantee that a rapidly cleaved protein is also a physiologically relevant substrate required for a signalling pathway to proceed, again indicating the importance of data validation. For example, several hundred caspase substrates have been identified. However, only for a handful of them it can be said with confidence that they are directly involved in apoptosis progression. Among these are the executioner caspase zymogens and Bid, which are activated, and PARP and ICAD (DFF45), which are inactivated (Timmer and Salvesen, 2007).
Another problem is posed by those substrates that are activated through the initial cleavage, but become degraded through subsequent processing events. Although these initial fragments play an active role in signalling, they are very difficult to identify due to their transient appearance, as they disappear from the sample.
As already mentioned above, proteases that bind their substrates through exosites represent an investigative challenge, as do multidomain proteases or proteases that change their properties upon binding to other proteins or other molecules. One example of the latter is thrombin that normally cleaves fibrinogen thereby finalizing blood clot formation. However, upon thrombomodulin binding, thrombin–thrombomodulin complex exhibits different specificity and instead activates the serine protease protein C, thereby acting as a negative feedback regulator of blood coagulation (Davie et al, 1991). The other example is glycosaminoglycans, which are known to substantially modify protease activities. When complexed to cathepsin K, chondroitin sulphate was shown to convert it into a very potent collagenase (Li et al, 2002), whereas heparin binding was found to substantially modify the properties of a number of serine proteases. Although these issues can be at least partially addressed using current proteomic technologies, no systematic studies have been performed so far.
Orphan enzymes represent another, though technologically less difficult challenge. Using proteomic methods and subsequently validating them in cellular or in-vivo models may help in elucidating their biological functions through the discovery of their true physiological substrates (Overall and Blobel, 2007).
In addition to qualitative assessment and identification of cleavage sites in substrates, a further challenge will be to quantitatively determine the extent of processing at a given site in a protein substrate. Although it should be possible to determine the extent of cleavage, its physiological relevance would remain questionable since for a physiological process to proceed, a complete processing at a single site is normally not required. A typical example can be seen with the executioner caspases such as caspase-3 that are never completely activated during apoptosis in a cellular system. Determining the actual threshold of a given physiological process therefore remains a major challenge for the future.
Protein degradation as a signalling pathway
Protease signalling can be a complex process, varying, at its simplest, from single proteolytic steps to sequential processing and at it most complex, signalling cascades. This latter, in addition, provides an excellent mechanism for system regulation (Turk, 2006). Although at first glance it may seem unlikely, sequential degradation of a protein can also lead to signal transduction. The best examples are antigen processing and presentation to MHC-I and MHC-II molecules; the essential components of adaptive immunity. There is, however, a fundamental difference between protein processing and degradation. Whereas during processing proteins are properly folded, during degradation such as observed during antigen processing they are at least partially unfolded as a consequence of the size restriction of the proteasome active site or of the acidic pH that is found in the endolysosomal system. This substantially increases the number of potential cleavage sites in a protein and thereby the chance of being processed.
It is the ubiquitin-proteasome system that plays the major role in MHC-I antigen processing. The proteasome is not a selective protease and the peptidyl fragments produced by the classical proteasome vary in size from 3 to 23 residues. They are rapidly degraded further by numerous other proteases such as thimet oligopeptidase and aminopeptidases. However, this size variation is too large for the MHC-I molecules, since their peptide binding groove can only accommodate peptides 8–12 residues long, with a typical antigenic peptide being 8–10 residues long. A proteasome species, called the ‘immunoproteasome’ in which the classical β1, β2, and β5 catalytic subunits are replaced by a special set of catalytic subunits, β1i (LMP2), β2i (MECL1), and β5i (LMP7), is much better suited for antigen processing as it generates longer peptides with the correct C-terminus due to cleavage after the hydrophobic and basic residues. This enables easier transport by the transporter associated with antigen processing (TAP) and more efficient binding to MHC class I molecules, although some trimming by aminopeptidases may still be required. Those peptidic fragments longer than 15 a.a. are trimmed by tripeptidyl peptidase II (TPP II) prior to aminopeptidase action. Antigenic peptides are then transported to the endoplasmic reticulum for final processing to the correct size (8–9 residues) by the ER aminopeptidases (ERAP-1; ERAP-2) and loaded onto the MHC-I molecules by a specific TAP, thereby escaping complete degradation (Wilkinson, 2000; Glickman and Ciechanover, 2002; Kloetzel and Ossendorp, 2004; Murata et al, 2009; Rock et al, 2010). The system is relatively inefficient, as it was found that on average only 1 in 2000 molecules degraded forms a stable complex with MHC class I molecules and becomes presented at the cell surface (Princiotta et al, 2003).
Processing of antigens by the proteases of the endosomal/lysosomal system for their presentation by the MHC class II molecules is somewhat different. Endosomes/lysosomes contain a large number of proteases, including cysteine cathepsins, the aspartic protease cathepsin D and aparaginyl endopeptidase (AEP), which are all involved in antigen processing. They are extremely well suited to this job as they are, with the exception of AEP, not very specific, and include endopeptidases (cathepsins B, L, S, and D) as well as exopeptidases (cathepsins X, H, C, and B). With the exception of the processing of some specific antigens, antigen processing is largely non-specific with a significant amount of redundancy, as none of the single knockouts of the proteases involved exhibited any problem with antigen processing (Zavas̆nik-Bergant and Turk, 2006; Vasiljeva et al, 2007; Bird et al, 2009). In contrast, the stepwise processing of the MHC class II chaperone invariant chain (Ii), which is required for the MHC class II molecules maturation, is a highly regulated process. In the first step, Ii is C-terminally processed into the 22-kDa fragment called leupeptin induced protein (LIP). Subsequent C-terminal processing of LIP results in the 18-kDa intermediate and in the ∼100 a.a. residues long N-terminal fragment called Iip 10 or small leupeptin induced protein (SLIP) that extends to the C-terminal residue of the CLIP fragment. This C-terminal processing can be accomplished by multiple aspartic or cysteine proteases. However, the final N-terminal processing of the Iip 10/SLIP into the CLIP fragment is solely accomplished by cathepsin S in bone marrow-derived immune cells and by cathepsin L (V in human) in thymocytes, as revealed by the in-vivo studies (Nakagawa et al, 1998, 1999; Shi et al, 1999; Tolosa et al, 2003). The CLIP fragment then protects the MHC class II molecules until they reach the endosomes. In the endosomes, the CLIP–MHC class II complex is recognized by the HLA-DM and HLA-DO molecules (in humans), which mediate substitution of CLIP for antigenic peptides. In contrast to the antigen processing occurring in the MHC-I response, where only peptides of the correct size can be loaded onto TAP, the larger peptides can be bound to MHC-II molecules first and trimmed to the correct size (12–15 residues) afterwards (Sercarz and Maverakis, 2003; Bird et al, 2009).
