Abstract
Most eukaryotic proteins are composed of two or more domains. These assemble in a modular manner to create new proteins usually by the acquisition of one or more domains to an existing protein. Promiscuous domains which are found embedded in a variety of proteins and co-exist with many other domains are of particular interest and were shown to have roles in signaling pathways and mediating network communication. The evolution of domain promiscuity is still an open problem, mostly due to the lack of sequenced ancestral genomes. Here we use inferred domain architectures of ancestral genomes to trace the evolution of domain promiscuity in eukaryotic genomes. We find an increase in average promiscuity along many branches of the eukaryotic tree. Moreover, domain promiscuity can proceed at almost a steady rate over long evolutionary time or exhibit lineage-specific acceleration. We also observe that many signaling and regulatory domains gained domain promiscuity around the Bilateria divergence. In addition we show that those domains that played a role in the creation of two body axes and existed before the divergence of the bilaterians from fungi/metazoan achieve a boost in their promiscuities during the bilaterian evolution.
Introduction
Protein domains are highly conserved sequence modules with specific structures and functions. Most eukaryotic proteins contain more than one domain and greater complexity of organisms is related to the ability to accrue new domains to an expanded repertoire of multidomain proteins.1–3 In many cases, acquisition of a new domain increases the protein’s connectivity in the protein interaction network through the interactions of the acquired domain. Such a modular nature of multidomain proteins allows them to acquire new properties and functions without interrupting their original function. Earlier studies showed that only a small number of all possible domain combinations are selected in evolution.4,5 Specifically, some combinations appear more frequently than others and some domains combine more often than others. It has been suggested that the creation of novel multidomain proteins is typically the result of an expansion of existing domain combinations, usually preserving the N to C sequential order of the domains, which is also known as the domain architecture of the protein.6,7 The acquisitions of new domains to existing architectures usually occur at the protein termini rather than by insertions between existing domains.8–10 The main molecular mechanisms which lead to new domain architectures and the propagation of the protein repertoire are gene duplication, divergence, recombination and gene fission and fusion.5,11
Several studies explored the evolution of domain architectures across species and characterized properties of multidomain architectures in different organisms. Fong et al.10 studied the evolution of domain architectures using maximum parsimony to infer architectures in ancestral genomes. Other studies used graph theoretical tools to explore co-occurrence of domains in a protein chain. For example, Przytycka et al.12 applied a graph theory approach to study the stability and independent gain of domain architectures. It has been also shown that clusters of co-occurring domains tend to have similar functions13–15 and that the sizes of highly connected domain sub-graphs grow with evolution.16 Recently, Yang and Bourne17 considered the role of horizontal gene transfer in the evolution of domain architectures. Domain architectures have been also used to detect homology between multidomain protein families and were shown to achieve a very good performance.18,19 For example, Krishnamurthy et al.20 clustered sequences sharing similar domain architectures to detect homologous proteins while the method introduced by Krishnadev et al.21 allowed identification of circular permutations in the evolution of multidomain protein families.
Recently Ekman et al.22 used data on domain evolutionary age and on common ancestors of domains’ architectures to calculate the rate of the emergence of new domain architectures and found an increased rate in metazoans accompanied by extensive domain shuffling. In particular, domain shuffling was found to have an important role in the evolution of some signaling systems of metazoans,23 in the development of typical characteristics of vertebrates and chordates,24 and in the evolution of innate immune systems in both vertebrates and invertebrates.25 A significantly large number of phylogenomic-specific domains and domain architectures were found in animals in general and in vertebrates in particular.26 It has been also shown that architectures in the human genome have accumulated twice as many domains compared to invertebrates’ genomes27 and that there is a positive correlation between the size of certain protein domain families and the organism complexity.28 It has been proposed previously that the higher rate of domain rearrangements in metazoans can be explained by the acquisition of new metazoan-specific domains, contributing to the formation of metazoan-specific domain combinations and functional diversification26,29 and by the presence of mobile promiscuous domains.22,30 It has been argued that such an increase is due to the gene structure and the large number of transposons in metazoans.22
We tried to delve into the evolution of eukaryotes to analyze domain promiscuity within their different lineages. Various methods have been used in previous studies to identify promiscuous or mobile domains. Some looked at the co-occurrence of domains on the same protein chain,12,13,16,31 and others considered the sequential order of domains as well.32 Weiner et al.33 explored domain promiscuity by accounting for the background domain frequency in the genomes. Their promiscuity measure was the highest for a domain occurring as single domain proteins and terminal domains. Recently, domain promiscuity and frequency in genomes have been used to measure the similarity between two domain architectures.34 Several studies have shown that promiscuous domains are predominantly involved in signal transduction, presumably by mediating various interactions with other proteins participating in the signaling pathways.13,35,36 Thus, it is not surprising that there is a substantial increase in promiscuous domains in eukaryotes compared to prokaryotes in general and in multicellular organisms as compared to unicellular.32,35 However, how promiscuity of domains changes in evolution is still unclear. Tracing the evolution of domain architectures is hampered by the absence of data on domain architectures in ancestral genomes. In the present study, we use data on domain architectures for 14 ancestral genomes inferred with a maximum parsimony method from our previous analysis.10 Using the data on branch lengths of taxonomic trees we also calculate the rates of the evolution of domain promiscuity. The reconstructed collections of ancestral domain architectures along with the estimated time line permit the exploration of domain promiscuity throughout evolutionary pathways with an increasing resolution.
