1. Introduction
A promising theory of the aging process holds that senescence is brought about by dysregulation of transcription factors governing central physiological processes, including energy metabolism, and immunological homeostasis (Vellanoweth et al. 1994; Supakar et al., 1995; Roy et al. 1996, 2002; Roy 1997; Chung et al. 2000, 2001, 2002, 2006, 2009; Lavrovsky et al. 2000; Giardina and Hubbard, 2002; Gosselin and Abbadie, 2003; Herbein et al. 2006; Yu & Chung, 2006; Salminen et al. 2008a&b). One such transcription factor that has come to the forefront is nuclear factor kappa B (NF-κB) and its associated signal transduction network (STN), thus giving rise to the concept of NF-κB-dependent senescence. The NF-κB STN has also been implicated in the pathophysiology of complex diseases associated with aging, including cancer (Dolcet et al. 2005; Karin 2006; Inoue et al. 2007; Maeda and Omata 2008), neurological disorders (Kaltschmidt et al. 2005; Mattson 2006; Mattson and Meffert 2006; Memet 2006), and the metabolic syndrome, including type 2 diabetes (T2D), obesity, and cardiovascular disease (CVD) (Sonnenberg et al. 2004; de Winther et al. 2005; Xanthoulea et al. 2005; Bastard et al. 2006; Schwartz and Reaven, 2006; Gil et al. 2007). Thus, studies on the genetic regulation of the NF-κB STN may contribute toward understanding the physiology of senescence and the pathophysiology of age-associated complex diseases.
In a current approach variously known as system- or systems-genetics it has been argued that the incorporation of a systems biology perspective can greatly aid efforts to delineate the genetic architecture underlying transcriptional regulatory networks (Galitski, 2004; Schadt et al. 2005; Drake et al. 2006; Kadarmideen et al. 2006; Sieberts and Schadt 2007; Werner 2007; Ayroles et al. 2009; Mackay et al. 2009). Here we employ these methods to study the genetic regulation of the NF-κB STN.
We first derive a network of gene expression variables intrinsic to the NF-κB STN using Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com) methods. We then perform principal components factor analysis (PCFA) to identify sets of highly correlated gene expression variables within the network, where we take the factors identified to be multivariate proxies of functional modules at the molecular level. This is similar to methods that exploit the inherent correlation structure in a transcriptional profiling array to detect modules within a system (Han et al. 2004; Ihmels et al. 2004; Xia et al. 2006; Xue et al. 2007; Zhan 2007; Han 2008; Kutalik et al. 2008; Wang et al. 2008). Lastly, using statistical genetic variance component methods (Blangero et al., 2001; Almasy and Blangero, 2008, 2010), we seek to identify and localize quantitative trait loci (QTLs).
2. Materials and Methods
2.1. Study Population
Our analyses were performed on data from the San Antonio Family Heart Study (SAFHS), which is a study of the genetic determinants of cardiovascular disease (CVD) in Mexican American families of San Antonio, Texas. The SAFHS population is comprised of large Mexican American extended families randomly ascertained with respect to CVD (MacCluer et al. 1999). The SAFHS protocols were approved by the Institutional Review Board at the University of Texas Health Science Center at San Antonio and all study participants provided written informed consent.
We note that the crude prevalence of CVD in our sample is 19 %, and so the extent to which our findings are applicable to “normal aging” is somewhat debatable. However, it is widely held that aging and age-associated complex diseases such as CVD have common underlying causes such as inflammation and oxidative stress (Chung et al. 2000, 2001, 2002, 2006, 2009; Brüünsgaard et al., 2001, 2003; Yu & Chung, 2006; Salminen et al. 2008a&b). Moreover, as detailed in more theoretical work by our group (Blangero et al., 2000, 2001; Almasy and Blangero, 2008), our statistical genetic approach is optimal for the detection of causal genes for complex diseases.