In addition, this pathway also indicates that even the very non-selective proteases such as cathepsins can accomplish very selective processing, such as stepwise processing of the invariant chain to the CLIP fragment. This suggests that the dogma that the cathepsins only degrade proteins applies only to partially unfolded proteins, such as those destined for antigen processing or recycling, whereas properly folded proteins are only processed at a very limited number of sites possibly leading to their activation. This argument is further strengthened by the role cathepsins play in the activation of several TLRs, such as TLR-9, where proteolytic processing of the N-terminal, luminal part of the receptors by the cathepsins is absolutely required for their activity (Asagiri et al, 2008; Park et al, 2008) and for their role in the release of T3 and T4 hormones from thyroglobulin (Brix et al, 2008).
Regulation of protease activity
Since the function of a protease is to cleave proteins and peptides, their action could be extremely harmful if not controlled. As a result, proteases are not always active in their physiological environment. They can exist as inactive zymogens or can remain trapped in an inactive complex with an inhibitor. However, only the active protease fraction can cleave substrates and thus participate in protease signalling, thus presenting additional difficulties for protease research. Moreover, a protease does not need to be constantly active in order to trigger a signalling pathway; it is sufficient to generate a significant amount of activated protein, which in turn carries on the signalling function, or to inactivate a regulatory protein to shift the balance. A typical example is the lysosomal cathepsins that are slowly inactivated at neutral pH, but can either trigger apoptosis through cleavage of Bid or degrade extracellular matrix proteins when secreted. Therefore, analysing protease activity in vivo is the current challenge and one of the hottest areas of current protease research (Paulick and Bogyo, 2008).
Protease activities are regulated at multiple levels and the complexity of regulation usually increases with the complexity of the organism. The first step is regulation of transcription. Not every protease can be found in every tissue and cell types: some are ubiquitous, such as the proteasome, the lysosomal cathepsins B, L, C, and D, and the caspases, whereas a number have an extremely stringent expression pattern, being found at a very few locations, often reflecting their specialized roles. One such example is cathepsin G that can be found only in the azurophilic granules of the neutrophils (Bird et al, 2009). In addition, some, like the housekeeping proteases, are expressed constitutively, whereas other groups can be differentially expressed during development, homeostasis or under other special conditions. An example of the latter is caspase-3. During development, it is highly expressed in the brain, where it has a major role in removal of defective neurons (Kuida et al, 1996), whereas during homeostasis and later during senescence, its expression levels in the brain are considerably downregulated, probably to prevent unwanted neurodegeneration (Yakovlev et al, 2001). Other examples are MMPs and cathepsins, which are generally heavily upregulated during inflammation (Coussens and Werb, 2002; Vasiljeva et al, 2007).
The next regulatory step for most proteases is their activation. Proteases are synthesized as inactive zymogens, which generally require limited proteolysis for their activation (Figure 2). In general, we discriminate between the zymogens, where the active site is already formed but sterically blocked by the propeptide (e.g., papain-like cysteine proteases, various aspartic and metalloproteases), and the ones, where the active site is not formed and require a conformational rearrangement to form it (e.g., chymotrypsin-like serine proteases, caspases) (Khan et al, 1999; Fuentes-Prior and Salvesen, 2004). Further, activation can be either autocatalytic or performed by other proteases. The autocatalytic mechanisms are somewhat distinct with a minor conformational rearrangement often being the first step. This is followed by a proteolytic step, which often proceeds in trans with one molecule of the same protease species cleaving another (Caglič et al, 2007). Sometimes, proteases are activated with the assistance of activation complexes, such as some upstream caspases that are activated with the help of DISC (death-inducing signalling complex; Kischkel et al, 1995) the apoptosome (Zou et al, 1999) and the inflammasome (Martinon et al, 2002), as well as a number of blood plasma serine proteases (Davie et al, 1991). The size of the cleaved fragment varies from short peptides such as those observed with the granzymes (Jenne and Tschopp, 1988) and trypsinogen (Davie and Neurath, 1955) to whole domains as in the case of cysteine cathepsins (Turk et al, 2001c) and furin (Seidah and Prat, 2007). Prodomains of the latter two are believed also to act as intramolecular chaperones and are required for proper trafficking of the enzymes. However, there are exceptions and proteolysis is not always needed for activation, as has been demonstrated for the allosteric activation of Factor VII by tissue factor that converts a poor protease into a very potent one (Dickinson et al, 1996), and the proapoptotic cysteine proteases caspases-8 and -9, where activation is driven by dimerization (Renatus et al, 2001; Boatright et al, 2003; Donepudi et al, 2003). The latter activity is also an important factor in determining enzymatic activity as some proteases are inactive as monomers or do not even exist as monomers, as is the case with the proteasome (Löwe et al, 1995), β-tryptase (Pereira et al, 1998), bleomycin hydrolase (Joshua-Tor et al, 1995), meprins (Ishmael et al, 2001), and cathepsin C (Turk et al, 2001b). A very special case is TIMP-2 (tissue inhibitor of metalloproteases) which can, in addition to its inhibitory function, also participate in the activation of MMP-2 through a formation of a trimolecular complex with the membrane type 1-MMP (MT1-MMP), where the TIMP-2–MT1-MMP complex serves as a receptor for regulating the activation of MMP-2 (Butler et al, 1998).
Figure 2.
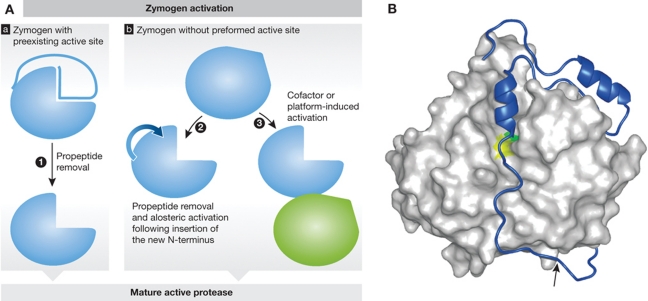
Protease activation. (A) Schematic representation of protease zymogen activation with proteases where active site is preexisting (a) or non-existing (b). (1) Propeptide removal in proteases with preexisting active site such as seen in cysteine cathepsins (see B as an example); (2) allosteric or conformational activation of proteases with distorted active site, such as chymotrypsin-like serine proteases (granzymes and trypsin), and some caspases; (3) conformational rearrangement and formation of the active site upon cofactor or platform-based activation such as seen in caspases and during Factor VII activation. (B) Crystal structure of a zymogen form of a protease offers a major support in understanding the mechanism of zymogen activation. Procathepsin B is shown as an example of a protease with preexisting active site. Propeptide region of cysteine cathepsins covers the active site cleft of the enzyme thereby blocking access to substrates. The mature part of procathepsin B (Podobnik et al, 1997; 3PBH) is shown as a white surface, whereas the active site residues C29 and H199 areas are in yellow and green, respectively. The propeptide is shown as a blue ribbon with the autocatalytic cleavage site (M56-F57) marked with an arrow. Figure was prepared with MAIN (Turk, 1992) and rendered with POV-Ray.