Our study leads to several major observations: First, we showed that for the majority of eukaryotic lineages the domain promiscuity averaged over all domains increases, especially the highest rate is observed along the branches leading to Homo sapiens, Oryza sativa, Deuterostomia and Ascomycota. Second, we found that for almost one-third of all domain families domain promiscuity positively correlates with the evolutionary time and shows a constant increase rate over long evolutionary time for eukaryotes. Moreover, we report the greatest increase in domain promiscuity around the time of the Bilateria divergence especially for those domains that play a role in the creation of two body axes.
Results
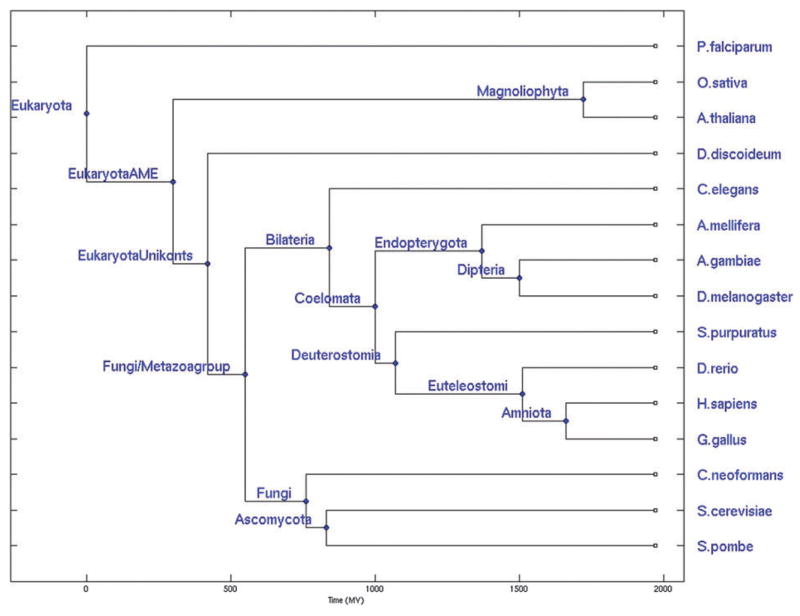
We calculated domain architectures for 15 complete genomes of eukaryotic species (Fig. 1 and Methods section). Here, leaves corresponded to the collection of protein domain architectures of contemporary genomes. Protein domain architectures were defined as the sequential order of domains on the protein chain and were taken from the NCBI Conserved Domain Architecture Retrieval Tool (CDART) database.37 Domain architectures for 14 internal nodes were inferred using the maximum parsimony method from our previous analysis10 using the NCBI taxonomy tree which included more genomes than shown in Fig. 1. However, the need for reliable estimated divergence times in the study of time dependent domain promiscuity constrained us to use a phylogenetic tree depicted in Fig. 1. We will refer hereafter to the collection of ancestral architectures as an “ancestral genome” (see Methods section for details). In total, we characterized 4384 unique domains and 9952 different domain architectures. EukaryotaAME and EukUnikonts internal nodes were not included in the analysis since these two nodes of the ancestral architectures were not very well resolved (the difference in their divergence times was rather ambiguous but crucial for our analysis) and were not defined in the NCBI taxonomy tree.
Fig. 1.
The phylogenetic tree of eukaryotes used in this study adopted from ref. 41. Ancestral architectures were reconstructed for all nodes except for EukaryotaAME and EukUnikonts.
Defining measures of domain promiscuity
Using the collection of domain architectures described above, domain promiscuity can be calculated for each external and internal (inferred) node of the tree. This allows us to study, for the first time, domain promiscuity not only for contemporary but also for ancestral genomes. Two promiscuity measures were used to investigate the tendency of each domain to combine with other domains. We started by calculating the abundance of each domain in the variety of different domain architectures. The abundance of each domain in a particular genome was defined as the number of different architectures in the genome containing that domain. The second measure of domain promiscuity was the degree of the domain in the bigram network. A bigram in this context is a pair of domains that are found adjacent on the protein chain. In the bigram network, two domains (nodes) are connected by an edge if they belong to one bigram (that is, if they are adjacent in at least one architecture in the genome). This means that highly promiscuous domains are found next to a variety of domains. It has been shown that the larger the number of bigrams the higher the organism complexity.32 Promiscuities measured by the abundance and bigram network degree are fairly correlated for each domain.
