Abstract
The ricinosome (synonym, precursor protease vesicle) is a novel organelle, found so far exclusively in plant cells. Electron microscopic studies suggest that it buds off from the endoplasmic reticulum in senescing tissues. Biochemical support for this unusual origin now comes from the composition of the purified organelle, which contains large amounts of a 45-kDa cysteine endoprotease precursor with a C-terminal KDEL motif and the endoplasmic reticulum lumen residents BiP (binding protein) and protein disulfide isomerase. Western blot analysis, peptide sequencing, and mass spectrometry demonstrate retention of KDEL in the protease proform. Acidification of isolated ricinosomes causes castor bean cysteine endopeptidase activation, with cleavage of the N-terminal propeptide and the C-terminal KDEL motif. We propose that ricinosomes accumulate during senescence by programmed cell death and are activated by release of protons from acidic vacuoles.
Keywords: Ricinus communis, papain-type KDEL peptidase
Programmed cell death (PCD) in higher plants and animals fulfills multiple functions in the differentiation or turnover of tissues and is triggered by pathogens at and around the site of infection as a mechanism to prevent proliferation of the disease (1, 2). Examples for elimination by PCD of organs that serve temporary functions are tadpole tails during metamorphosis and the endosperm of the castor bean during germination. In plants, the PCD of whole organs such as leaves, flowers, and fruits is designated as senescence (3).
Biochemical pathways accompanying PCD or apoptosis in mammals have been elucidated in detail (reviewed in ref. 4). Plant cells undergoing senescence by PCD express papain-type cysteine endoproteases with a C-terminal KDEL sequence (5). They attract attention because they are present in the senescing endosperm cells of castor bean (Ricinus communis) as relatively inactive 45-kDa proenzymes until the final stages of PCD, at which time they are processed into the 35-kDa mature form with a 50–100 times higher activity (6).
The cysteine endopeptidase from senescing castor bean endosperm (CysEP) is homologous to papain-type KDEL cysteine endopeptidases from senescing flower petals of daylily, the maturing French bean pods, and the cotyledons of germinating mung bean and vetch (6). In castor bean, daylily, and mung bean, the KDEL-tailed propeptidase is stored in a special organelle, called ricinosome (5, 6) or KDEL-tailed cysteine proteinase-accumulating vesicles (KV) (7). Ricinosomes were first described and ultrastructurally characterized in the endosperm of R. communis (8, 9), whereas KV were described as cytoplasmic “foci” in Vigna radiata (10). The mature CysEP was originally purified from germinating castor bean endosperm (11). Sequencing of cDNA clones from the endosperm of germinating seedlings and from developing seeds established the presence of a presequence for cotranslational targeting into the lumen of the endoplasmic reticulum (ER), an N-terminal propeptide and a C-terminal KDEL motif. Immunogold labeling with antibodies raised against the purified CysEP occurred exclusively in the ricinosomes (6).
Castor beans store oil and protein in a living endosperm, which is laterally attached to the cotyledons. From days 1 to 4 of germination, the storage material is mobilized for transfer into the cotyledons. On days 5 and 6 when the oil and protein reserves are depleted, the endosperm becomes increasingly slimy and detaches from the cotyledons. As first recognized by Vigil (9) at 4 to 6 days after germination, all developmental changes in the fine structure of the endosperm during protein and lipid mobilization and subsequent senescence can be seen along a cross-section extending from the seed coat inward to the cotyledons in the center of the seed. The final stages of cellular breakdown are very sudden, resulting in complete disruption of both cytoplasm and cell walls, which produces a line of demarcation between living and non-living cells.
The temporal separation of storage mobilization from ricinosome formation and PCD in the senescing endosperm of germinating castor bean shows that development of ricinosomes is concomitant with nuclear DNA fragmentation (5). Intact ricinosomes accumulate the 45-kDa proCysEP as shown by Western blot analysis and by in situ fluorescence labeling with antibodies directed against the propeptide. Disintegration of ricinosomes during the final cell collapse releases the mature enzyme, giving a diffuse labeling of collapsed cells near the cotyledons (5).
As a first step in analyzing the cell biological function of the ricinosome and the biochemical function of the CysEP in senescence, we have studied in the present investigation the biogenesis of the ricinosomes. We have prepared highly purified ricinosomes and analyzed their contents. The C-terminal KDEL tailed proCysEP is the dominant protein in the matrix of the ricinosome. The C-terminal KDEL-signal motif remains attached to the proenzyme in the ricinosome, as shown by Western blot analysis, double labeling immuno-fluorescence microscopy, and mass spectroscopy and peptide sequencing of CNBr cleaved peptides. The ER lumen proteins BiP (Hsp70) and protein disulfide isomerase (PDI) were detected in the ricinosomes by Western blot analysis and by immunofluorescence microscopy using double labeling of cysteine endopeptidase and BiP or PDI. Acidification of purified ricinosomes leads to removal of the propeptide and KDEL motif in vitro.