Once activated the other major system for regulation of protease activity is their inhibitors, either endogenous or exogenous. By definition, the function of a protease is to cleave a protein substrate, while the function of an inhibitor is to prevent it. Here, nature has obviously been economical as the number of endogenous human inhibitors identified is substantially smaller than the number of proteases identified, with only a slightly higher number found in the mouse (Rawlings et al, 2010). The main reason for this seems to be the substantially reduced necessity for specificity of the inhibitors for their target proteases, as one inhibitor can inhibit several proteases. In order to achieve that the inhibitors use several common mechanisms to inhibit their targets (Figure 3). The main inhibitors of all the metallo-endopeptidases are only four proteins, TIMP-1 TIMP-2, TIMP-3, and TIMP-4. Moreover, no mammalian protein inhibitors of aspartic proteases have, as yet, been identified. The majority of the known inhibitors thus inhibit serine proteases (e.g., serpins, serine protease inhibitors, and Kunitz type inhibitor; Figure 3) and some fewer cysteine proteases (e.g., cystatins and thyropins; Figure 3). In addition, α2-macroblobulin, circulating in blood plasma, is a very special inhibitor capable of inhibiting various classes of proteases. Several inhibitors have exhibited cross-class inhibition, that is, inhibiting targets from a different protease class. This was first observed with the serpin CrmA (cowpox response modifier A from pox viruses) inhibiting cysteine proteases from the caspase family (Ray et al, 1992). This cross-class inhibition was later confirmed for some additional serpins, such as hurpin, squamous cell carcinoma antigen 1 (SCCA), and endopin 2, which were found to inhibit cysteine proteases from the papain family and/or serine proteases (Schick et al, 1998; Welss et al, 2003; Hwang et al, 2005).
Figure 3.
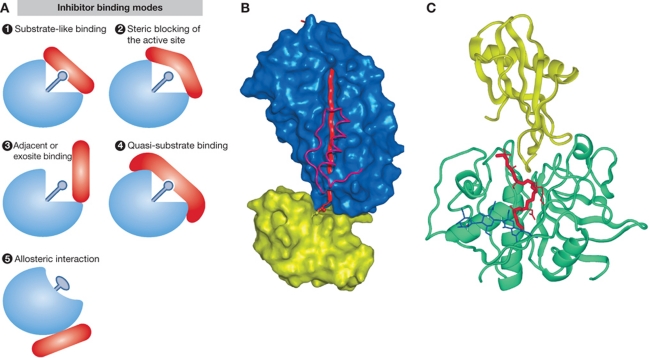
Inhibitors, major regulators of protease activities. (A) Schematic representations of different modes of inhibitor binding (adapted from Bode and Huber, 2000). (1) Substrate-like binding with direct blockade of the protease active site, such as seen with most inhibitors of trypsin-like serine proteases (see B for trypsin–antitrypsin interaction as an example); (2) steric blocking of the active site such as seen with a number of cathepsin inhibitors (see C for stefin A–cathepsin H interaction as an example); (3) exosite binding such as seen with a number of thrombin inhibitors and in a more extreme case with XIAP binding to caspases-3 and -7, which is a combination of sterical blocking of the active site and exosite binding; (4) quasi-substrate-like binding such as seen with TIMP binding to MMPs; (5) allosteric inhibition through distortion of the active site such as seen with propeptides of chymotrypsin-like proteases. (B) Inhibition of serine proteases by a serpin as an example of substrate-like binding with direct blockade of the protease active site (A1). The crystal structure of antitrypsin is shown in complex with trypsin (Huntington et al, 2000; 1EZX) is shown as an example. In the first step of inhibition, the RSL of the serpin is cleaved and the P1 residue M358 remains covalently bound to the reactive site S195 of trypsin. In the next step, 15 residues of the reactive loop (orange strand) carrying trypsin are inserted into the middle of the β-sheet of the serpin molecule (blue surface), resulting in a movement of the trypsin molecule (yellow surface) from the top to the bottom of the serpin. Trypsin catalytic site is thereby distorted and partially unfolded. The inserted part of the RSL of serpin and helix F were removed from the serpin surface in order to make the insertion visible behind the scaffold of the helix F. The ester link atoms between the side chain trypsin S195 and carbonyl group serpin M358 are shown in ball-and-stick representation. (C) Inhibition of cysteine cathepsins by endogenous inhibitors. The crystal structure of stefin A in complex with cathepsin H (Jenko et al, 2003; 1NB3) is shown as an example of steric blocking of the active site (A2). The folds of stefin A and cathepsin H are shown in ribbon presentation. Stefin A is on the top and is shown in yellow, while cathepsin H is shown in cyan. The mini-chain of cathepsin H is shown as a red stick model, while the three visible carbohydrate rings are shown in blue.
When describing the physiological relevance of inhibition one has to taken into account a number of different aspects: temporal and spatial colocalization of a protease and its inhibitor (as for the substrates), physiological concentration of the inhibitor(s), kinetics of their interaction (in vitro as an approximation since in vivo is still not possible due to the complexity of the system), and possible effects of other cofactors on the interaction. Two of these properties, binding kinetics and physiological concentration, account for the inhibitory potential of an inhibitor, that is, the quantity of a protease(s), which can be inhibited at a certain time by the inhibitor. It is primarily on this basis that the inhibitors can be grouped into their two major categories, the emergency inhibitors and the regulatory inhibitors. The major difference between these two groups is in their localization, which is also reflected in their function: the emergency inhibitors are normally localized in different cellular compartments from their target proteases in order to prevent any activity of the protease on proteins inadvertently translocating into an inappropriate compartment. They also provide the first line of defense against invading organisms that use proteases. On the other hand, the regulatory inhibitors, further subdivided into the buffer, threshold, delay and feedback type, are often colocalized together with their targets thus allowing them a much more subtle regulation (fine-tuning) of the system (Turk et al, 2002; Turk, 2006).
In addition to direct protease inhibition, inhibitors exhibit many other important cellular and physiological functions, which are often of comparable or even of greater importance to their innate inhibitory function. One of the first such examples discovered were kininogens, which were long known to be critically involved in blood pressure regulation as precursors of bradykinin and in blood coagulation, before their function as inhibitors of cysteine proteases was discovered (Müller-Esterl et al, 1985). Another example is the p41 fragment of the Ii that under normal conditions serves as a chaperone for the MHC-II molecules and prevents antigenic peptide binding to the MHC-II molecules. However, following proteolytic processing, the p41 fragment can serve as an inhibitor of its processing proteases, cathepsins L, V, and S (Bevec et al, 1996; Mihelič et al, 2008).