We show that the distributions of the promiscuity values are well approximated by a power-law in the various genomes (Fig. S1, ESI†). These results are in congruence with previous studies, where the number of combination partners for domain families is power law distributed as well as the number of unique domain pairs.3,32 We also defined the promiscuity profile of each domain as a vector of promiscuity values in different genomes (contemporary and inferred genomes). Then, similar promiscuity profiles were clustered, using hierarchical clustering (see Methods section for details).
The evolutionary rates of domain promiscuities
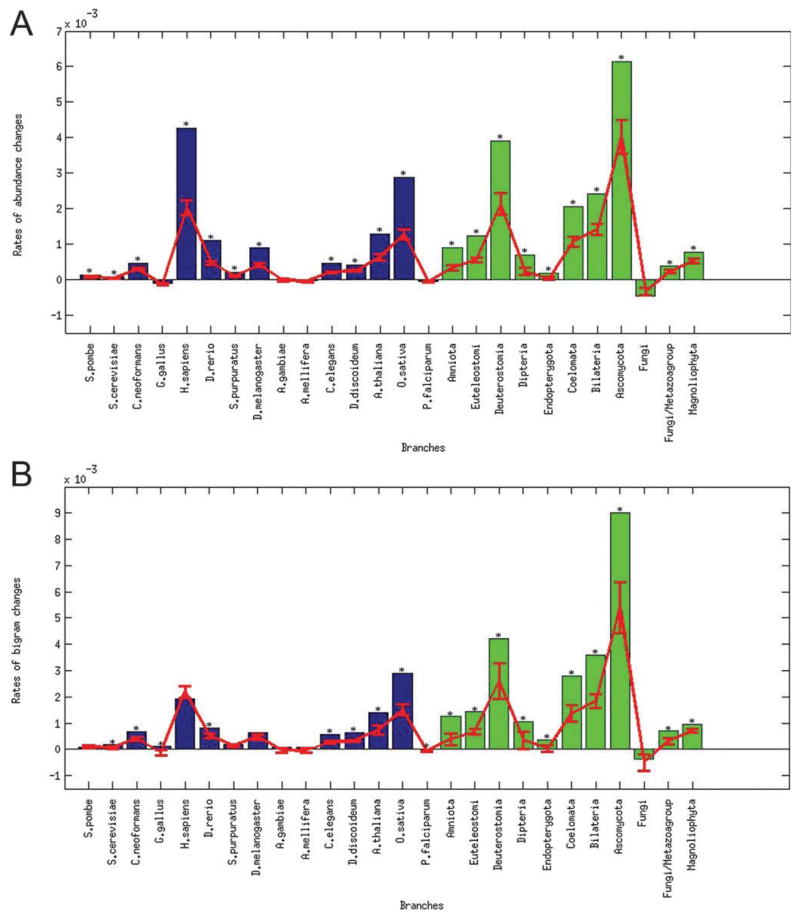
In order to address the question of the evolution of domain promiscuity, we first checked how rapidly domains gain or lose promiscuity along the different branches of the phylogenetic tree (Fig. 1). We calculated the domain promiscuity for each genome (i.e. for each contemporary and ancestral genome). Domain promiscuity was measured using the two methods described above, abundance and degree in bigram network. Then, for each branch of the tree, the rate of promiscuity change was calculated as the mean difference between the domain promiscuities of the descendant and the ancestral nodes, divided by the branch length provided by ref. 41. In the next step we calculated the expected rate of promiscuity change, herein the change that is expected by chance, if all architectures were assigned randomly on the phylogenetic tree, preserving the original number of architectures for each genome. The results were congruent for promiscuity measured by the abundance and the bigram network degree (Fig. 2), showing high positive rates considerably higher than expected by chance along several branches including H. sapiens, O. sativa, Bilateria, Deuterostomia and Ascomycota and negative rate in the branch leading to the Fungi node. For example, for branch leading to H. sapiens the increase in domain promiscuity per domain type was estimated to be approximately 0.004 abundance or 0.002 bigrams gained on average per MY.
Fig. 2.
Histogram of domain promiscuity rates averaged over domain families. Each bar represents a branch, and is labeled with the symbol corresponding to the descendant node on this branch. The rate of promiscuity change is the mean difference between the promiscuity of the descendant and the promiscuity of the ancestral node, divided by the corresponding branch length. Ancestral and contemporary genomes are colored with green and blue, correspondingly. The expected values and standard errors obtained from the permutation test are plotted in red and those branches which had p-value estimated from the permutation test less than 0.01 are marked by the asterisk.