This result corroborates the suggestion from the ultrastructural observations (9) that the ricinosomes bud from the ER. Our results indicate further that this plant system may employ a very different cell biological strategy for PCD than the caspase cascade used by mammalian cells during apoptosis.
Materials and Methods
Preparation of Highly Purified Ricinosomes.
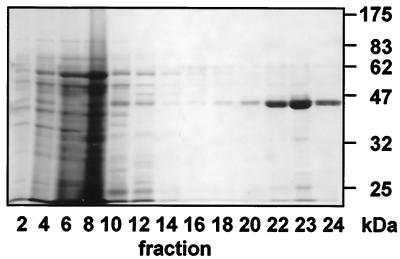
Ricinosomes were isolated from 5-day-old germinating castor bean endosperm (11) because, at this stage of development, the yield is highest and protein bodies are almost completely degraded (5). Ricinosomes isolated on a discontinuous sucrose gradient (10% to 57% sucrose) copurify with glyoxysomes at the interface between the 50 and 57% sucrose steps (cf. refs. 6 and 11). A discontinuous 44% to 65% sucrose gradient (see additional text, which is published as supplemental data on the PNAS web site, www.pnas.org) separated ricinosomes from glyoxysomes and other organelles in one step. Twenty-four 1.25-ml fractions were harvested and analyzed by SDS/PAGE and electron microscopy. Coomassie blue staining of the acrylamide gel revealed that the 3 lowermost fractions 22–24 contained a major band of 45 kDa (Fig. 1), which was recognized by antibodies against the CysEP (Fig. 2). The three fractions bordering the 57/65% sucrose interface formed a visible white band of organelles. Their content of CysEP identified them as ricinosomes.
Figure 1.
Purification of ricinosomes on a 44–65% sucrose gradient and analysis (fraction 1, upper part, and fraction 24, lower part, of the gradient) by SDS/PAGE and Coomassie blue staining. Ricinosomes are in fractions 22–24 as demonstrated by the presence of the 45-kDa pro-CysEP.
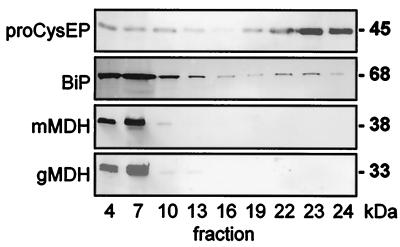
Figure 2.
Purification of ricinosomes on a 44–65% sucrose gradient and Western blot analysis (fraction 1, upper part, and fraction 24, lower part, of the gradient) by using antibodies against the ricinosome marker enzyme CysEP, against the ER-resident BiP (Hsp70) and against marker enzymes for glyoxysomes (gMDH) and mitochondria (mMDH). Fractions 22–24 contain ricinosomes with no contamination by mitochondria or glyoxysomes. Anti-BiP antibodies give a strong signal with the ER in the upper part of the gradient, and a smaller peak in fractions 22–24 containing ricinosomes.
Biochemical Procedures.
SDS/PAGE (12% gels) was performed followed by either Coomassie staining or Western blotting using the primary antibodies α-CysEP (6), α-BiP (12), α-glyoxysomal malate dehydrogenase (α-gMDH), α-mitochondrial malate dehydrogenase (α-mMDH) (13), and α-KDEL (MAC256; ref. 14). Primary antibodies were decorated with alkaline phosphatase-coupled goat anti-rabbit IgG (α-CysEP, α-BiP, α-gMDH, and α-mMDH) and goat anti-rat IgG (MAC256) and developed as described (5).
Cleavage of the Propeptide and the KDEL Motif from ProCysEP in Vitro.
Purified ricinosomes (40 μl; Fig. 1, fraction 24) were diluted 10-fold, by using either 10 mM Hepes, 10 mM DTT (pH 7.5) or 10 mM Mes, 10 mM DTT (pH 5.5) and incubated at 20°C. Fifty-microliter aliquots were withdrawn at different time points (see Results). The 50-μl aliquots were mixed with loading buffer, heated at 95°C for 5 min, and analyzed by SDS/PAGE and Coomassie staining or Western blotting.
CNBr-Cleavage of Isolated Ricinosomes and Peptide Analysis.
