Abstract
Biobanks are essential tools in diagnostics and therapeutics research and development related to personalized medicine. Several international recommendations, standards and guidelines exist that discuss the legal, ethical, technological, and management requirements of biobanks. Today's biobanks are much more than just collections of biospecimens. They also store a huge amount of data related to biological samples which can be either clinical data or data coming from biochemical experiments. A well-designed biobank software system also provides the possibility of finding associations between stored elements. Modern research biobanks are able to manage multicenter sample collections while fulfilling all requirements of data protection and security. While developing several biobanks and analyzing the data stored in them, our research group recognized the need for a well-organized, easy-to-check requirements guideline that can be used to develop biobank software systems. International best practices along with relevant ICT standards were integrated into a comprehensive guideline: The Model Requirements for the Management of Biological Repositories (BioReq), which covers the full range of activities related to biobank development. The guideline is freely available on the Internet for the research community.
Availability
The database is available for free at http://bioreq.astridbio.com/bioreq_v2.0.pdf
Keywords: Biobank Software System, guideline, model requirement, personalized medicine
Background
Research biobanks are among the most indispensable tools in the era of personalized medicine. A decade ago, biobanks were just simply repositories of biospecimens. Clinical data of limited quantity and quality had to be retrieved from hospital information systems retrospectively.
This trend has significantly changed recently. The number of patients involved in projects increased, sample and data collection has also become multicentred. Molecular biological experiments based on high-throughput technologies (e.g. microarray, Next Generation Sequencing) serve as primary sources of the increasing amount of data. Consequently, biobanks store huge amounts of data of different origin that need to be managed and analyzed by combining them with each other while supporting scientific needs. Besides data protection and security, the legal and ethical regulation of patients' rights also receives more pronounced attention. All this amounts to a strong need for a biobank software system supporting data collection, management and analysis.
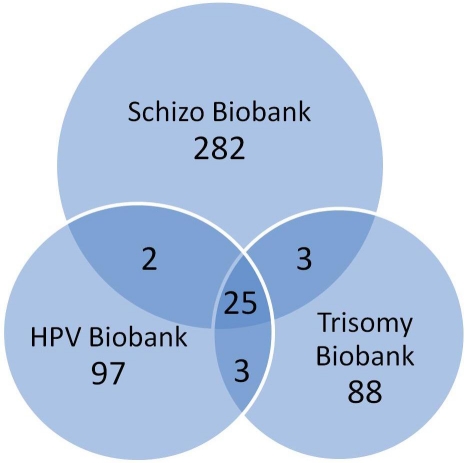
The fact that most research biobanks are disease and projectspecific makes it unfeasible to develop standard software systems solutions. Biobank projects differ in study design, applied methods of data collection and analysis in terms of collected biospecimens, personal and health information (e.g. health records, family history, lifestyle and genetic information). These differences are demonstrated by comparing the clinical data content of three biobanks developed by the research team. (Figure 1) illustrates the results, showing that there is little overlap between the clinical data features stored in them. According to this comparison, as many as 500 features are stored in the three biobanks but the center overlap between them is just 5%, which means approximately 25 features [1–3]; the majority of the matching data originate from blood samples. The significant diversity of the 95% of features can be explained by the project-specific nature of diagnostics and therapeutics research required for specified questionnaires. In spite of the variety of biobanks mentioned above, there are standards and regulations that should be taken into consideration during the development of a biobank software system, like data protection and security. Other aspects should also be considered during the development process.
Figure 1.
A Venn diagram showing the intersection of features represented in SCHIZO (schizophrenia patients), HPV (HPVinfected human tissue), and Trisomy (numerical chromosomal aberrations) Biobanks (numbers refer to clinical data per patient)
Methodology
The objective of the work was to prepare a comprehensive guideline, Model Requirements for the Management of Biological Repositories (BioReq) covering ethical, technical, management, and ICT requirements applicable to the development of biobank software systems. The guideline also references the best-known international standards and recommendations, such as Best Practices for Repositories I: Collection, Storage, and Retrieval of Human Biological Materials for Research or OECD Best Practice Guidelines for Biological Resource Centers [4, 5]. Its novelty is the well-structured and precise collection of ICT requirements, which can be a practical help for research groups with little experience in the field of biobank software system development.
The first version of BioReq was finished in 2011. In the same year, using this guideline, our research group developed a new biobank software system. The project proved the feasibility of the guideline, and its success served as a further motivation for developing and releasing version 2.0.