To detect strong promiscuity signals we also looked at the evolution of individual domains rather than their average and calculated a domain promiscuity rate for each domain family and for each branch. Then for each domain we found the branch where the highest rate was observed, and for each branch on the tree counted the fraction of domain families which have a maximum promiscuity gain on the branch (Fig. S2A and S2C, ESI†). It should be noted that in this case the branches with the highest promiscuity increase will probably correspond to branches with the highest number of architectures. Remarkably, our permutation test indicated that for Bilateria and some other branches this number is significantly higher than that would be expected based on the number of architectures alone.
Although we used a non-redundant set of protein domain families, there can be inter-dependencies between different domains in terms of their preference to evolve together in a correlated fashion.38 Such background correlations mostly come from the underlined common phylogenetic history. To account for this we clustered domain families together based on the similarity of their domain promiscuity profiles (see Methods section). Then we reanalyzed the data using clusters of domains instead of individual domains. The promiscuity of each cluster in a node is defined as the mean promiscuity of the domains composing that cluster in that node. As can be seen from Fig. S2 (B and D) (ESI†), clustering the domain promiscuity profiles allowed us to account for the background phylogenetic signal and indeed refined the results, showing remarkable increases in promiscuity along several branches including branches leading to H. sapiens, O. sativa, Bilateria, Deuterostomia, and Ascomycota nodes.
Change of domain promiscuity along different evolutionary pathways
Next, we studied how domain promiscuity changes along different evolutionary pathways on the eukaryotic phylogenetic tree. We followed all possible pathways and looked for those domains whose promiscuity profiles were significantly correlated with evolutionary time from the root of the tree. We found 1597 domain families with a significant correlation between domain promiscuity (at least one promiscuity measure produced significant correlation) and evolutionary time (p-value ≪ 0.05). Out of these domain families, 1574 and 23 were positively and negatively correlated, respectively. In 623 domain families, this dependence was well described by the linear regression with multiple correlation coefficient R2 higher than 0.8. It should be mentioned that detection of significant correlation between domain promiscuity and time requires a sufficient number of internal nodes on the path. Plants, protists and amoeba did not have enough internal nodes on their root-to-leaf path to allow statistical tests, so none of their domains had a significant correlation with time.
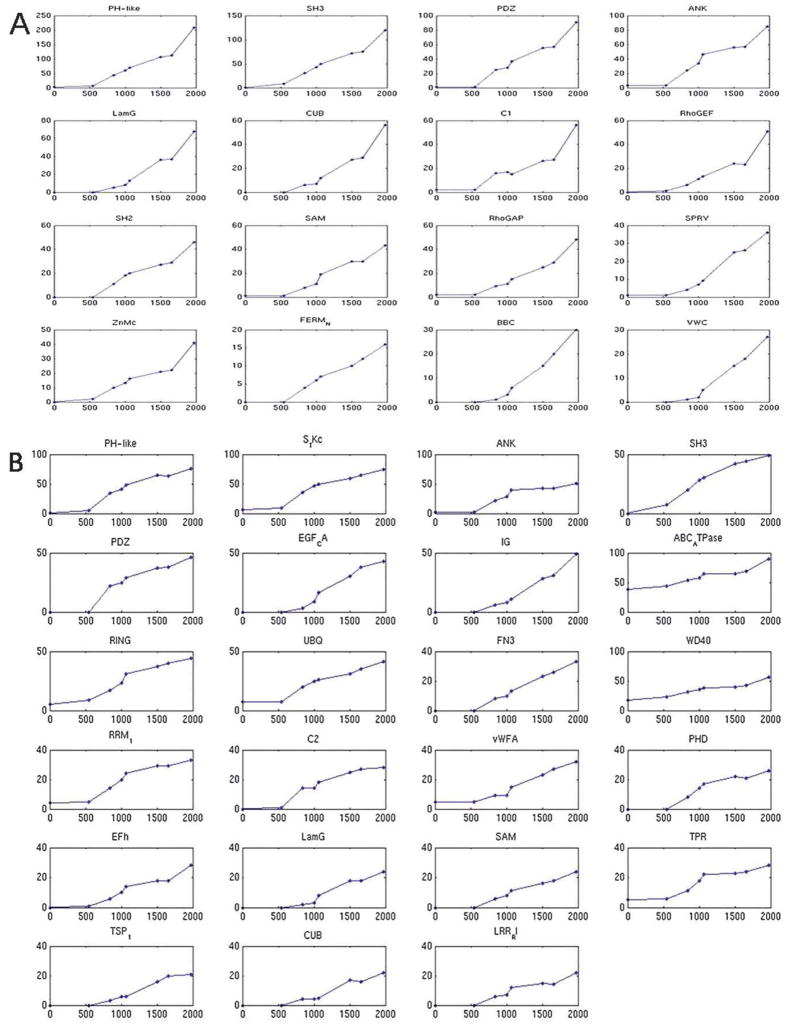