2.2. Genotype Data
Fasting blood samples were obtained from study participants at a clinic exam and transported daily to the Southwest Foundation for Biomedical Research (SFBR), San Antonio, Texas. Plasma and serum were isolated by low-speed centrifugation and the buffy coat was harvested for DNA extraction. DNA extracted from lymphocytes was used in polymerase chain reactions (PCRs) for the amplification of individual DNA (N = 1339) at 432 dinucleotide repeat microsatellite loci (STRs), spaced approximately 10 centiMorgan (cM) intervals apart across the 22 autosomes, with fluorescently-labeled primers from the MapPairs Human Screening set, versions 6 and 8 (Research Genetics, Hunstsville, AL). PCRs were performed separately according to manufacturer specifications in Applied Biosystems 9700 thermocyclers (Applied Biosystems, Foster City, CA). The products of separate PCRs, for each individual, were pooled using the Robbins Hydra-96 Microdispenser, and a labeled size standard was added to each pool. The pooled PCR products were loaded into an ABI PRISM 377 or 3100 Genetic Analyzer for laser-based automated genotyping. The STRs and standards were detected and quantified, and genotypes were scored using the Genotyper software package (Applied Biosystems).
Mistyping analyses were performed on the preliminary genotype marker data using SimWalk2, following the recommendations of the program developers for accounting for mistyping error (Sobel and Lange 1996; Sobel et al. 2002). Our overall rate of blanking mistyped markers was 1.37%. These mistyping analyses allow investigators to account for Mendelian errors and spurious double recombinants, both of which can severely reduce the power of a linkage analysis if not accounted for (Sobel et al. 2002). On addressing mistyping error, these genotype data were then used to compute maximum likelihood estimates of allele frequencies in SOLAR (Almasy and Blangero 1998). Empirical estimates of identity-by-descent (IBD) allele sharing at points throughout the genome for every relative pair were computed using the Loki package (Heath 1997). The multipoint IBD estimates are required under our variance components modeling approach. The Simwalk II and Loki programs both require chromosomal maps. We used the set of high-resolution chromosomal maps provided by the research group at deCODE genetics, Reykjavik, Iceland, which are available online as a supplemental table to the primary article (Kong et al. 2002).
2.3. Microarray Gene Expression Data
2.3.1. Expression profiling
The expression profiling methodology is described, in detail, in Göring et al. (2007). In brief, frozen lymphocyte samples were available from 1,280 individuals, collected during their first clinic visit between 1991 and 1995, after an overnight fast, in EDTA tubes. Lymphocytes were isolated from a 10ml sample using Histopaque (Sigma Chemical Co., St. Louis, MO), following the suggested protocol of the manufacturer, washed, and stored in a freeze media in liquid nitrogen.
Total RNA was isolated using a modified procedure of the QIAGEN RNeasy® 96 protocol for isolation of total RNA from animal cells using spin technology (QIAGEN Inc., Valencia, CA), and a total of 500ng total RNA dried down and stored at −20°C. Anti-sense RNA (aRNA) was synthesized, amplified and purified using the Ambion MessageAmp II Amplification Kit (Ambion, Austin, TX) following the Illumina Sentrix Array Matrix 96-well expression protocol (Illumina Inc., San Diego, CA). Synthesized cDNA samples were purified using QIAGEN’s QIAquick 96 PCR purification supplementary protocol for spin technology (QIAGEN document QQ01.doc, October 2001). Biotin-16-UTP (Roche, Germany) labeled aRNA was synthesized using Ambion’s proprietary MEGAscript® in vitro transcription (IVT) technology and T7 RNA Polymerase. Purification of aRNA samples was performed using QIAGEN’s RNeasy® 96 protocol for RNA cleanup using spin technology, and a total of 1.5μg aRNA was dried and stored at −20°C prior to sample hybridization.
