Abstract
Diabetes is one of the major life threatening diseases worldwide. It creates major health problems in urban India. Glycogen Synthase Kinase-3 (GSK-3) protein of human is known for phosphorylating and inactivating glycogen synthase which also acts as a negative regulator in the hormonal control of glucose homeostasis. In traditional medicine, Momordica charantia is used as antidiabetic plant because of its hypoglycemic effect. Hence to block the active site of the GSK-3 protein three anti-diabetic compounds namely, charantin, momordenol & momordicilin were taken from Momordica charantia for docking study and calculation of binding energy. The aim of present investigation is to find the binding energy of three major insulin-like active compounds against glycogen synthase kinase-3 (GSK-3), one of the key proteins involved in carbohydrate metabolism, with the help of molecular docking using ExomeTM Horizon suite. The study recorded minimum binding energy by momordicilin in comparison to the others.
Keywords: anti-diabetic, docking, Exome™ Horizon suite, GSK-3, Momordica charantia:
Background
Diabetes is a heterogeneous metabolic disorder characterized by altered carbohydrate, lipid and protein metabolism [1]. This can cause severe short-term and long-term consequences ranging from brain damage to amputations and heart disease [2]. Traditional plant remedies have always provided sources of useful hypoglycemic agents and therefore, should continue to be investigated for possible drug alternatives [3]. The increase in demand for the use of plant based medicines to treat diabetes may be due to the side effects associated with the use of orthodox drugs such as insulin and oral hypoglycemic agents. Effective control of blood glucose level is a key step in preventing or reversing diabetic complications and improving the quality of life in both type 1 and type 2 diabetic patients [4]. In our present study we have selected glycogen synthase kinase-3 (GSK-3) as a target for the inhibitors because of its unique multifunctional serine/threonine kinase activity and inactivation by phosphorylation. In response to insulin binding, PKB/AKT phosphorylates GSK-3 on serine 9, which prevents the enzyme from phosphorylating glycogen synthase [5–7]. Unphosphorylated glycogen synthase is active and able to synthesize glycogen. Thus it plays a key role in the transduction of regulatory and proliferative signals arising out at the cell membrane in the insulin signaling pathway, leading to potential modulation of blood glucose levels [8]. GSK-3 is also unique and it requires a substrate that has been phosphorylated by a distinct kinase before it can phosphorylate the substrate. In traditional medicine Momordica charantia, under the family cucurbitaceae is used as anti-diabetic plant because of its hypoglycemic effect. Polypeptide p, isolated from fruit, seeds and tissues of M. charantia showed significant hypoglycemic effect when administered subcutaneously to langurs and humans [9]. The acetone extract of whole fruit powder of Momordica charantia in doses 25, 50, 75 mg/100g body weight lowered the blood glucose from 13.30 to 50% after 8 to 30 days treatment in alloxan diabetic rats, confirming anti hyperglycemic activity of this plant in diabetic animals [10]. Oral administration of M. charantia could lead to the secretion of insulin from endocrine pancreatic beta cells [11–14]. Charantin, momordenol & momordicilin are some of the major active compounds, believed to possess insulin-like chemical structure and properties. In our present study glycogen synthase kinase-3 (GSK-3), considered as a target for binding energy and docking study analysis against three of the active compounds using docking software, Exome™ Horizon. The validation procedures are carried out and the interactions and scores are generated.
Methodology
Protein Retrieval and Receptor Preparation:
Glycogen synthase kinase-3 (GSK-3) was downloaded from RCSB protein data bank having PDB ID 1Q5K (Figure 2) and selected for docking analysis. The water molecule and ligands attached with the protein were removed by Swiss PDV viewer. After removing the ligands and water molecules the refined protein was saved in PDB format.
Figure 2.