Protease signalling: it is not all proteolysis
When considering protease signalling, the initial consideration might be the proteolytic processing of a substrate and the consequences of that step. However, a closer analysis of most of the physiological processes shows that the proteolytic and non-proteolytic steps are strongly intertwined and that the different steps cannot be readily separated without affecting the whole process. One such example is apoptosis. A general belief is that apoptosis is a typical proteolytic cascade with the initiator caspases (-2, -8, -9, and -10) proteolytically activating the executioner caspases (primarily caspases-3 and -7), thereby leading to the controlled proteolysis of a number of critical cellular substrates and ultimately to cell death (Figure 4). The initiator caspases thus provide the fine-tuning of the system and the executioners the brute force. It is widely accepted that activation of the initiators is the critical step in triggering apoptosis. However, this is largely not a proteolytic event. In the death receptor pathway, such a non-proteolytic event is the formation of the DISC complex, which is initiated by a death ligand (e.g., TNF, TRAIL, and Fas-L) binding to its receptor (e.g., Fas, TNF-R1, DR-4, and DR-5) and oligomerization of the latter, followed by recruitment of the adaptor protein (e.g., FADD; Kischkel et al, 1995; Scott et al, 2009). In the next step, the DISC complex recruits caspase-8 molecules (Muzio et al, 1996), which are then activated by the combination of dimerization-induced conformational change and autoproteolytic cleavage of the interdomain linker (Boatright et al, 2003; Donepudi et al, 2003; Oberst et al, 2010). All of these steps are, with the partial exception of the last one, non-proteolytic. The first completely proteolytic step is the processing of caspase-3/-7, directly leading to the execution of cell death, and/or processing of Bid, and subsequently to the engagement of the mitochondrial pathway. A similar situation is seen in the mitochondrial pathway. Mitochondrial membrane permeabilization occurring through a mechanism in which the Bak and Bax proteins play a major role and which is critical for apoptosome formation, is essentially non-proteolytic. Bax activation, a critical step in intrinsic apoptosis, is a combination of a conformational change, translocation to mitochondria outer membrane and oligomerization (Tait and Green, 2010). Although cathepsin D was suggested to be involved in Bax activation during staurosporine-induced cell death of activated T lymphocytes, the molecular mechanism of the process as well as the cathepsin D targets remain elusive (Bidere et al, 2003). Moreover, apoptosome formation, that is, cytochrome c-mediated APAF-1 conformational change and its subsequent oligomerization (Zou et al, 1999; Acehan et al, 2002), also lacks proteolysis entirely. The same is true for the subsequent steps, recruitment of procaspase-9 to the complex and its dimerization-induced activation (Renatus et al, 2001). Again, proteolysis can only be seen in the subsequent step(s) with executioner caspase activation.
Figure 4.
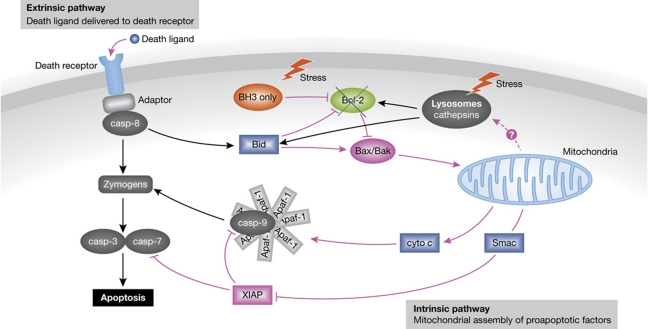
The apoptotic cascade. The two major apoptotic pathways, the extrinsic one (the death receptor pathway) and the intrinsic one (the mitochondrial pathway) are shown schematically. All the major proteolytic (black) and non-proteolytic steps (magenta) are marked with arrows, whereas the proteases (caspases and cathepsins) are shown in white characters in dark grey field. The lysosomal amplification loop is also marked, although the link between mitochondria and lysosomes has not been established yet. The direct destabilization of lysosomes (marked with a lightning) seems the only way that the lysosomes destabilization can trigger the apoptotic cascade, although it also ends up in self-amplification through the engagement of the mitochondrial pathway.
In addition to caspases, lysosomal proteases have also been linked with the apoptotic pathway, although the exact molecular mechanisms are less clear. However, it has been clearly demonstrated that after their release from the lysosomes, a non-proteolytic step, the cathepsins proteolytically process Bid and simultaneously degrade the antiapoptotic Bcl-2 family members, thereby engaging the mitochondrial pathway (Stoka et al, 2001; Cirman et al, 2004; Droga-Mazovec et al, 2008). Moreover, several recent studies suggest that the link between the lysosomes and mitochondria exists even in those systems where lysosomes are not directly targeted, such as in the death receptor pathway where mitochondrial integrity has been found to be lost prior to lysosomal destabilization (Bojič et al, 2007; Wattiaux et al, 2007; Oberle et al, 2010). The latter is therefore just a consequence of mitochondrial destabilization, suggesting that lysosomes and lysosomal proteases are only involved in an amplification loop enhancing the mitochondrial signal (Stoka et al, 2007; Turk and Turk, 2009; Repnik and Turk, 2010; Schrader et al, 2010). Although the link between mitochondria and lysosomes has not yet been identified at the molecular level, it is unlikely to be a direct protease effect, as proteases cannot cleave membrane lipids. Whether such lysosomal amplification is also functional in other pathways targeting mitochondria, distinct from the death receptor pathway, remains to be established.
Another, perhaps even more striking example than apoptosis, is autophagy, the major cellular mechanism for degradation of cytoplasmic proteins and organelles. In this process, lysosomes and lysosomal hydrolases play a major role in degrading the material engulfed by the autophagosomes following their fusion with lysosomes into autolysosomes. However, among over 20 proteins involved in the formation of autophagosome, there is a single protease, as revealed in the yeast system. This protease, called ATG4 or autophagin, removes a few residues from the C-terminus of another protein, ATG8, exposing a Gly residue that is conjugated to phosphatidylethanolamine in the subsequent step, thereby enabling prolongation of the autophagosomal membrane (Levine and Kroemer, 2008; Mizushima et al, 2008). In humans and some other organisms, the situation is apparently more complicated, as several autophagins (four in humans) have been identified (Marino et al, 2003). However, subsequent validation studies have revealed that only one of them, ATG4b or autophagin-1 in humans and mice, has a major role in the initiation of autophagy, whereas the biological functions of the others are less clear, although they may provide some low level of basal autophagy that enables ATG4b-deficient mice to survive (Marino et al, 2010). The same is true also for mammalian ATG8, where several ATG8 orthologues have been identified with microtubule-associated protein 1 light chain 3 (LC-3) having the major role (Tanida et al, 2001). The process is, however, remarkably conserved and the ATG8 conjugation system was identified in organisms as low as unicellular parasites (Alvarez et al, 2008; Duszenko et al, 2011).