Hybridization of aRNA to Illumina® Sentrix® Human Whole Genome (WG-6) Series I BeadChips and subsequent washing, blocking and detecting were performed using Illumina’s BeadChip 6×2 protocol. Samples were scanned on the Illumina® BeadArray™ 500GX Reader using Illumina® BeadScan image data acquisition software (version 2.3.0.13). Illumina® BeadStudio software (version 1.5.0.34) was used for preliminary data analysis, with a standard background normalization, to generate an output file for statistical analysis. In total we interrogated 47,289 unique transcripts: 22,151 probes (47%) are targeted at Reference Sequence (RefSeq) transcripts, and the remaining 25,128 probes (53%) correspond to other, generally less well characterized transcripts (including predicted genes).
2.3.2. Identification and standardization of expressed transcripts
In order to identify transcripts that exhibited sufficient quantitative expression in lymphocytes, the distribution of expression values for a given transcript was compared to the distribution of the expression values of the controls that are imbedded in each chip. For each transcript, we performed a χ2 “tail” test of whether there was a significant excess of samples with values above the 95th percentile of the control null distribution. This test was used because it allows detection of even those transcripts that are clearly present above baseline levels in only a subset of individuals, while not being detectable above baseline levels in most individuals. Using a false discovery rate of 0.05, we identified 20,413 transcripts that exhibited significant expression by this criterion.
To minimize effects due to RNA quantity and quality we performed within and across sample normalization. This conservative procedure results in normalized expression phenotypes that are comparable between individuals and across transcripts.
2.4. Network Analysis Methods
We used Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) to identify published gene/gene product interactions between NF-κB and the genes detected in our transcriptional profiling. Our genes of interest were overlaid onto a global molecular network developed from the literature on reported connectivity recorded in the Ingenuity Pathways Knowledge Base. This allows the generation of gene networks, and graphical representation of the molecular relationships between genes/gene products.
2.5. Principal Component Factor Analysis (PCFA)
To aid in the discovery of QTLs controlling gene expression networks, we use the approach of PCFA, which has been widely used in genetic studies of the metabolic syndrome (Arya et al. 2002; Liu et al. 2003; North et al. 2003, 2005; Cai et al. 2004; Lin et al. 2005; Edwards et al. 2008). We note also that PC analysis (PCA), which is the first step of PCFA, is widely used in gene expression analyses, usually in combination with other statistical approaches (Selaru et al. 2004; Wang and Gehan 2005; Roden et al. 2006; Wang et al. 2007; Ringnér 2008). Moreover, PCA has been used to identify functional modules of gene expression variables that manifest at the molecular level (Alter et al. 2000; Selaru et al. 2004; Janes and Yaffe 2006; Roden et al. 2006; Alter 2007). Generally, PCFA is a data-reduction technique that produces representative subsets in multivariate space—termed factors—of the original set of variables that explain a disproportionately higher portion of the total variance in the data. PCFA first uses PCA to find “raw” factors, and then uses a factor rotation procedure, such as varimax rotation, to produce factors that may be easier to interpret (Dunteman 1989; Kachigan 1991; Manly 1994; Jolliffe 2002). Factors with eigenvalues >1 were retained for subsequent analysis, and, to aid in factor interpretation, variables with factor loadings >|0.4| were taken to be the defining variables of the factor (Edwards et al. 1994; Arya et al. 2002; Stevens 2002; Cai et al. 2004; Lin et al. 2005).
2.6. Variance Components (VC) Models
Consider a data vector of N individuals, yN×1, which we assume follows the multivariate normal distribution, and is well-described by the following linear model:
| Eq. 1 |
where XN×k is a matrix of individual fixed effects (including a column of “1s” followed by the covariate effects), βk×1 is a vector of the grand mean and (k−1) beta-coefficients, and gN×1 and eN×1 are unobservable vectors of random genetic and environmental effects. The model for the multivariate mean is given by:
| Eq. 2 |
Eq. 2 where we have dropped the dimension subscripts for simplicity. Let the residuals vector between the data and the mean vector be given as: Δ = y − Xβ. Assuming independent genetic and environmental effects, the covariance matrix of the data vector, denoted by Σ (of dimensions N × N), is modeled as:
| Eq. 3 |
where the kinship matrix, Φ, and the identity matrix, I, partition the total variance into shared genetic and random environmental variance components, respectively. We note that y, φxz, and δxz are the scalar versions of y, Φ, and I. From this equation, we get the polygenic heritability, given as .