Fig. 3 shows the top 10th percentile of the domains crossing these filters, with the domain promiscuities of the internal nodes on the pathway leading to H. sapiens node plotted versus the evolutionary time from the root of the tree. The promiscuity-versus-time dependences for other animals’ genomes are shown in Fig. S3 (ESI†). In human, we found 16 domains with a significant linear correlation (and R2>0.8) using the domain abundance measure and 23 domains using the bigram network degree. The following 7 domains were found using both measures: PDZ, PH-like, SAM, SH3, LamG, ANK and CUB (domain identifiers: cl00117, cl00273, cl00131, cl09950, cl00102, cl02529 and cl00049, respectively). The overall linear dependence of promiscuity on evolutionary time points to a gradual change of domain promiscuity in evolution and possible constant evolutionary pressure on these domains. Interestingly, we also found that the majority of these domains showed acceleration in domain promiscuity around the time when Bilateria diverged from the Fungi/Metazoan group (as evident in Fig. 3) (discussed in the next section). This implies that the rate of promiscuity change remained almost constant along branches except for the branch separating Bilateria from Fungi/Metazoan. We used the Gene Ontology39 biological process terms for functional annotation of those domains. We found that in many organisms, especially in the animals, these domains are enriched with regulatory and signaling functions (chi-square contingency test, FDR (false discovery rate correction for multiple comparisons) corrected, p-value ≪ 0.01, Table S2, ESI†). For example, most of the seven domains mentioned above, that were found to be significantly time correlated in the human genome using both promiscuity measures, had signaling functions. Tables S1 and S3 (ESI†) summarize the results obtained in this analysis.
Fig. 3.
Evolution of domain promiscuity along the pathway leading to H. sapiens. X-axis represents cumulative time from the root. Y-axis represents the domain promiscuity in the corresponding ancestral genome. Shown are top tenth percentile of domains with significant correlation between promiscuity and evolution time. Functional analysis reveals that these domains are enriched in signal transduction and regulatory functions. (A) Abundance and (B) bigram network number of adjacent domains. See Table S2 (ESI†) for functional enrichment of these domains.
Evolution of domain promiscuity in bilaterians
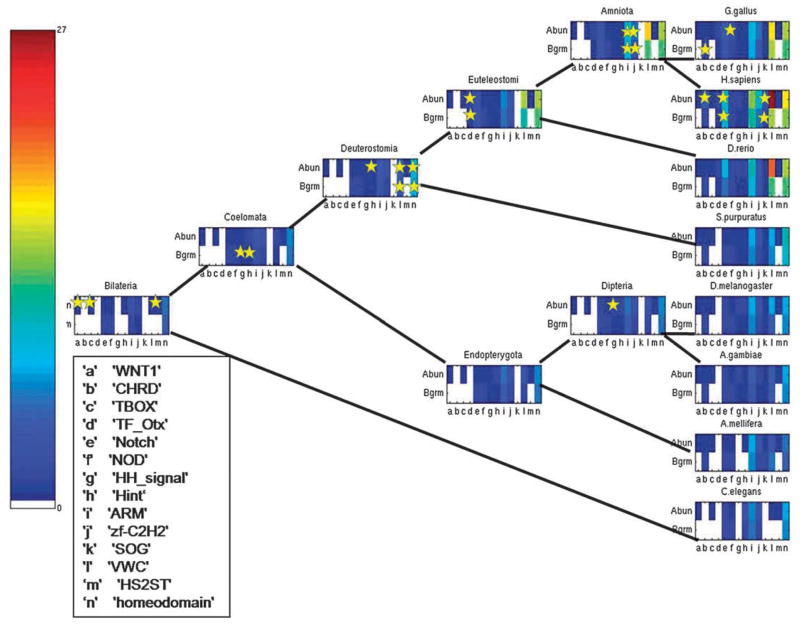
Among those domains which showed significant increase of promiscuity over time in eukaryotic evolution, many also showed acceleration in domain promiscuity around the time of the Bilateria divergence from the Fungi/Metazoa group. These domains could have played a role in the development of species with the bilaterian symmetry. Indeed, a major mile-stone in the evolution of bilaterians is the creation of two body axes: the anterior–posterior (AP) and the dorso-ventral (DV) axes. Studies on radial symmetry (single axis) organisms, such as cnidaria, have revealed that many of the components involved in the creation of the two body axes in bilaterians are already present in cnidaria. The creation of two body axes was proposed to be the result of rearrangements and expansion of an existing functional signaling system rather than by an invention of a new, bilaterian signaling system. The expansion and improvement were mainly in the proteins composing two signaling systems; the WNT and the Chordin/BMP systems, which originally created the only existing axis in hydra.40 Thus, it would be interesting to follow the evolution of promiscuity of domains that manipulate the process of body axes formation during the development of the embryo. Here, we looked at a representative set of 14 domains that are known to play a role in the creation of an embryonic body pattern in various organisms.