Purified ricinosomes were reduced with 3% β-mercaptoethanol in 100 mM Tris⋅HCl (pH 8.5), 1 mM EDTA, and 6 M guanidine HCl for 3 h at 50°C and then pyridylethylated with 3% 4-vinylpyridine (Sigma) in the same solution (15). The protein was desalted by using a Centricon 10 cartridge (Millipore). To cleave the methionyl bonds, the desalted protein was incubated with 10% CNBr (wt/vol) in 100 μl of 70% formic acid at 20°C for 2 h in the dark. The resulting peptides were separated for matrix-supported laser desorption ionization (MALDI) mass spectrometry and Edman degradation by reversed phase HPLC on a capillary liquid chromatography (LC) column (150 × 1 mm, 5 μm, 300 Å, RP C18 from Maisch, Ammerbuch, Germany). Collected fractions were subjected to N-terminal sequence analysis by using a model 492 protein sequencer (PE Biosystems, Weiterstadt, Germany). A Bruker Reflex III time-of-flight mass spectrometer (Bruker-Franzen, Bremen, Germany) was used for MALDI. The matrix was either α-cyano-4-hydroxy-cinnamic acid at 5 mg/ml in 50% acetonitrile, 0.1% trifluoroacetic acid (TFA) or 2,5-dihydroxybenzoic acid at 20 mg/ml in 30% acetonitrile. A Hewlett–Packard 1100 was coupled directly to an electrospray ionization (ESI) source of an LCQ ion trap (Thermoquest, San Jose, CA). For the HPLC of LC-MS-runs, separation of cyanogen bromide fragments was performed on a capillary LC column (150 × 0.5 mm, 3 μm, 300 Å, RP C18).
Electron Microscopy.
Isolated ricinosomes (1.5 ml) were mixed with an equal volume of double strength fixative [4% (vol/vol) formaldehyde/2% (vol/vol) glutaraldehyde/50 mM Hepes, pH 7.5/500 mM sucrose/10 mM KCl/1 mM MgCl2] at 4°C. Three milliliters of fixed ricinosomes were gradually diluted within 1 h with 4 × 0.75 ml single strength fixative [2% (vol/vol) formaldehyde/1% (vol/vol) glutaraldehyde/80 mM Pipes, pH 8.0/1 mM MgCl2] and spun down at 10,000 rpm (Beckman, Ti75) at 4°C for 15 min, followed by embedding, sectioning, and immunocytochemistry (6).
Direct Labeling of Primary Antibodies with Fluorochromes, Immunofluorescence Staining, and Confocal Laser Scanning Microscopy.
Rabbit IgGs were purified from antisera raised against castor bean CysEP, barley BiP, and watermelon mMDH by using protein A agarose affinity chromatography (Boehringer Mannheim) and labeled either with the dye AlexarFluor 546 or AlexaFluor 488 protein labeling kit (Molecular Probes). Castor bean endosperm (3-day-old) was fixed and embedded in Paraplast Plus (Sigma), and prepared for immunocytochemistry (5). At least one direct labeled primary antibody was used in all double-labeling experiments. Unlabeled antibodies against castor bean PDI and the C-terminal KDEL epitope were used in combination with FITC-labeled goat anti-rabbit and goat anti-rat Fab′ fragments. Fluorochrome-labeled sections were excited at 488 nm and 543 nM by using a model LSM 510 scanning confocal microscope (Zeiss). (For details, see supplemental material.)
Results
The 45-kDa ProCysEP Is the Major Protein in the Matrix of Ricinosomes.
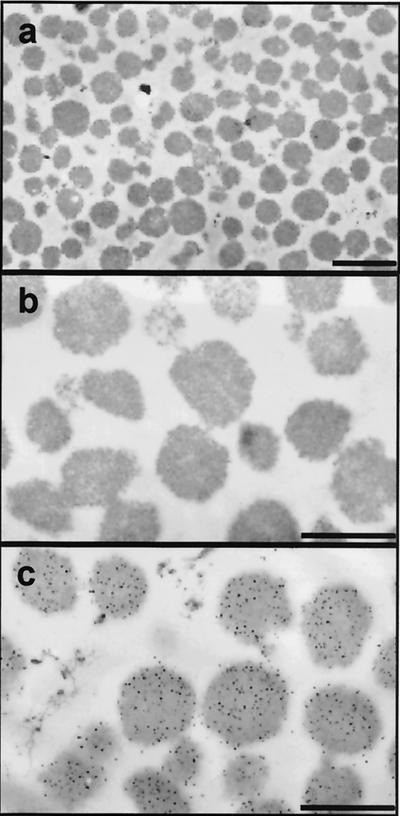
Highly purified ricinosomes were prepared from 5-day-old germinating castor bean endosperm on a discontinuous sucrose gradient. Western blots of the sucrose gradient fractions by using antibodies against gMDH and mMDH revealed that ricinosomes were well separated from glyoxysomes and mitochondria; their marker enzymes gMDH and mMDH could be detected only in the upper part of the gradient around fractions 4 to 7 (Fig. 2). No glyoxysomes or mitochondria were detected in fractions 22 to 24, where the ricinosomes were found. Electron microscopy confirmed that the ricinosome fraction was not contaminated with other organelles or microsomal membranes (Fig. 3 a and b). Immunocytochemistry with α-CysEP antibodies showed that all of the organelles were ricinosomes (Fig. 3c).
Figure 3.