The guideline consists of four chapters: (1) Ethical Requirements; (2) Technical Requirements; (3) Management Requirements; and (4) Biobank Software System Requirements. Each chapter is divided into sections containing the requirements presented in a table format. In this way, the guideline is a collection of carefully organized, easy-to-check requirements as opposed to other standards, where the recommendations are provided in the form of unstructured narrative paragraphs. During the guideline preparation, internationally-accepted recommendations and standards have been considered that were judged as applicable to researchpurpose biorepositories. Since the various standards show a significant overlap in scope, repetitions have been removed, and conflicting requirements have been resolved. While preparing BioReq, we have adapted the requirements of MoReq2 [6] (a joint European specification of electronic records management) to serve the needs of biobanks.
The content of the first three chapters have been analyzed and translated into software system requirements so that the Biobank Software System (BSS) Requirements part (chapter 4) can be used on its own for design purposes.
BSS Requirements
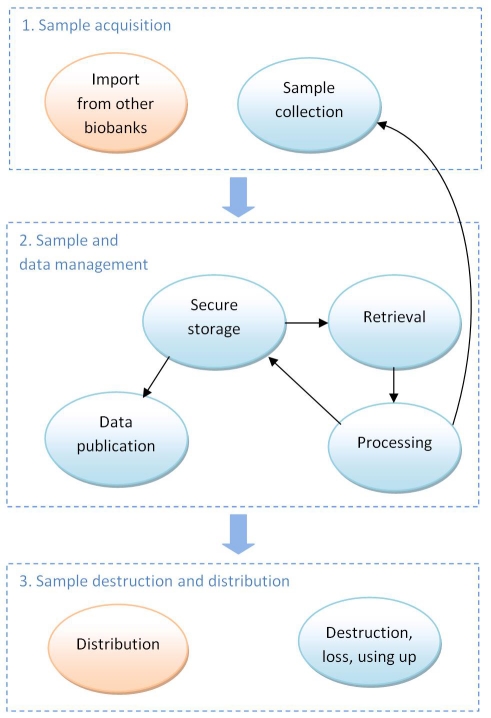
The BSS Requirements (last chapter of the guideline) chapter first covers metadata requirements [7] and the outline of entity types managed by a BSS [8, 9], then discusses general samplerelated processes requirements [10].The flow chart displayed on (Figure 2) presents the biological sample lifecycle processes from sample acquisition through sample and data management to sample destruction and distribution. Depending on the size and needs of the operating organization, the BSS may be required to track laboratory processes.
Figure 2.
Biological sample life cycle processes
BBS contains purely technical requirements not strictly related to biobank-specific processes (e.g. user management, search, logging), and supports biobank audits with features to monitor the system and provide a number of standard reports capable of being configured by authorized users. Non-functional requirements contributing to security, usability, performance, and scalability are also important attributes of a successful BSS implementation. External interface requirements specify the characteristics of the system's interaction with the hardware platform and with other software components outside the boundary of the BSS and they provide guidance on user interface design consideration as well.
Conclusion
In this paper we have given an overview of the Model Requirements for the Management of Biological Repositories guideline. We have shown that the guideline successfully incorporates the best practices of international standards on biobanks, giving users a single and consistent document to review. The novelty of the guideline lies (1) in its comprehensive coverage of topics from legal regulations to ICT considerations, and (2) in the strict format used in describing the requirements, which can guide implementation and can serve as a feature checklist. Note however that for any specific biobank software system care must be taken to adhere to all national regulations; this lies outside the scope of BioReq. Based on the presented content, the Model Requirements for the Management of Biological Repositories guideline provides a comprehensive aid in operating biobanks and conducting research involving biospecimens.
Acknowledgments
The authors sincerely thank KOVEX-Computer Ltd. for their help in adapting the specification of electronic records management of MoReq2 for the needs of biobanks. The authors are also grateful to the SCHIZOBANK Consortium and EU Pharma Ltd. for the possibility of participating in their biobank projects and to gain experience in biobank software system development.
Footnotes
Citation:Tukacs et al, Bioinformation 8(6): 290-292 (2012)
References
- 1. http://www.schizobank.hu/schizobank/home.html.
- 2. http://biobank.hpvdiagnosztika.hu/hpvbiobank/login.
- 3. http://triszomia.hu.
- 4. http://www.isber.org/bp/BestPractices2008.pdf.
- 5. http://www.oecd.org/dataoecd/7/13/38777417.pdf.
- 6. http://www.moreq2.eu/home.
- 7. http://www.iso.org/iso/iso_catalogue/catalogue_tc/cat alogue_detail.htm?csnumber=40874.
- 8. http://www.coe.int/t/dg3/healthbioethic/texts_and_do cuments/Rec_2006_4.pdf.
- 9. http://www.iso.org/iso/iso_catalogue/catalogue_tc/cat alogue_detail.htm?csnumber=39883.
- 10. http://www.gpo.gov/fdsys/pkg/FR-2006-04-28/pdf/06- 3997.pdf.