We followed the evolution of their promiscuity throughout the bilaterian sub-tree and illustrated the two promiscuity measures using a color scale (Fig. 4). These pathways include the ancestral nodes Bilateria, Coelomata, Deuterostomia, Euteleostomi, Amniota, Endopterygota and Dipteria and the contemporary animals C. elegans, S. purpuratus, A. mellifera, D. rerio, G. gallus, H. sapiens, D. melanogaster and A. gambiae. Then, for each domain, we marked the branch where its domain promiscuity rate was maximal. The stars on the colored domain promiscuity bar in a particular genome denote a domain that showed the highest rate on the branch leading to this external node. We found that there is a statistically significant tendency for domains which participate in the creation of the embryonic body pattern to be more actively reshuffled in different proteins during the evolution of bilaterians (the hypergeometric test and Fischer exact test p-values < 0.005 for both abundance and bigram-based promiscuities). Table 1 shows branches on the tree with the maximum rates of the domains from the above-mentioned set. Fig. S4 (ESI†) shows the correlation between the two promiscuity measures and the cumulative time from the root of the tree, for all genomes in the bilaterian sub-tree.
Fig. 4.
Evolution of promiscuity for domains participating in the formation of embryonic body pattern for bilaterian subtree. For each genome, the domain promiscuity values of two different measures are presented. Empty bars correspond to domain promiscuity measures equal to zero. Small stars show that the domain’s promiscuity had the highest rate on the branch leading to the marked genome.
Table 1.
Change of promiscuity for domains participating in the creation of embryonic body pattern. For each domain, the branch where its domain promiscuity rate was maximal is shown. Empty cells indicate that the domain had no changes in the promiscuity rate
| Abundance | Bigram | |
|---|---|---|
| WNT1 | Bilateria | — |
| CHRD | H. sapiens | G. gallus |
| TBOX | Bilateria | — |
| TF_Otx | Euteleostomi | Euteleostomi |
| Notch | H. sapiens | H. sapiens |
| NOD | G. gallus | Bilateria |
| HH_signal | Dipteria | Coelomata |
| Hint | Deuterostomia | Coelomata |
| ARM | Amniota | Amniota |
| Zf-C2H2 | Amniota | Amniota |
| SOG | H. sapiens | H. sapiens |
| VWC | Deuterostomia | Deuterostomia |
| HS2ST | Bilateria | — |
| homeodomain | Deuterostomia | Deuterostomia |
Discussion
Using 15 contemporary and 14 inferred ancestral collections of domain architectures, along with estimated branch lengths of the eukaryotic tree, we were able to conduct the first study to trace the evolution of domain promiscuity along the different evolutionary pathways. To investigate the tendency of domains to combine with other domains, we used two promiscuity measures, the domain abundance in different domain architectures and the degree of the domain in the bigram network. Both measures were congruent in most of the performed analyses. Tracing the evolution of domain promiscuity across ancestral genomes enabled us to address the rate of gain/loss of promiscuity and to point to specific branches where promiscuity was elevated compared to other nodes on the tree. Some branches consistently showed the highest increase using the averaged domain promiscuity and promiscuity of individual or clusters of domains (e.g.: branches leading to H. sapiens, O. sativa, Bilateria, Deuterostomia and Ascomycota). Previously it was shown that domains whose boundaries are located close to the boundaries of their encoding exons are common in the human genome and may account for the increase in domain promiscuity.30
Of particular note, we identified promiscuous domains with different patterns of evolution: some have lineage-specific acceleration while others gain promiscuity at steady rate over a long evolutionary period starting from the common ancestor of all eukaryotes (almost one-third of all domains). Examination of the dependencies of the promiscuity values of the domains on evolutionary time reveals a fascinating observation on the evolution of animals: most of the highly time-correlated domains showed acceleration in domain promiscuity around Bilateria divergence. The bilaterian divergence was accompanied by an expansion of many signaling systems, among them the system that determines the two axes of body symmetry. We then investigated a set of domains that are known to have a role in the creation of a single body axis in radial symmetry organisms. Interestingly, we found that some of these ancient domains, that were present in distant genomes such as amoeba and fungi, achieved a boost in their promiscuities during the evolution of bilaterians. Thus, this leads us to propose that the creation of two body axes was a result of an expansion of existing signaling systems partly by acceleration of the rate of domain promiscuity gains in those systems.