Ultrastructure of ricinosomes purified from 5-day-old castor bean endosperm and immunolocalization of their marker enzyme CysEP. Electron micrographs (a, ×4,400; b, ×12,000), and immunocytochemistry by using α-CysEP (c, ×12,000). Scale bar: a = 1.0 μm; b and c = 0.5 μm.
Isolated ricinosomes were diluted with water to disrupt the organelles, and membranes were separated from the matrix proteins by high speed centrifugation and analyzed by SDS/PAGE and Coomassie blue staining (not shown). Among the matrix proteins, only the 45-kDa proCysEP could be detected. Most of the faint protein bands in intact ricinosomes (Fig. 1, fractions 22 to 24) were enriched in the membrane fractions. ER-lumen proteins such as BiP (Hsp70) were also present, but at such low levels that they could only be detected by Western blot analysis of intact ricinosomes (Fig. 2).
The Proform of the CysEP in Ricinosomes Retains the C-Terminal KDEL Motif.
The C-terminal KDEL motif functions as a Golgi retrieval signal for the 5% escaping ER lumen proteins (16). In terms of ricinosome biogenesis, it is relevant if ricinosomal proCysEP retains its C-terminal KDEL motif, thus indicating a direct development of ricinosomes from the ER. Cleavage of the KDEL motif after cotranslational import into the ER would imply transport of proCysEP out of the ER via the Golgi. We demonstrate the retention of the C-terminal KDEL motif in ricinosomal proCysEP by three independent methods.
Western blot analysis of isolated ricinosomes by using the α-KDEL antibody MAC256 (14).
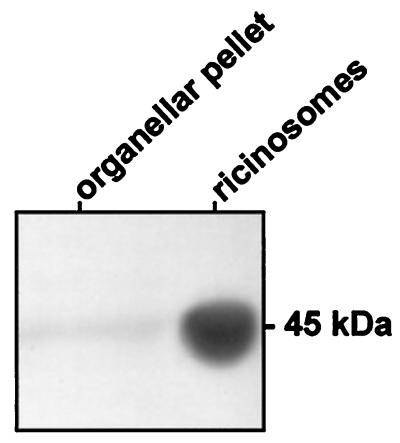
Isolated ricinosomes gave a strong signal at 45 kDa, which represents the dominant matrix protein proCysEP in ricinosomes and a weaker signal was also found in crude organelles (Fig. 4).
Figure 4.
Western blot of a crude organellar pellet and isolated ricinosomes from 5-day-old germinating castor bean endosperm by using α-KDEL antibodies (MAC256).
Analysis of CNBr cleavage products by LC-MS or HPLC fractionation followed by MALDI, and N-terminal peptide sequencing of the cleavage products.
CNBr cleavage of purified ricinosomes composed of more than 90% proCysEP was carried out, with or without reduction and alkylation of the disulfide bridges, and the cleavage products were analyzed. The proCysEP sequence (amino acids 21–360) contains 8 methionines (6), giving rise to nine cleavage products for the reduced proCysEP. The C-terminal peptide has a predicted mass of 2,577.0 if the KDEL motif is still attached (Table 1). A peptide with a mass of 2,577.0 was found and identified additionally by its N-terminal sequence (EASYPIKKSSNNPS) as the C-terminal CNBr peptide. We also found a small peak with the mass 1,819.8 with the N terminus EASYP; the C terminus SGIK of this peptide was determined by ladder sequencing with MALDI (Table 1). Comparison of the peaks indicates that 80–90% of the procysEP still has a C-terminal KDEL. It is not known whether the loss of KDEL from 20% of the enzyme occurs within intact ricinosomes. We found CNBr fragments covering amino acids 21–360 with the expected masses (see supplemental Table 2). We conclude that the potential N-glycosylation site NGT (amino acids 115–117) within the propeptide was not glycosylated. Furthermore, there was no indication of O-glycosylation. The calculated mass for proCysEP is 37,945.2 Da, whereas the migration behavior on SDS/PAGE suggests a molecular mass of 45 kDa. The mature protease has a predicted mass of 25,233.8 Da and migrates in SDS/PAGE with a molecular mass between 32 kDa and 35 kDa. This discrepancy is apparently not due to glycosylation.
Table 1.