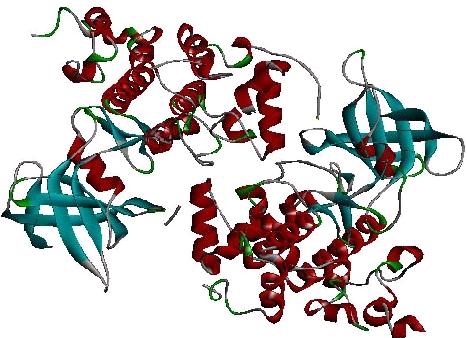
Structure of the target (1Q5K)
Ligand Preparation and ADMET properties Analysis:
The refined protein structure was taken for Docking with selected ligands. All the ligands Table 1 (see supplementary material). Were drawn using MOLDRAW tool of Exome TM Horizon [15]. Then ligands were filtered using ADMET tools of Exome TM Horizon software for checking drug likeliness by different rules of lipinski rule [16]. All the ligands that satisfy lipinki's rule is also provided Table 1 (see supplementary material).
Binding Site Analysis and Grid Preparation:
The active site and grid map of the receptor was done by of Exome TM Horizon and total eight active sites were identified. Residues of the active sites were shown both in molecule as well graphical structure and grid map result were shown in Table 2 (see supplementary material). The spacing of grid was set as 1.00 A° and the no. of grid point was taken as 60 × 60 × 60 A°.
Protein Ligand Interaction studies:
The protein-ligand docking was performed using Lamarckian genetic algorithm using default parameter [17, 18]. This is the same as the standard genetic algorithm except that, before scoring, each conformation (gene) is subjected to energy minimization. The next population is then originated by members of energy-minimized population. The name “Lamarckian” refers to the failed genetic theory of Jean-Baptiste Lamarck, who held that an organism could pass on changed experienced in its lifetime to its offspring. This theory was eventually abandoned in favor of Mendel's now familiar laws of inheritance. The LGA is faster than both simulated annealing and the standard genetic algorithm, and it allows the docking of ligands with more degrees of freedom. The number of automated docking runs, number of individuals in the population, maximum number of energy evaluations and number of generations were set to 10, 50, 2500 and 3000 respectively. All the molecules successfully docked to the binding site.
Discussion
In insulin signaling pathway, the phosphorylation of glycogen synthase by GSK-3 involves the priming phosphorylation by a distinct kinase to facilitate the binding of the substrate to the kinase. The importance of protein phosphorylation as a main regulatory mechanism used by cells to regulate enzymes and other proteins and the association of many maladies with its aberrations make kinases to be important targets and the hunt for kinase inhibitors has attracted a great attention in drug discovery in recent years. Hence to block the phosphorylation activity of GSK-3 (1Q5K) three active compounds of Momordica charantia were used. The protein structure was having 689 groups, 5515 atoms and 5695 bonds. The ligands (Figure 1) selected for docking studies and the corresponding interaction energies are given in Table 2 (see supplementary material).
Figure 1.
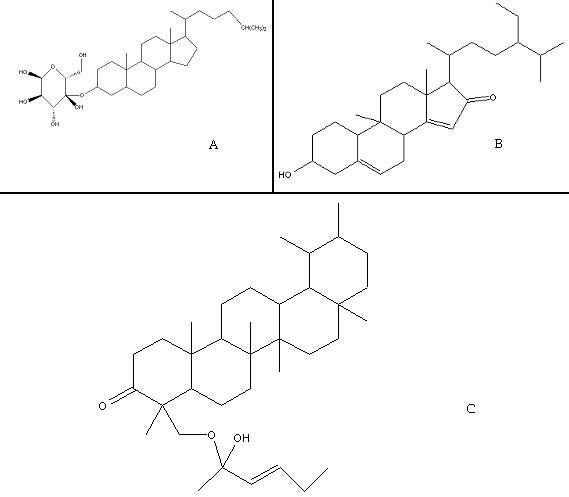
Structures of the ligands (A) Charantin; (B) Momordenol; (C) Momordicilin
The active site of 1Q5K offers many different binding modes for these compounds. The active sites from H1 to H8 were found to be IGNGGVVKQNCD, LDPVDPPALHAR, QPIFEKVHKVF, LHTSSIVRLTPP, VLGQWCSRLEY, FKLQKNRELFGS, VYRARHSRYGQPE and PIFGGVEF. All the ligands were docked into the active sites of the protein and the resulted binding energy scores that ranges from -2.42 to -5.48 kcal/mol. After docking study it was observed that these compounds successfully bind with GSK-3 as a result it may prevent the phosphorylation of glycogen synthase for which the production of glycogen will continue and maintain the glucose level. From the calculation of binding energy it was found that the compound momordicilin was found to be the most active compound and having minimum binding energy in all the active sites. The ADMET prediction of momordicilin showed good result with least number of violations Table 1 (see supplementary material). From this result, we can suggest that momordicilin is a potent inhibitor of glycogen synthase kinase- 3 and can be used as the major anti-diabetic compound form M. charantia. The H7 active site of GSK-3 containing VYRARHSRYGQPE residues was identified as the most favorable binding site for all the ligands, so H7 site can be used as the major target site for the anti-diabetic compounds.