Methods
The data set
The data set consists of 15 contemporary and 14 ancestral eukaryotic species with completely sequenced contemporary genomes according to the NCBI Entrez Genomes. The contemporary species include eight animals (Caenorhabditis elegans, Strongylocentrotus purpuratus, Apis mellifera, Danio rerio, Gallus gallus, Homo sapiens, Drosophila melanogaster, Anopheles gambiae); three fungi (Cryptococcus neoformans, Schizosaccharomyces pombe, Saccharomyces cerevisiae); two plants (Arabidopsis thaliana, Oryza sativa); the protist Plasmodium falciparum; and the amoebozoan Dictyostelium discoideum. We adopt the phylogenetic tree topology described by Carmel et al.41 Briefly, a traditional “crown-group” tree topology is assumed, where the root of the tree is positioned between the protists and the common ancestor of multicellular eukaryotes (AME). Additionally, Deuterostomia and insects are grouped together in the Coelomata ancestor, excluding the nematodes. The divergence times are taken from ref. 41.
The leaf nodes of the tree are composed of the contemporary organisms and their corresponding domain architectures. The architectures are taken from the NCBI CDART database.37 Briefly, the domain architectures of all proteins in these organisms are calculated by applying the domain definitions from the Conserved Domain Database (CDD)42 at the level of domain superfamilies, using the RPS-BLAST algorithm.43 Similar domains in CDD, including NCBI-curated domains and domains imported from Pfam44 and SMART,45 have been clustered into superfamilies by identifying overlapping sequence matches to the NCBI non-redundant sequence database.
Inferring ancestral domain architectures
Data on domain architectures in ancestral nodes are taken from Fong et al.10 Briefly, the maximum parsimony modified Fitch algorithm46 is implemented to populate internal nodes with architectures. Each architecture is marked as ‘present’ in a parent node if found in more than half of the children. Similarly, if the architecture is found in less than half of the children it is marked as ‘absent’ in the parent node and if found in exactly half, is marked as ‘unknown’. Traversal of the tree from the root to leaf removes unknown labels by assigning each node the same label as its parent. We break ties at balanced trees, i.e. trees with unknown root, by setting the root to present. At the very end of this process, each ancestral genome is represented by a collection of architectures. Labeling of internal nodes was performed using the more extensive list of complete genomes from NCBI taxonomy from Fong et al.10
Assigning GO annotations to domains
The domain annotation by GO terms was based on the mappings of Pfam and SMART domains to GO terms from the Gene Ontology Annotation (GOA) database.39 First, NCBI-curated domains in CDD were mapped to the closest Pfam or SMART domain, defined as having the largest number of shared non-identical sequences. Then, each superfamily, or cluster of similar domains, was assigned the GO terms associated with Pfam, SMART, or NCBI-curated domain in the cluster.
Chi-square contingency test for functional enrichment
We implemented the chi-square contingency test to check for the association between domains which showed a significant correlation with the evolutionary time of eukaryotes and their functional annotation as follows. For each species, we defined a contingency table to be the presence and absence of function X in the set of domains and in the complementary set of domains that did not present a significant correlation with time. Then a p-value was calculated using a chi-square test followed by a FDR correction to correct for multiple comparisons.
Clustering of promiscuity profiles
A promiscuity profile of a domain was defined as a vector of its promiscuity values in different genomes along the tree. Similar promiscuity profiles were clustered together using hierarchical clustering and Euclidean distance, considering only profiles having non-zero promiscuity values in at least five genomes.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the National Library of Medicine at National Institutes of Health/DHHS. RS was supported by a research grant from the Israel Science Foundation (grant no. 385/06). ICG is a fellow of the Edmond J. Safra Bioinformatics Program and of the Ela Kodesz Research and Scholarship Fund at Tel Aviv University.
Footnotes
Electronic supplementary information (ESI) available: distributions of promiscuity scores and evolution of promiscuity rates across species. See DOI: 10.1039/c0mb00182a
Contributor Information
Teresa M. Przytycka, Email: przytyck@ncbi.nlm.nih.gov.
Anna R. Panchenko, Email: panch@ncbi.nlm.nih.gov.