C-terminal CNBr cleavage peptides of proCysEP: Predicted and found amino acid sequences and masses (Da)
| Mass, Da | N-terminus | Peptide | |
|---|---|---|---|
| Predicted | 2,576.84 | EASY… | EASYPIKKSSNNPSGIKSSPKDEL |
| Found (MALDI) | 2,576.70 | EASY… | EASYPIKKSSNNPSGIKSSPKDEL |
| 1,819.82 | EASY… | EASYPIKKSSNNPSGIK |
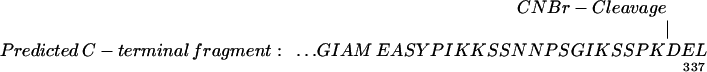
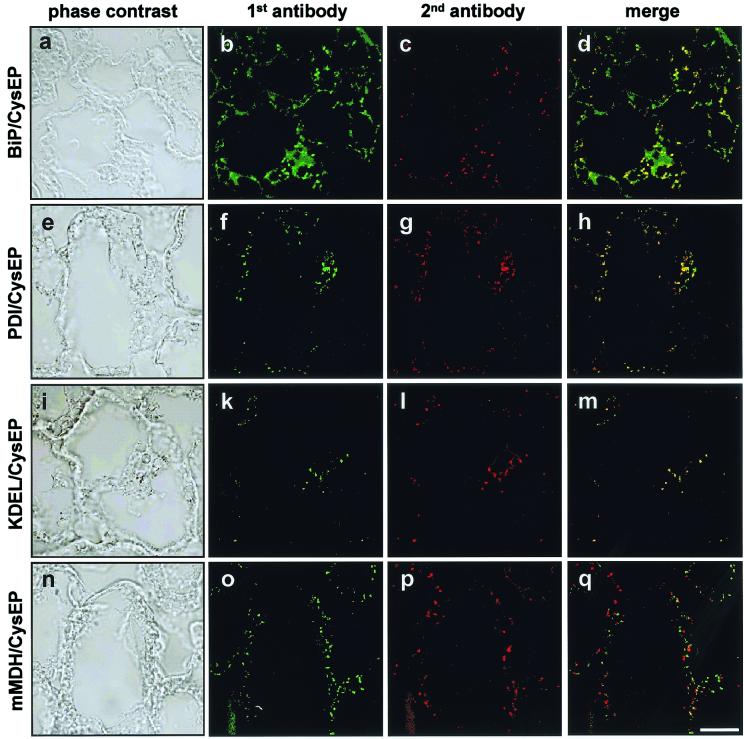
Double immunofluorescence labeling by using antibodies against the KDEL epitope and CysEP.
Tissue from 3-day-old germinating castor bean endosperm was used for immunofluorescence studies, because ricinosomes were easier to detect at this stage (5). The tissue was reacted with the α-KDEL antibody and subsequently FITC-conjugated α-rat antibody, which produces a green fluorescence signal (Fig. 5k), and ricinosomes were then identified by incubation with α-CysEP antibody (Fig. 5l, red color). Both signals localize to the same organelle as shown by the yellow color of the merged image (Fig. 5m). The α-KDEL antibody also recognizes ER-lumen proteins, such as PDI, which are also present in ricinosomes (see below), but these are minor components in ricinosomes (Fig. 2).
Figure 5.
Double immunofluorescence labeling of 3-day-old germinating castor bean endosperm. (a, e, i, and n) Phase contrast. Sections were first incubated with α-BiP (Hsp70), α-PDI, α-KDEL (MAC256), or α-mMDH (b, f, k, and o; green color), with α-CysEP as the second antibody (c, g, l, and p; red). The merged images (d, h, m, and q) are yellow for BiP, PDI, and KDEL because of colocalization with CysEP, whereas for CysEP and mMDH the red and green dots are separated because of localization in different organelles.
The ER Resident Proteins BiP (Hsp70) and PDI Are Found Within Ricinosomes.
Colocalization of CysEP with BiP (Fig. 5 a–d) and of CysEP with PDI (Fig. 5 e–h) in ricinosomes was demonstrated, whereas the control labeling showed that CysEP and mMDH are localized in different organelles (Fig. 5 n–q). BiP and PDI are highly enriched in ricinosomes giving rise to a distinct signal. BiP within the ER gave a weak diffuse signal. The concentration of PDI within the ER in situ, however, seems to be below the detection level. The presence of ER-lumen proteins in ricinosomes is further evidence for the direct development of ricinosomes from the ER.
The presence of BiP (Fig. 2) and PDI (data not shown) in ricinosomes was also demonstrated by Western blotting. Analysis of the sucrose gradient fractions revealed a strong signal with α-BiP antibodies in the upper part of the gradient (around fractions 4–7), where the ER is found, a weaker signal in the middle of the gradient (fractions 16–19), and again a peak where ricinosomes are found (fractions 22–24). As controls, gMDH and mMDH were used and did not react with fractions containing ricinosomes.
In Vitro Cleavage of the Propeptide and the KDEL Motif of ProCysEP by Acidification of Isolated Ricinosomes.
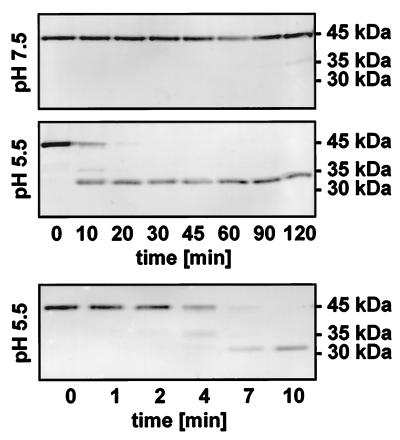
ProCysEP with the C-terminal KDEL motif is accumulated within ricinosomes. During the final stages of PCD, the ricinosomes disintegrate, and proCysEP is processed to the mature fully active peptidase. Loss of ricinosome integrity could result from a change in the permeability of the tonoplast, leading to acidification of the cytoplasm. As a consequence, low pH could trigger cleavage of the CysEP propeptide. To test this hypothesis, we incubated isolated ricinosomes at pH 5.5 and pH 7.5 as a control.