To model a QTL effect, we add it to the linear model in Equation 1 as an unobservable random effect, and its variance component to the model for the covariance matrix in Equation 3 as follows:
| Eq. 4 |
where is the QTL variance, and Φ̂ is the matrix of pair-wise estimates of identity-by-descent at a given point or locus in the genome based on flanking marker information.
2.7. Inferential Procedures: Likelihood ratio and LOD score
Our inferences are either directly formed from the results of the likelihood ratio test (LRT) or indirectly in that, under the VC approach, the LOD score is derived as the ratio of the LRT statistic, denoted by Λ, to twice ln(10). Let the parameters under the full polygenic model (Equations 1–3) be collected in a column vector: . Under this model, the likelihood function of the parameter vector conditional on the data is then given by:
| Eq. 5 |
Using this equation, maximum likelihood estimates are computed in SOLAR (Almasy and Blangero, 1996), and Λ is given as minus twice the difference of likelihoods between the null and alternative hypotheses:
| Eq. 6 |
where θH0 and θHA represent the parameter vectors under the null (H0) and alternative (HA) hypotheses, respectively. In general, Λ is distributed as a chi-square random variable with degrees of freedom (d.f.) given by the difference in the number of estimated parameters under the null and alternative hypotheses. For variances and/or standard deviations, however, the null hypothesis is on a boundary of the parameter space, in which case Λ has been shown to be distributed as a 50:50 mixture of point-mass at 0 and a chi-square variate with 1 d.f (Self and Liang 1987).
Inferences for multipoint genome-wide linkage scans are traditionally made on the basis of LOD scores. To provide a conservative corrective for the number of tests under a typical genome-wide linkage scan, a LOD score greater than or equal to 3.0 is taken to indicate significance of linkage (MacCluer et al. 1999; Ott 1999).
3. Results
3.1. Network Analysis
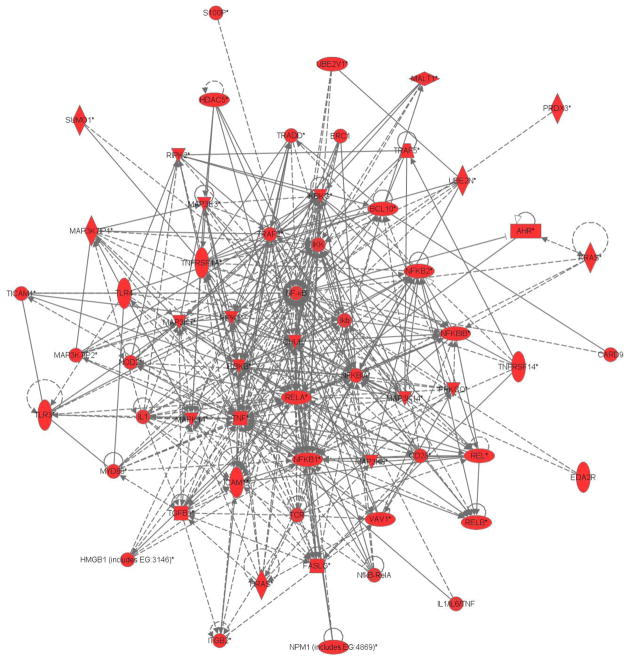
We used IPA to construct a network based solely on evidence of a first degree relationship (no intermediary gene/product) between each of the 20,413 transcripts that had detectable expression levels and NF-κB. We identified 60 genes with published first-degree gene/gene product interactions with NF-κB in our dataset, as shown in Figure 1.
Figure 1.
Gene expression network of the heritable transcripts in the nuclear factor kappa B signaling pathway.