Protease signalling: processing, signal transduction, and more
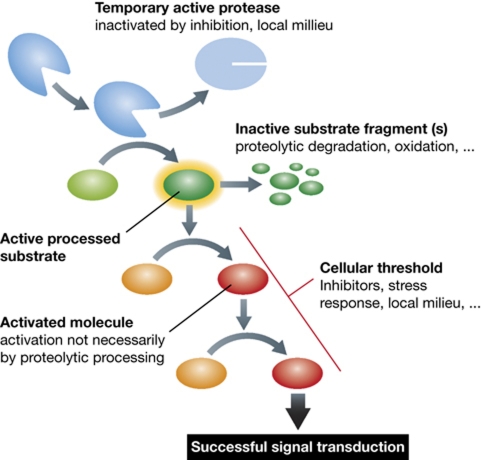
Once a protease becomes activated, it starts processing its substrates and initiates signal transduction. In principle, even a single active protease molecule could turn the processes on and off. However, in reality this is not the case. So why does this not happen? There are a number of reasons. Continuing with apoptosis as an example, one might imagine that with a single activated caspase molecule, the affected cell would ultimately die, and this would mean that organisms would be extremely vulnerable to any kind of stress and be impaired in their survival and development. To prevent this, all organisms have developed different regulatory mechanisms of various complexities, which all contribute to the robustness of the system. These include transcriptional regulation, activation mechanisms, inhibitors, and other regulatory molecules (e.g., Bcl-2 molecules), as well as protection mechanisms such as autophagy. Collectively, they constitute a damage threshold of a cell or of an organism. Signal transduction can only initiate when this threshold has been overcome, the signal dying out in other cases. From this it may be seen that a temporarily active protease, which can become inactive at a later stage of signal transduction, can nonetheless trigger a signalling pathway as long as the threshold has been overcome. The same holds true for a protease substrate, which is initially activated by proteolytic processing but becomes inactivated/degraded through subsequent proteolytic steps (Figure 5). Very good examples are the cathepsins which when functioning outside lysosomes or extracellularly may become rapidly inactivated at neutral pH (Turk et al, 1995; Turk and Turk, 2009).
Figure 5.
Signal transduction in protease signalling. A hypothetical signal transduction pathway is shown with the first step being a proteolytic processing of its substrate leading to the activation of the latter. This triggers a chain reaction of events, leading to transfer of the signal. However, if the cellular or reaction threshold (inhibitors, autophagy, …) is not overcome, the signal is not transmitted. If a protease is inactivated/degraded during the process, the signal can be transmitted if the overall threshold is overcome. Similar is true for subsequent processing/degradation or milieu-mediated inactivation of the protease substrate(s).
Although protease webs may be considered as entirely dependent on proteolytic processing of substrates and further signal transduction by these modified substrates (Doucet et al, 2008; Krüger, 2009), it can be seen from the above that a full understanding of these complex systems is rather more complex Physiological processes occur through a combination of proteolytic and non-proteolytic steps leading to their progression, proteases and their substrates representing an integral part of the whole system. However, they cannot drive the processes in the absence of other factors and a step forward in our understanding has been the link between proteases and kinases that has been established for malignant disease progression (Lopez-Otin and Hunter, 2010), but it is clear that a more systemic approach will be required before we can safely say that we fully understand protease signalling.
Acknowledgments
The work has been supported by grants from the Slovene Research Agency (P1-0140 and J1-3602 to BT, J1-2307 to VT and P1-0048 to DT) and by the FP7 projects 241919 LIVIMODE and 201279 MICROENVIMET (to BT). We would like to thank David Pim for critical reading of the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW (2002) Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell 9: 423–432 [DOI] [PubMed] [Google Scholar]
- Al-Khunaizi M, Luke CJ, Askew YS, Pak SC, Askew DJ, Cataltepe S, Miller D, Mills DR, Tsu C, Bromme D, Irving JA, Whisstock JC, Silverman GA (2002) The serpin SQN-5 is a dual mechanistic-class inhibitor of serine and cysteine proteinases. Biochemistry 41: 3189–3199 [DOI] [PubMed] [Google Scholar]
- Alvarez VE, Kosec G, Sant’Anna C, Turk V, Cazzulo JJ, Turk B (2008) Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem 283: 3454–3464 [DOI] [PubMed] [Google Scholar]
- Asagiri M, Hirai T, Kunigami T, Kamano S, Gober HJ, Okamoto K, Nishikawa K, Latz E, Golenbock DT, Aoki K, Ohya K, Imai Y, Morishita Y, Miyazono K, Kato S, Saftig P, Takayanagi H (2008) Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science 319: 624–627 [DOI] [PubMed] [Google Scholar]
- Bar-Shavit R, Kahn A, Wilner GD, Fenton JW II (1983) Monocyte chemotaxis: stimulation by specific exosite region in thrombin. Science 220: 728–731 [DOI] [PubMed] [Google Scholar]
- Barkan DT, Hostetter DR, Mahrus S, Pieper U, Wells JA, Craik CS, Sali A (2010) Prediction of protease substrates using sequence and structure features. Bioinformatics 26: 1714–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevec T, Stoka V, Pungerc̆ic̆ G, Dolenc I, Turk V (1996) Major histocompatibility complex class II-associated p41 invariant chain fragment is a strong inhibitor of lysosomal cathepsin L. J Exp Med 183: 1331–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidere N, Lorenzo HK, Carmona S, Laforge M, Harper F, Dumont C, Senik A (2003) Cathepsin D triggers Bax activation, resulting in selective apoptosis-inducing factor (AIF) relocation in T lymphocytes entering the early commitment phase to apoptosis. J Biol Chem 278: 31401–31411 [DOI] [PubMed] [Google Scholar]
- Bird PI, Trapani JA, Villadangos JA (2009) Endolysosomal proteases and their inhibitors in immunity. Nat Rev Immunol 9: 871–882 [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS (2003) A unified model for apical caspase activation. Mol Cell 11: 529–541 [DOI] [PubMed] [Google Scholar]
- Bode W, Huber R (2000) Structural basis of the endoproteinase-protein inhibitor interaction. Biochim Biophys Acta 1477: 241–252 [DOI] [PubMed] [Google Scholar]
- Bojič L, Petelin A, Stoka V, Reinheckel T, Peters C, Turk V, Turk B (2007) Cysteine cathepsins are not involved in Fas/CD95 signalling in primary skin fibroblasts. FEBS Lett 581: 5185–5190 [DOI] [PubMed] [Google Scholar]
- Bredemeyer AJ, Lewis RM, Malone JP, Davis AE, Gross J, Townsend RR, Ley TJ (2004) A proteomic approach for the discovery of protease substrates. Proc Natl Acad Sci USA 101: 11785–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix K, Dunkhorst A, Mayer K, Jordans S (2008) Cysteine cathepsins: cellular roadmap to different functions. Biochimie 90: 194–207 [DOI] [PubMed] [Google Scholar]
- Brockstedt E, Rickers A, Kostka S, Laubersheimer A, Dorken B, Wittmann-Liebold B, Bommert K, Otto A (1998) Identification of apoptosis-associated proteins in a human Burkitt lymphoma cell line. Cleavage of heterogeneous nuclear ribonucleoprotein A1 by caspase 3. J Biol Chem 273: 28057–28064 [DOI] [PubMed] [Google Scholar]
- Butler GS, Butler MJ, Atkinson SJ, Will H, Tamura T, Schade van Westrum S, Crabbe T, Clements J, d’Ortho MP, Murphy G (1998) The TIMP2 membrane type 1 metalloproteinase “receptor” regulates the concentration and efficient activation of progelatinase A. A kinetic study. J Biol Chem 273: 871–880 [DOI] [PubMed] [Google Scholar]