The 45-kDa proCysEP was stable at pH 7.5 for at least 2 h (Fig. 6 Top). At pH 5.5, however, the propeptide was cleaved off within the first 10 min, and the resulting mature protein was stable for at least 2 h (Fig. 6 Middle). Analysis of the cleavage time course within the first 10 min (Fig. 6 Bottom) showed that propeptide removal was rapid, occurring within 4 min. The N terminus of this mature protease was sequenced and revealed the expected VPASV sequence, showing that the propeptide was removed in vitro at the correct cleavage site, giving rise to the fully activated protease.
Figure 6.
Processing of the propeptide from proCysEP in vitro by acidification of isolated ricinosomes. Ricinosomes were incubated either at pH 5.5 or pH 7.5 at room temperature. Aliquots were withdrawn at the time points indicated and analyzed by Western blot by using α-CysEP. ProCysEP is stable at pH 7.5 at least for 2 h, whereas the propeptide is cleaved off at pH 5.5 giving rise to the stable mature protease.
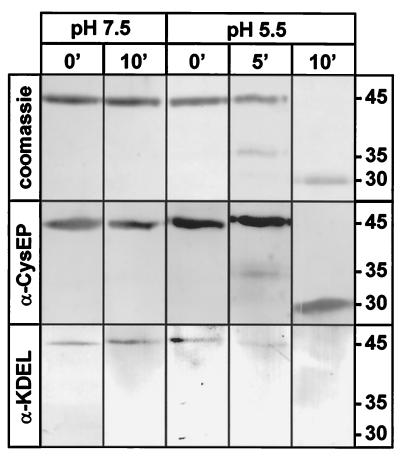
We also wanted to know whether the KDEL motif was also removed from proCysEP during activation in vitro. We incubated the isolated ricinosomes at pH 5.5 and at pH 7.5 as a control. Aliquots were withdrawn immediately after dilution into the buffer (0 min) and after 5 min and 10 min. The products were analyzed by SDS/PAGE, followed by Coomassie staining or by Western blot analysis using either α-CysEP or α-KDEL antibodies (Fig. 7). Coomassie staining and incubation with α-KDEL demonstrated that the KDEL motif was present in the pro-enzyme, but was not detectable after activation induced by low pH in vitro (Fig. 7, bottom part). We conclude that low pH triggers activation of the protease and removal of the propeptide and the KDEL motif. The weak 35-kDa intermediate band constitutes a processing intermediate of unknown structure presumably lacking the KDEL motif.
Figure 7.
Acidification of isolated ricinosomes in vitro leads to cleavage of the propeptide and the KDEL motif. Isolated ricinosomes were incubated at pH 7.5 or pH 5.5 for 10 min. Aliquots were withdrawn immediately (0 min), after 5 min (only at pH 5.5), and after 10 min. Anti-CysEP reacted with the proenzyme and the mature protease, whereas α-KDEL reacted with the proenzyme, but not with the mature protease.
Discussion
The C-terminal KDEL Motif Is Associated with the Accumulation of ProCysEP in an ER-Derived Organelle.
The 45-kDa proCysEP is the dominant protein in the matrix of highly purified ricinosomes. The C-terminal KDEL motif is not cleaved and probably functions as an ER-retention signal. The ER-resident proteins BiP and PDI are present at low levels within ricinosomes. These results are supporting evidence that proCysEP goes directly from the ER lumen to ricinosomes, and the secretory pathway via the Golgi apparatus is not involved in ricinosome biogenesis. The direct biogenesis of ricinosomes from the ER would appear to be similar to the formation of protein storage bodies in maize (17) and rice (18).
An experimental demonstration for the function of the C-terminal KDEL as an ER retention signal has been provided with a recombinant pea seed vicilin (19). The storage proteins vicilin and legumin normally pass through the Golgi apparatus on their way to the protein storage vacuole (20). The gene for vicilin was modified by the addition of a sequence coding for the C-terminal KDEL motif. Vicilin-KDEL, but not unmodified vicilin, was found in large aggregates (≈0.5 μm) in the ER of transgenic tobacco and lucerne leaves. It is not known how proCysEP is sorted away from other ER-subcompartments and what causes the ER to dilate and pinch off to form ricinosomes.
Maturation of the Propeptidase.