References
- 1.Koonin EV, Aravind L, Kondrashov AS. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- 2.Ekman D, Bjorklund AK, Frey-Skott J, Elofsson A. J Mol Biol. 2005;348:231–243. doi: 10.1016/j.jmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Apic G, Gough J, Teichmann SA. J Mol Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 4.Apic G, Huber W, Teichmann SA. J Struct Funct Genomics. 2003;4:67–78. doi: 10.1023/a:1026113408773. [DOI] [PubMed] [Google Scholar]
- 5.Vogel C, Teichmann SA, Pereira-Leal J. J Mol Biol. 2005;346:355–365. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 6.Gough J. Bioinformatics. 2005;21:1464–1471. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- 7.Weiner J, 3rd, Beaussart F, Bornberg-Bauer E. FEBS J. 2006;273:2037–2047. doi: 10.1111/j.1742-4658.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 8.Bjorklund AK, Ekman D, Light S, Frey-Skott J, Elofsson A. J Mol Biol. 2005;353:911–923. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 9.Moore AD, Bjorklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Trends Biochem Sci. 2008;33:444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Fong JH, Geer LY, Panchenko AR, Bryant SH. J Mol Biol. 2007;366:307–315. doi: 10.1016/j.jmb.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chothia C, Gough J, Vogel C, Teichmann SA. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 12.Przytycka T, Davis G, Song N, Durand D. J Comput Biol. 2006;13:351–363. doi: 10.1089/cmb.2006.13.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Godzik A. Genome Res. 2004;14:343–353. doi: 10.1101/gr.1610504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kummerfeld SK, Teichmann SA. BMC Bioinf. 2009;10:39. doi: 10.1186/1471-2105-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen-Gihon I, Nussinov R, Sharan R. BMC Genomics. 2007;8:161. doi: 10.1186/1471-2164-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wuchty S, Almaas E. BMC Evol Biol. 2005;5:24. doi: 10.1186/1471-2148-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S, Bourne PE. PLoS One. 2009;4:e8378. doi: 10.1371/journal.pone.0008378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song N, Joseph JM, Davis GB, Durand D. PLoS Comput Biol. 2008;4:e1000063. doi: 10.1371/journal.pcbi.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeats C, Redfern OC, Orengo C. Bioinformatics. 26:745–751. doi: 10.1093/bioinformatics/btq034. [DOI] [PubMed] [Google Scholar]
- 20.Krishnamurthy N, Brown D, Sjolander K. BMC Evol Biol. 2007;7(Suppl 1):S12. doi: 10.1186/1471-2148-7-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnadev O, Rekha N, Pandit SB, Abhiman S, Mohanty S, Swapna LS, Gore S, Srinivasan N. Nucleic Acids Res. 2005;33:W126–W129. doi: 10.1093/nar/gki474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekman D, Bjorklund AK, Elofsson A. J Mol Biol. 2007;372:1337–1348. doi: 10.1016/j.jmb.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 23.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawashima T, Kawashima S, Tanaka C, Murai M, Yoneda M, Putnam NH, Rokhsar DS, Kanehisa M, Satoh N, Wada H. Genome Res. 2009;19:1393–1403. doi: 10.1101/gr.087072.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Zmasek CM, Dishaw LJ, Mueller MG, Ye Y, Litman GW, Godzik A. Genome Biol. 2008;9:R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh M, Nacher JC, Kuma K, Goto S, Kanehisa M. Genome Biol. 2007;8:R121. doi: 10.1186/gb-2007-8-6-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichler EE. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- 28.Vogel C, Chothia C. PLoS Comput Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen-Gihon I, Lancet D, Yanai I. Trends Genet. 2005;21:210–213. doi: 10.1016/j.tig.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Liu M, Walch H, Wu S, Grigoriev A. Nucleic Acids Res. 2005;33:95–105. doi: 10.1093/nar/gki152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wuchty S. Mol Biol Evol. 2001;18:1694–1702. doi: 10.1093/oxfordjournals.molbev.a003957. [DOI] [PubMed] [Google Scholar]
- 32.Basu MK, Carmel L, Rogozin IB, Koonin EV. Genome Res. 2008;18:449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiner J, 3rd, Moore AD, Bornberg-Bauer E. BMC Evol Biol. 2008;8:285. doi: 10.1186/1471-2148-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee B, Lee D. BMC Bioinf. 2009;10(Suppl 15):S5. doi: 10.1186/1471-2105-10-S15-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu MK, Poliakov E, Rogozin IB. Briefings Bioinf. 2009;10:205–216. doi: 10.1093/bib/bbn057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tordai H, Nagy A, Farkas K, Banyai L, Patthy L. FEBS J. 2005;272:5064–5078. doi: 10.1111/j.1742-4658.2005.04917.x. [DOI] [PubMed] [Google Scholar]
- 37.Geer LY, Domrachev M, Lipman DJ, Bryant SH. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel C, Berzuini C, Bashton M, Gough J, Teichmann SA. J Mol Biol. 2004;336:809–823. doi: 10.1016/j.jmb.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 39.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meinhardt H. Curr Top Dev Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- 41.Carmel L, Wolf YI, Rogozin IB, Koonin EV. Genome Res. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A, Bryant SH. Nucleic Acids Res. 2004;32(Web server issue):W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz J, Milpetz F, Bork P, Ponting CP. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitch WM. Syst Zool. 1971;20:406–416. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.