Conclusion
In the present work we docked the ligands, charantin, momordenol & momordicilin from the medicinal plant Momordica charantia with the protein glycogen synthase kinase- 3, one of the key targets for Type 2 diabetes and momordicilin found as the most active compound in the respective target site. Further investigation in this line of research may include docking studies with other targets involved in carbohydrate metabolism. The predicted information is hoped to help for understanding the mechanisms of these plant based three antidiabetic compounds for the treatment of diabetics cost effectively. Based on this successful identification of plant-derived anti-diabetic drug candidates, we can look forward to further successes in this research area in the future.
Supplementary material
Acknowledgments
The authors gratefully acknowledge Bioinformatics Infrastructure Facility (BIF) funded by Department of Biotechnology, Govt. of India, at Centre for Studies in Biotechnology, Dibrugarh University.
Footnotes
Citation:Hazarika et al, Bioinformation 8(6): 251-254 (2012)
References
- 1.S Mutalik, et al. Indian J Exp Biol. 2003;41:316. [PubMed] [Google Scholar]
- 2.American Diabetes Association. Diabetes Care. 2007;1:S4. doi: 10.2337/dc07-S004. [DOI] [PubMed] [Google Scholar]
- 3.GO Alade1, et al. J Chem Pharm Res. 2011;3:506. [Google Scholar]
- 4.A Shaheed, et al. J Chem Pharm Res. 2011;3:204. [Google Scholar]
- 5.S Frame, et al. Mol Cell. 2001;7:1321. [Google Scholar]
- 6.F Mukai, et al. J Neurochem. 2002;81:1073. [Google Scholar]
- 7.BW Doble, J Woodgett. R J Cell Sci. 2003;116:1175. [Google Scholar]
- 8.E McManus, et al. EMBO J. 2005;24:1571. doi: 10.1038/sj.emboj.7600633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.P Khanna, et al. J Nat Prod. 1981;44:648. doi: 10.1021/np50018a002. [DOI] [PubMed] [Google Scholar]
- 10.N Singh, M Gupta. Indian J Exp Biol. 2007;45:1055. [PubMed] [Google Scholar]
- 11.E Cummings, et al. Mol Cell Biochem. 2004;261:99. doi: 10.1023/b:mcbi.0000028743.75669.ab. [DOI] [PubMed] [Google Scholar]
- 12.I Ahmed, et al. Diabetes Res Clin Pract. 1998;40:145. doi: 10.1016/s0168-8227(98)00022-9. [DOI] [PubMed] [Google Scholar]
- 13.I Ahmed, et al. Mol Cell Biochem. 2004;261:63. doi: 10.1023/b:mcbi.0000028738.95518.90. [DOI] [PubMed] [Google Scholar]
- 14.I Ahmed, et al. Int J Diabetes. 1999;7:110. [Google Scholar]
- 15.VN Viswanadhan, et al. J Chem Inf Comput Sci. 1989;29:163. [Google Scholar]
- 16.C Lipinski, A Hopkins. Nature. 2004;432:855. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 17.GM Morris, et al. J Comput Chem. 1998;19:1639. [Google Scholar]
- 18.GM Morris, et al. J Comput Aided Mol Des. 1996;10:293. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