KDEL-tailed papain-type cysteine endopeptidases form a distinct class within the plant cysteine endopeptidases (6) and are predominately expressed in senescing tissue (5). The most extensively studied CysEP with a C-terminal KDEL motif is SH-EP (cysteine endopeptidase from Vigna mungo) (21). It is synthesized as a 45-kDa inactive precursor, which is cotranslationally processed to a 43-kDa intermediate after cleavage of the signal peptide. Inactive intermediates of 39 and 36 kDa have also been observed in vitro before the formation of the mature 33 kDa protease in V. mungo cotyledon extracts. Expression of SH-EP in insect Sf-9 cells revealed the transient presence of a 42-kDa form, which was not recognized by an antibody against the KDEL sequence (21). These authors also showed that the C-terminal KDEL was not required for correct folding and that the mature enzyme was not glycosylated at any of the three potential N-glycosylation sites. In addition, processing of an enzymatically inactive form of SH-EP (C26G, numbering of the mature protein) to the 33-kDa mature polypeptide occurred much more slowly, without the presence of the 39- and 36-kDa intermediate forms. These results may imply that SH-EP maturation involves autocatalytic self-processing. It should be noted, however, that in vitro processing experiments were performed with SH-EP isolated from crude extracts, and the possibility of proteolytic attack by proteases and peptidases from other cellular compartments cannot be excluded.
Intact ricinosomes accumulate the 45-kDa proCysEP, whereas the mature form is released from disintegrating ricinosomes during the final stages of PCD (5). We have shown that acidification of ricinosomes in vitro results in removal of the N-terminal propeptide within a few minutes, leading to the mature protease with the correct amino terminus and cleavage of the C terminus, removing the KDEL motif. We suggest that this maturation is due to autocleavage, as is the case for papain and other cysteine proteases (22), but cannot exclude the possibility of activation by low levels of another endoprotease. Loss of ricinosome integrity in vivo may occur with the acidification of the cytoplasm resulting from a change in tonoplast permeability at a late stage of PCD. Only 7-aa residues are lost by autocleavage from the C terminus of Ricinus CysEP, whereas 10 and 12 residues are cleaved from V. mungo and Vicia sativa SH-EPs, respectively. This removal may indicate the involvement of carboxypeptidases or other processing proteases in situ.
Cysteine Endoproteases in Storage Protein Mobilization.
The role of cysteine endoproteases in storage protein degradation in Vicia and Vigna is the subject of some debate. The protease(s) must be synthesized and targeted to the globulins in protein storage vacuoles, either during seed maturation or during germination. Identification of a protease essential for storage globulin degradation should satisfy the following prerequisites: (i) it must be present with its natural substrate in the same cell compartment; (ii) the time course of protease activity must coincide with that of globulin degradation; and (iii) it has to be able to degrade its natural substrate in vitro (23). Most cysteine endoproteases in legumes are synthesized during germination, but the corresponding increase in proteolytic activity occurs after globulin degradation has started. V. sativa protease A, a 29-kDa ortholog of V. mungo SH-EP, is capable of hydrolyzing globulins in vitro, but could not be localized in the protein storage bodies isolated from the cotyledons of germinating seeds. The active form of the protease was first detected when globulin degradation was almost complete (24), and the same is true for R. communis endosperm (6).
In contrast, SH-EP accumulates in V. mungo cotyledons whereas storage globulins are still abundant (25). It is proposed that SH-EP is involved in storage globulin degradation, based on protease activity levels during germination and capacity for in vitro degradation of globulins, even though SH-EP has not been found in isolated storage protein vacuoles. SH-EP accumulates in ER-derived vesicles (KV) comparable to ricinosomes (7) and the cytoplasmic “foci” of V. radiata (10), also designated precursor protease vesicles (26). Measurement of endoprotease activity in V. mungo involves an extraction at pH 7.4, followed by an incubation overnight at pH 5.4 (27). Treatment at pH 5.4 would ensure autoactivation of the pro-enzyme, and, when this treatment is omitted, the pro-enzyme is the major form in 3-day-old seedlings (27). It follows that SH-EP cannot be involved in globulin degradation in vivo. Globulin degradation may be carried out by asparaginyl endoproteases and other cysteine endoproteases (without a KDEL terminus), made early during germination (28), possibly together with asparaginyl endoproteases accumulated during maturation and localized in storage protein vacuoles (29). We suggest that, in contrast to the observable temporal separation of storage protein mobilization and proliferation of ricinosomes in R. communis, storage protein degradation in V. mungo and V. radiata is concurrent with precursor protease vesicle proliferation in preparation for PCD, as may also be the case in other systems of cell content mobilization in connection with senescence in plant tissue lacking storage proteins.
Supplementary Material
Acknowledgments
We thank Dr. F. Kalousek for castor bean α-CysEP antibodies and Dr. S. J. Coughlan for castor bean α-PDI antibodies. This work was supported by the Deutsche Forschungsgemeinschaft (Gi 154/9-2,3 and/11-1).