- Caglič D, Rozman-Pungerc̆ar J, Pejler G, Turk V, Turk B (2007) Glycosaminoglycans facilitate procathepsin B activation through disruption of propeptide-mature enzyme interactions. J Biol Chem 282: 33076–33085 [DOI] [PubMed] [Google Scholar]
- Canals F, Colome N, Ferrer C, Plaza-Calonge Mdel C, Rodriguez-Manzaneque JC (2006) Identification of substrates of the extracellular protease ADAMTS1 by DIGE proteomic analysis. Proteomics 6 (Suppl 1): S28–S35 [DOI] [PubMed] [Google Scholar]
- Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G (1999) Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell 96: 615–624 [DOI] [PubMed] [Google Scholar]
- Cirman T, Ores̆ić K, Mazovec GD, Turk V, Reed JC, Myers RM, Salvesen GS, Turk B (2004) Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J Biol Chem 279: 3578–3587 [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420: 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie EW, Fujikawa K, Kisiel W (1991) The coagulation cascade: initiation, maintenance, and regulation. Biochemistry 30: 10363–10370 [DOI] [PubMed] [Google Scholar]
- Davie EW, Neurath H (1955) Identification of a peptide released during autocatalytic activation of trypsinogen. J Biol Chem 212: 515–529 [PubMed] [Google Scholar]
- Davie EW, Ratnoff OD (1964) Waterfall sequence for intrinsic blood clotting. Science 145: 1310–1312 [DOI] [PubMed] [Google Scholar]
- Dean RA, Overall CM (2007) Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics 6: 611–623 [DOI] [PubMed] [Google Scholar]
- Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J, Helsens K, Impens F, Wejda M, Schymkowitz J, Rousseau F, Madder A, Vandekerckhove J, Declercq W, Gevaert K, Vandenabeele P (2009) Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol Cell Proteomics 8: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth HU, McIntosh CH, Pederson RA (2005) Type 2 diabetes – therapy with dipeptidyl peptidase IV inhibitors. Biochim Biophys Acta 1751: 33–44 [DOI] [PubMed] [Google Scholar]
- Dickinson CD, Kelly CR, Ruf W (1996) Identification of surface residues mediating tissue factor binding and catalytic function of the serine protease factor VIIa. Proc Natl Acad Sci USA 93: 14379–14384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donepudi M, Mac Sweeney A, Briand C, Grutter MG (2003) Insights into the regulatory mechanism for caspase-8 activation. Mol Cell 11: 543–549 [DOI] [PubMed] [Google Scholar]
- Doucet A, Butler GS, Rodriguez D, Prudova A, Overall CM (2008) Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteomics 7: 1925–1951 [DOI] [PubMed] [Google Scholar]
- Drag M, Salvesen GS (2010) Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9: 690–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droga-Mazovec G, Bojic̆ L, Petelin A, Ivanova S, Romih R, Repnik U, Salvesen GS, Stoka V, Turk V, Turk B (2008) Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J Biol Chem 283: 19140–19150 [DOI] [PubMed] [Google Scholar]
- Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, Allis CD (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135: 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszenko M, Ginger ML, Brennand A, Gualdron-Lopez M, Colombo MI, Coombs GH, Coppens I, Jayabalasingham B, Langsley G, de Castro SL, Menna-Barreto R, Mottram JC, Navarro M, Rigden DJ, Romano PS, Stoka V, Turk B, Michels PA (2011) Autophagy in protists. Autophagy 7: 127–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S (1998) A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391: 43–50 [DOI] [PubMed] [Google Scholar]
- Enoksson M, Li J, Ivancic MM, Timmer JC, Wildfang E, Eroshkin A, Salvesen GS, Tao WA (2007) Identification of proteolytic cleavage sites by quantitative proteomics. J Proteome Res 6: 2850–2858 [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS (2004) The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 384: 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gerner C, Frohwein U, Gotzmann J, Bayer E, Gelbmann D, Bursch W, Schulte-Hermann R (2000) The Fas-induced apoptosis analyzed by high throughput proteome analysis. J Biol Chem 275: 39018–39026 [DOI] [PubMed] [Google Scholar]
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol 21: 566–569 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Huntington JA, Read RJ, Carrell RW (2000) Structure of a serpin-protease complex shows inhibition by deformation. Nature 407: 923–926 [DOI] [PubMed] [Google Scholar]
- Hwang IK, Park SM, Kim SY, Lee ST (2004) A proteomic approach to identify substrates of matrix metalloproteinase-14 in human plasma. Biochim Biophys Acta 1702: 79–87 [DOI] [PubMed] [Google Scholar]
- Hwang SR, Stoka V, Turk V, Hook VY (2005) The novel bovine serpin endopin 2C demonstrates selective inhibition of the cysteine protease cathepsin L compared to the serine protease elastase, in cross-class inhibition. Biochemistry 44: 7757–7767 [DOI] [PubMed] [Google Scholar]
- Ishmael FT, Norcum MT, Benkovic SJ, Bond JS (2001) Multimeric structure of the secreted meprin A metalloproteinase and characterization of the functional protomer. J Biol Chem 276: 23207–23211 [DOI] [PubMed] [Google Scholar]
- Janin J, Seraphin B (2003) Genome-wide studies of protein-protein interaction. Curr Opin Struct Biol 13: 383–388 [DOI] [PubMed] [Google Scholar]
- Jenko S, Dolenc I, Gunćar G, Dobers̆ek A, Podobnik M, Turk D (2003) Crystal structure of Stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases. J Mol Biol 326: 875–885 [DOI] [PubMed] [Google Scholar]
- Jenne DE, Tschopp J (1988) Granzymes, a family of serine proteases released from granules of cytolytic T lymphocytes upon T cell receptor stimulation. Immunol Rev 103: 53–71 [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L, Xu HE, Johnston SA, Rees DC (1995) Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6. Science 269: 945–950 [DOI] [PubMed] [Google Scholar]
- Khan AR, Khazanovich-Bernstein N, Bergmann EM, James MN (1999) Structural aspects of activation pathways of aspartic protease zymogens and viral 3C protease precursors. Proc Natl Acad Sci USA 96: 10968–10975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 14: 5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleifeld O, Doucet A, auf dem Keller U, Prudova A, Schilling O, Kainthan RK, Starr AE, Foster LJ, Kizhakkedathu JN, Overall CM (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol 28: 281–288 [DOI] [PubMed] [Google Scholar]
- Kloetzel PM, Ossendorp F (2004) Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol 16: 76–81 [DOI] [PubMed] [Google Scholar]
- Krüger A (2009) Functional genetic mouse models: promising tools for investigation of the proteolytic internet. Biol Chem 390: 91–97 [DOI] [PubMed] [Google Scholar]
- Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA (1996) Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature 384: 368–372 [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hou WS, Escalante-Torres CR, Gelb BD, Bromme D (2002) Collagenase activity of cathepsin K depends on complex formation with chondroitin sulfate. J Biol Chem 277: 28669–28676 [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Bond JS (2008) Proteases: multifunctional enzymes in life and disease. J Biol Chem 283: 30433–30437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Hunter T (2010) The regulatory crosstalk between kinases and proteases in cancer. Nat Rev Cancer 10: 278–292 [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Overall CM (2002) Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol 3: 509–519 [DOI] [PubMed] [Google Scholar]
- Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268: 533–539 [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134: 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, Salvador-Montoliu N, Vega JA, Germana A, Fueyo A, Freije JM, Lopez-Otin C (2010) Autophagy is essential for mouse sense of balance. J Clin Invest 120: 2331–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C (2003) Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem 278: 3671–3678 [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10: 417–426 [DOI] [PubMed] [Google Scholar]
- Matthews DJ, Wells JA (1993) Substrate phage: selection of protease substrates by monovalent phage display. Science 260: 1113–1117 [DOI] [PubMed] [Google Scholar]
- Mihelič M, Dobers̆ek A, Gunc̆ar G, Turk D (2008) Inhibitory fragment from the p41 form of invariant chain can regulate activity of cysteine cathepsins in antigen presentation. J Biol Chem 283: 14453–14460 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison CJ, Butler GS, Rodriguez D, Overall CM (2009) Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr Opin Cell Biol 21: 645–653 [DOI] [PubMed] [Google Scholar]
- Müller-Esterl W, Fritz H, Machleidt W, Ritonja A, Brzin J, Kotnik M, Turk V, Kellermann J, Lottspeich F (1985) Human plasma kininogens are identical with alpha-cysteine proteinase inhibitors. Evidence from immunological, enzymological and sequence data. FEBS Lett 182: 310–314 [DOI] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K (2009) Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10: 104–115 [DOI] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death--inducing signaling complex. Cell 85: 817–827 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos JA, Ploegh H, Peters C, Rudensky AY (1998) Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science 280: 450–453 [DOI] [PubMed] [Google Scholar]
- Nakagawa TY, Brissette WH, Lira PD, Griffiths RJ, Petrushova N, Stock J, McNeish JD, Eastman SE, Howard ED, Clarke SR, Rosloniec EF, Elliott EA, Rudensky AY (1999) Impaired invariant chain degradation and antigen presentation and diminished collagen-induced arthritis in cathepsin S null mice. Immunity 10: 207–217 [DOI] [PubMed] [Google Scholar]
- Nicholson DW (2000) From bench to clinic with apoptosis-based therapeutic agents. Nature 407: 810–816 [DOI] [PubMed] [Google Scholar]
- Oberle C, Huai J, Reinheckel T, Tacke M, Rassner M, Ekert PG, Buellesbach J, Borner C (2010) Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death Differ 17: 1167–1178 [DOI] [PubMed] [Google Scholar]
- Oberst A, Pop C, Tremblay AG, Blais V, Denault JB, Salvesen GS, Green DR (2010) Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J Biol Chem 285: 16632–16642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall CM, Blobel CP (2007) In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol 8: 245–257 [DOI] [PubMed] [Google Scholar]
- Overall CM, McQuibban GA, Clark-Lewis I (2002) Discovery of chemokine substrates for matrix metalloproteinases by exosite scanning: a new tool for degradomics. Biol Chem 383: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Page IH, Helmer OM (1940) Angiotonin-activator, renin- and angiotonin-inhibitor, and the mechanism of angiotonin tachyphylaxis in normal, hypertensive, and nephrectomized animals. J Exp Med 71: 495–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL (2008) Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol 9: 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulick MG, Bogyo M (2008) Application of activity-based probes to the study of enzymes involved in cancer progression. Curr Opin Genet Dev 18: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PJ, Bergner A, Macedo-Ribeiro S, Huber R, Matschiner G, Fritz H, Sommerhoff CP, Bode W (1998) Human beta-tryptase is a ring-like tetramer with active sites facing a central pore. Nature 392: 306–311 [DOI] [PubMed] [Google Scholar]
- Plasman K, Van Damme P, Kaiserman D, Impens F, Demeyer K, Helsens K, Goethals M, Bird PI, Vandekerckhove J, Gevaert K (2011) Probing the efficiency of proteolytic events by positional proteomics. Mol Cell Proteomics 10: M110.003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podobnik M, Kuhelj R, Turk V, Turk D (1997) Crystal structure of the wild-type human procathepsin B at 2.5 A resolution reveals the native active site of a papain-like cysteine protease zymogen. J Mol Biol 271: 774–788 [DOI] [PubMed] [Google Scholar]
- Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW (2003) Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18: 343–354 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A (2010) MEROPS: the peptidase database. Nucleic Acids Res 38: D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, Pickup DJ (1992) Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell 69: 597–604 [DOI] [PubMed] [Google Scholar]
- Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS (2001) Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci USA 98: 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repnik U, Turk B (2010) Lysosomal-mitochondrial cross-talk during cell death. Mitochondrion 10: 662–669 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Farfan-Arribas DJ, Shen L (2010) Proteases in MHC class I presentation and cross-presentation. J Immunol 184: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozman-Pungerčar J, Kopitar-Jerala N, Bogyo M, Turk D, Vasiljeva O, S̆tefe I, Vandenabeele P, Brömme D, Puizdar V, Fonović M, Trstenjak-Prebanda M, Dolenc I, Turk V, Turk B (2003) Inhibition of papain-like cysteine proteases and legumain by caspase-specific inhibitors: when reaction mechanism is more important than specificity. Cell Death Differ 10: 881–888 [DOI] [PubMed] [Google Scholar]
- Schick C, Pemberton PA, Shi GP, Kamachi Y, Cataltepe S, Bartuski AJ, Gornstein ER, Bromme D, Chapman HA, Silverman GA (1998) Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry 37: 5258–5266 [DOI] [PubMed] [Google Scholar]
- Schilling O, Barre O, Huesgen PF, Overall CM (2010) Proteome-wide analysis of protein carboxy termini: C terminomics. Nat Methods 7: 508–511 [DOI] [PubMed] [Google Scholar]
- Schilling O, Overall CM (2008) Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol 26: 685–694 [DOI] [PubMed] [Google Scholar]
- Schrader K, Huai J, Jockel L, Oberle C, Borner C (2010) Non-caspase proteases: triggers or amplifiers of apoptosis? Cell Mol Life Sci 67: 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze AJ, Baumann U, Knof S, Jaeger E, Huber R, Laurell CB (1990) Structural transition of alpha 1-antitrypsin by a peptide sequentially similar to beta-strand s4A. Eur J Biochem 194: 51–56 [DOI] [PubMed] [Google Scholar]
- Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, Robinson H, Salvesen GS, Schwarzenbacher R, Riedl SJ (2009) The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature 457: 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Prat A (2007) The proprotein convertases are potential targets in the treatment of dyslipidemia. J Mol Med 85: 685–696 [DOI] [PubMed] [Google Scholar]
- Sercarz EE, Maverakis E (2003) Mhc-guided processing: binding of large antigen fragments. Nat Rev Immunol 3: 621–629 [DOI] [PubMed] [Google Scholar]
- Shi GP, Villadangos JA, Dranoff G, Small C, Gu L, Haley KJ, Riese R, Ploegh HL, Chapman HA (1999) Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity 10: 197–206 [DOI] [PubMed] [Google Scholar]
- Skeggs LT Jr, Kahn JR, Shumway NP (1956) The preparation and function of the hypertensin-converting enzyme. J Exp Med 103: 295–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoka V, Turk B, Schendel SL, Kim TH, Cirman T, Snipas SJ, Ellerby LM, Bredesen D, Freeze H, Abrahamson M, Bromme D, Krajewski S, Reed JC, Yin XM, Turk V, Salvesen GS (2001) Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J Biol Chem 276: 3149–3157 [DOI] [PubMed] [Google Scholar]
- Stoka V, Turk V, Turk B (2007) Lysosomal cysteine cathepsins: signaling pathways in apoptosis. Biol Chem 388: 555–560 [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11: 621–632 [DOI] [PubMed] [Google Scholar]
- Tam EM, Morrison CJ, Wu YI, Stack MS, Overall CM (2004) Membrane protease proteomics: isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc Natl Acad Sci USA 101: 6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E (2001) The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem 276: 1701–1706 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, Elliston KO, Ayala JM, Casano FJ, Chin J, Ding GJ-F, Egger LA, Gaffney EP, Limjuco G, Palyha OC, Raju SM et al. (1992) A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356: 768–774 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 272: 17907–17911 [DOI] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS (2007) Caspase substrates. Cell Death Differ 14: 66–72 [DOI] [PubMed] [Google Scholar]
- Timmer JC, Zhu W, Pop C, Regan T, Snipas SJ, Eroshkin AM, Riedl SJ, Salvesen GS (2009) Structural and kinetic determinants of protease substrates. Nat Struct Mol Biol 16: 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolosa E, Li W, Yasuda Y, Wienhold W, Denzin LK, Lautwein A, Driessen C, Schnorrer P, Weber E, Stevanovic S, Kurek R, Melms A, Bromme D (2003) Cathepsin V is involved in the degradation of invariant chain in human thymus and is overexpressed in myasthenia gravis. J Clin Invest 112: 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B (2006) Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov 5: 785–799 [DOI] [PubMed] [Google Scholar]
- Turk B, Bieth JG, Bjork I, Dolenc I, Turk D, Cimerman N, Kos J, Čolić A, Stoka V, Turk V (1995) Regulation of the activity of lysosomal cysteine proteinases by pH-induced inactivation and/or endogenous protein inhibitors, cystatins. Biol Chem Hoppe Seyler 376: 225–230 [DOI] [PubMed] [Google Scholar]
- Turk B, Turk D, Salvesen GS (2002) Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Curr Pharm Des 8: 1623–1637 [DOI] [PubMed] [Google Scholar]
- Turk B, Turk D, Turk V (2000) Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta 1477: 98–111 [DOI] [PubMed] [Google Scholar]
- Turk B, Turk V (2009) Lysosomes as “suicide bags” in cell death: myth or reality? J Biol Chem 284: 21783–21787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk BE, Huang LL, Piro ET, Cantley LC (2001a) Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol 19: 661–667 [DOI] [PubMed] [Google Scholar]
- Turk D (1992) Weiterentwicklung eines Programms fuer Molekuelgraphik und Elektrondichte-Manipulation and Seine Anwendung auf Verschiedene Protein-Strukturaufklerungen. PhD Thesis, Technische Universitaet Muenchen, Munich
- Turk D, Janjić V, S̆tern I, Podobnik M, Lamba D, Dahl SW, Lauritzen C, Pedersen J, Turk V, Turk B (2001b) Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J 20: 6570–6582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk V, Turk B, Turk D (2001c) Lysosomal cysteine proteases: facts and opportunities. EMBO J 20: 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K (2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods 2: 771–777 [DOI] [PubMed] [Google Scholar]
- Van Damme P, Maurer-Stroh S, Plasman K, Van Durme J, Colaert N, Timmerman E, De Bock PJ, Goethals M, Rousseau F, Schymkowitz J, Vandekerckhove J, Gevaert K (2009) Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol Cell Proteomics 8: 258–272 [DOI] [PubMed] [Google Scholar]
- Van Damme P, Staes A, Bronsoms S, Helsens K, Colaert N, Timmerman E, Aviles FX, Vandekerckhove J, Gevaert K (2010) Complementary positional proteomics for screening substrates of endo- and exoproteases. Nat Methods 7: 512–515 [DOI] [PubMed] [Google Scholar]
- Vasiljeva O, Reinheckel T, Peters C, Turk D, Turk V, Turk B (2007) Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr Pharm Des 13: 387–403 [DOI] [PubMed] [Google Scholar]
- Verspurten J, Gevaert K, Declercq W, Vandenabeele P (2009) SitePredicting the cleavage of proteinase substrates. Trends Biochem Sci 34: 319–323 [DOI] [PubMed] [Google Scholar]
- Wattiaux R, Wattiaux-de Coninck S, Thirion J, Gasingirwa MC, Jadot M (2007) Lysosomes and Fas-mediated liver cell death. Biochem J 403: 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Martoglio B (2003) Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol 13: 71–78 [DOI] [PubMed] [Google Scholar]
- Welss T, Sun J, Irving JA, Blum R, Smith AI, Whisstock JC, Pike RN, von Mikecz A, Ruzicka T, Bird PI, Abts HF (2003) Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis. Biochemistry 42: 7381–7389 [DOI] [PubMed] [Google Scholar]
- Wilkinson KD (2000) Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol 11: 141–148 [DOI] [PubMed] [Google Scholar]
- Wilson KP, Black JA, Thomson JA, Kim EE, Griffith JP, Navia MA, Murcko MA, Chambers SP, Aldape RA, Raybuck SA, Livingston DJ (1994) Structure and mechanism of interleukin-1 beta converting enzyme. Nature 370: 270–275 [DOI] [PubMed] [Google Scholar]
- Wolfe MS (2009) Intramembrane proteolysis. Chem Rev 109: 1599–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev AG, Ota K, Wang G, Movsesyan V, Bao WL, Yoshihara K, Faden AI (2001) Differential expression of apoptotic protease-activating factor-1 and caspase-3 genes and susceptibility to apoptosis during brain development and after traumatic brain injury. J Neurosci 21: 7439–7446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavas̆nik-Bergant T, Turk B (2006) Cysteine cathepsins in the immune response. Tissue Antigens 67: 349–355 [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X (1999) An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem 274: 11549–11556 [DOI] [PubMed] [Google Scholar]