Abstract
With the advent of next-generation sequencing, -omics fields such as transcriptomics have experienced increases in data throughput on the order of magnitudes. In terms of analyzing and visually representing these huge datasets, an intuitive and computationally tractable approach is to map quantified transcript expression onto biochemical pathways while employing datamining and visualization principles to accelerate knowledge discovery. We present two cross-platform tools: MAPT (Mapping and Analysis of Pathways through Time) and PAICE (Pathway Analysis and Integrated Coloring of Experiments), an easy to use analysis suite to facilitate time series and single time point transcriptomics analysis. In unison, MAPT and PAICE serve as a visual workbench for transcriptomics knowledge discovery, data-mining and functional annotation. Both PAICE and MAPT are two distinct but yet inextricably linked tools. The former is specifically designed to map EC accessions onto KEGG pathways while handling multiple gene copies, detection-call analysis, as well as UN/annotated EC accessions lacking quantifiable expression. The latter tool integrates PAICE datasets to drive visualization, annotation, and data-mining.
Availability
The database is available for free at http://sourceforge.net/projects/paice/http://sourceforge.net/projects/mapt/
Background
With next-generation sequencing becoming a mainstay in molecular biology, transcriptomics research will continue to make ever-growing leaps and bounds. Genomic coverage, not to mention advances in gene expression and gene copies are now at our fingertips. Just as our knowledge of highthroughput experiments continues to progress, so too will our understanding of annotated biochemical pathways. Databases such as KEGG [1] and Reactome provide a visual means of exploring functional enzyme activity within biological pathways. Numerous tools are actively in use which interface - omics data with KEGG: Paintomics [2], Genoscape [3], and KEGGanim [4]. The Caleydo software [5] utilizes KEGG to provide a means of visualizing gene expression in a 3D manner, equipped with capabilities such as hierarchal clustering and a user-driven GUI to assist pathway exploration and analysis. The above tools provide useful features and are built with solid capabilities, however we found that these tools are organism dependent or have minimal features for processing time series data and handling of multiple gene copies. We present MAPT and PAICE, tools to provide an organism independent transcriptomics workbench. Equipped with time series analysis, visualization and data-mining capabilities, both tools provide a low-resource and user friendly environment to drive knowledge discovery, data-mining and time-series analysis.
Software input/output
PAICE and MAPT are cross-platform standalone applications built using Python 2.7. The former tool requires the Python ‘suds’ SOAP client to facilitate KEGG pathway querying, while the latter tool requires ‘PyQt’ and ‘matplotlib’ to enable GUI and graphing capabilities respectively. Running PAICE is the first step to initiate analysis within this suite. In order to do so, a populated four-column tab-delimited text file is required. Each row in this file represents the necessary values for each of the four columns: an EC accession, a numerical experimental and control expression value, and a unique reference identifier (i.e. gene loci or chromosomal coordinates). PAICE utilizes the KEGG web-service to map EC accessions onto biochemical pathways, a service heavily studied with numerous resultant manuscripts and tools. PAICE however introduces additional features designed to deal with the complexities of todays -omics datasets. First is its handling of multiple EC gene copies: if a set of isoforms differ in expression such that some copies are induced while others are suppressed, each member in this set will be flagged. This feature provides insight into individual isoform quantification, useful when investigating gene duplication or alternative splicing as some copies may differ in expression more than others. Secondly, rather than adopting static coloring schemes whereby green and red represent induced and suppressed respectively, isoform expression is statistically stratified (lightly expressed, moderately expressed, heavily expressed).
This stratification translates to color gradients whereby each stratum has a unique color. Lastly, two additional strata are further allocated, one for accessions failing to pass a userdefined fold-change cutoff, and another for annotated accessions that lack expression. This latter strata serves the goal of highlighting accessions which are annotated but do not have quantifiable expression, hence failing to map onto any pathway.
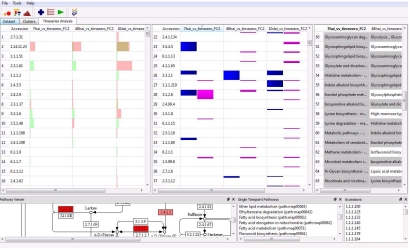
Upon PAICE completion, a collection of KEGG pathways will be generated whereby all mapped EC accessions are colored based on their applicable strata. These pathways are then fed into MAPT, a graphical interface for sifting through expression- Overlaid pathways. Numerous analytical tools like MAPT have been developed: CPTRA [6], GeneVestigator [7], and TRAM [8]. MAPT differs from the above tools by bundling biological pathways with quantified expression whilst providing an organism-independent data-mining and transcriptomics analysis platform. There are two analytical views to make such analysis possible: single and multi time point view. The single time point view within MAPT is ideal for analyzing a single timepoint or PAICE dataset, equipped with features such as functional annotation, k-Means clustering and pathway similarity analysis. On the contrary, multiple timepoint view (Figure 1) visualizes gene copy expression per time point as well as additional analyses into gene copy expression levels; useful in cases where X copies are induced but Y copies are suppressed across differing loci.
Figure 1.
MAPT time series analysis and viewer. The three tables above represent isoform expression levels, minimum and maximum expression levels per isoform, and an image viewer to visualize all pathways and their expression side by side; driven by PAICE-generated KEGG pathways. Any individual time point can honed in and analyzed independently in conjunction with additional built-in data-mining tools.
Conclusions
MAPT and PAICE are two tools designed for visualization and analysis of transcriptomics datasets. PAICE utilizes the proven and successful KEGG web-service to map numerical expression onto biochemical pathways, while MAPT provides an analytical framework to dissect such datasets and ultimately accelerate knowledge discovery through visualization and data-mining. Both MAPT and PAICE are actively in use throughout numerous research projects, e.g. in understanding the hostpathogen interactions within Soybean (Glycine max).
Future Improvement
PAICE and MAPT are continuously being worked on and improved. We welcome user feedback and suggestions as we strive to make them easier to use and intuitive in nature.
Acknowledgments
The authors wish to thank Dr. Vincent Klink and Heba Ibrahim for advice on initial application prototypes. We also wish to thank the United Soybean Board for their funding.
Footnotes
Citation:Hosseini et al, Bioinformation 8(6): 287-289 (2012)
References
- 1.M Kanehisa, et al. Nucleic Acids Res. 2008;36:D480. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.F García-Alcalde, et al. Bioinformatics. 2011;27:137. [Google Scholar]
- 3.M Clément-Ziza, et al. Bioinformatics. 2009;25:2617. doi: 10.1093/bioinformatics/btp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.P Adler, et al. Bioinformatics. 2008;24:588. [Google Scholar]
- 5.M Streit, et al. Bioinformatics. 2009;25:2760. doi: 10.1093/bioinformatics/btp432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.X Zhou, et al. BMC Bioinformatics. 2009;10:S16. doi: 10.1186/1471-2105-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.T Hruz, et al. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.L Lenzi, et al. BMC Genomics. 2011;12:121. doi: 10.1186/1471-2164-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]