Abbreviations
- BiP
binding protein
- CysEP
castor bean cysteine endopeptidase
- ER
endoplasmic reticulum
- gMDH
glyoxysomal malate dehydrogenase
- mMDH
mitochondrial malate dehydrogenase
- PCD
programmed cell death
- PDI
protein disulfide isomerase
- MALDI
matrix-supported laser desorption ionization
- LC
liquid chromatography
- SH-EP
mung bean cysteine endopeptidase
References
- 1.Jacobsen M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Pennell R I, Lamb C. Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadfield K A, Bennett A B. Cell Death Differ. 1997;4:662–670. doi: 10.1038/sj.cdd.4400308. [DOI] [PubMed] [Google Scholar]
- 4.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 5.Schmid M, Simpson D, Gietl C. Proc Natl Acad Sci USA. 1999;96:14159–14164. doi: 10.1073/pnas.96.24.14159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid M, Simpson D, Kalousek F, Gietl C. Planta. 1998;206:466–475. doi: 10.1007/s004250050423. [DOI] [PubMed] [Google Scholar]
- 7.Toyooka K, Okamoto T, Minamikawa T. J Cell Biol. 2000;148:453–463. doi: 10.1083/jcb.148.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mollenhauer H H, Totten C. Plant Physiol. 1997;46:794–799. doi: 10.1104/pp.46.6.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vigil E L. J Cell Biol. 1970;46:435–454. doi: 10.1083/jcb.46.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumgartner B, Tokuyasu K T, Chrispeels M J. J Cell Biol. 1978;79:10–19. doi: 10.1083/jcb.79.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietl C, Wimmer B, Adamec J, Kalousek F. Plant Physiol. 1997;113:863–871. doi: 10.1104/pp.113.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Møgelsvang S, Simpson D J. Plant Mol Biol. 1998;36:541–552. doi: 10.1023/a:1005916427024. [DOI] [PubMed] [Google Scholar]
- 13.Gietl C, Seidl C, Svendsen I. Biochim Biophys Acta. 1996;1274:48–58. doi: 10.1016/0005-2728(96)00009-6. [DOI] [PubMed] [Google Scholar]
- 14.Napier R M, Fowke L C, Hawes C, Lewis M, Pelham H R. J Cell Sci. 1992;102:261–271. doi: 10.1242/jcs.102.2.261. [DOI] [PubMed] [Google Scholar]
- 15.Raftery M, Cole R, J. Biol Chem. 1966;241:3457–3461. [PubMed] [Google Scholar]
- 16.Vitale A, Denecke J. Plant Cell. 1999;11:615–628. doi: 10.1105/tpc.11.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lending C R, Larkins B A. Plant Cell. 1989;1:1011–1123. doi: 10.1105/tpc.1.10.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Franceschi V R, Okita T W. Cell. 1993;72:869–879. doi: 10.1016/0092-8674(93)90576-c. [DOI] [PubMed] [Google Scholar]
- 19.Wandelt C I, Khan M R I, Craig S, Schroeder H E, Spencer D, Higgins T J V. Plant J. 1992;2:181–192. doi: 10.1046/j.1365-313x.1992.t01-41-00999.x. [DOI] [PubMed] [Google Scholar]
- 20.Robinson D G, Baeumer M, Hinz G, Hohl I. J Plant Physiol. 1998;152:659–667. [Google Scholar]
- 21.Okamoto T, Minamikawa T, Edward G, Vakharia V, Herman E. J Biol Chem. 1999;274:11390–11398. doi: 10.1074/jbc.274.16.11390. [DOI] [PubMed] [Google Scholar]
- 22.Vernet T, Bert P J, de Motigny C, Musil R, Tessier DC, Menard R, Magny M C, Storer AC, Thomas D Y. J Biol Chem. 1995;270:10838–10846. doi: 10.1074/jbc.270.18.10838. [DOI] [PubMed] [Google Scholar]
- 23.Shutov A D, Vaintraub I A. Phytochemistry. 1989;26:1557–1566. [Google Scholar]
- 24.Becker C, Senyuk V I, Shutov A D, Nong V H, Fischer J, Horstmann C, Müntz K. Eur J Biochem. 1997;248:304–312. doi: 10.1111/j.1432-1033.1997.00304.x. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto T, Minamikawa T. J Plant Physiol. 1998;152:675–682. [Google Scholar]
- 26.Chrispeels M J, Herman EM. Plant Physiol. 2000;123:1227–1233. doi: 10.1104/pp.123.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsuhashi W, Minamikawa T. Plant Physiol. 1989;89:274–279. doi: 10.1104/pp.89.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senyuk V, Rotari V, Becker C, Zakharov A, Horstmann C, Müntz K, Vaintraub I. Eur J Biochem. 1998;258:546–558. doi: 10.1046/j.1432-1327.1998.2580546.x. [DOI] [PubMed] [Google Scholar]
- 29.Hara-Nishimura I, Takeuchi Y, Nishimura M. Plant Cell. 1993;5:1651–1659. doi: 10.1105/tpc.5.11.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.