3.2. Pathway Analysis
The core of the NF-κB STN is made up five proteins of the Rel family, RelA (p65), RelB, c-Rel, NF-κB1 (p50/p105), and NF-κB2 (p52/p100), which form homodimers or heterodimers that are present in bound form in the cytoplasm, but upon activation may translocate to the nucleus to stimulate transcription (Ghosh and Karin 2002; Li and Verma 2002; Hayden et al. 2006; Xiao et al. 2006; Hoffmann and Baltimore 2006; Vallabhapurapu and Karin 2009). In the cytoplasm, the NF-κB core proteins are bound by inhibitors of NF-κB (IκB), and are activated by phosphorylation of IκB by IκB kinases (IKK) (Ghosh and Karin 2002; Liou 2002; Hoffmann and Baltimore 2006; Xiao et al. 2006; Perkins 2007; Vallabhapurapu and Karin 2009). Once activated by IKK mediated phosphorylation of the NF-κB/IκB complex, NF-κB can take either of two main pathways, the canonical or classical pathway and the noncanonical or alternative pathway (Bonizzi and Karin 2003; Hayden et al. 2006; Xiao et al. 2006; Vallabhapurapu and Karin 2009).
Using information available in the literature, we limited further analyses to a subset of the NF-κB STN to 19 transcripts. First of all, we selected a core component comprised of the transcripts central to the NF-κB STN, which included four of the five core proteins, namely Rel, RelA, RelB, and NF-κB1, one inhibitor, namely IκBα, and two members of the activator kinases, namely IKKβ and IKKγ. We also included tumor necrosis factor α (TNFα) because of its ubiquitous involvement in upregulating inflammatory pathways, mainly through the NF-κB STN, in relation to senescence (Brüünsgaard et al. 2001; Brüünsgaard and Pedersen 2003; Krabbe et al. 2004; Sonnenberg et al. 2004; Tedgui and Mallat 2006). An additional 12 transcripts were selected because they were either a cell-surface receptor protein or an important adaptor protein that interacts with a receptor protein. These include toll-like receptors 3 and 4 (TLR3 and TLR4), three members of the TNF receptor superfamily (TNFRSF), namely TNFRSF1A, TNFRSF6, TNFRSF14, two TNF receptor associated factors (TRAF), namely TRAF2 and TRAF5, two transcripts of TNF receptor 1 associated death domain protein (TRADD), the adaptor protein myeloid differentiation factor-88 (MyD88), and two transcripts of toll/interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF).
3.2. Principal Components Factor Analysis (PCFA)
Currently, TLR signaling pathways are classified as being either MyD88-dependent or MyD88-independent/TRIF-dependent (Akira et al. 2001; Heine and Lien 2003; Akira and Takeda 2004; West et al. 2006; Atkinson 2008). Therefore, we decided to perform PCFA on the 19 transcripts named above, but also on two subsets, one excluding just the MyD88 transcript, and the other excluding the two TRIF transcripts. We call the set of 19 transcripts the “All-Transcripts” (AT) set, the subset excluding MyD88 the “TRIF-dependent” (TD) set, and the subset excluding the TRIF transcripts the “MyD88-dependent” (MD) set. The latter two variable sets were considered in order to take advantage of the fact that PCFA finds factors that are made up of variables that are correlated within-factor but are orthogonal across factors, which would potentially accentuate effects that are truly independent of either MyD88 or TRIF, respectively. Conversely, the AT set was considered because of the potential for synergistic effects involving both MyD88 and TRIF.
For the AT set, eigenvalues and percent variance explained for all factors with an eigenvalue greater than 1 are presented in Table 1. The factor loadings after varimax rotation for the AT set are presented in Table 2. Similarly, for the TD and MD sets, the eigenvalues and percent variance explained for all factors with an eigenvalue greater than 1 and the factor loadings are respectively presented in Tables 3–6.
Table 1.
Factors, Eigenvalues, and Variance Explained: All Transcription Variables
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | Factor 7 | |
|---|---|---|---|---|---|---|---|
| Eigenvalue | 3.879 | 2.172 | 2.016 | 1.591 | 1.267 | 1.131 | 1.034 |
| % Variance Explained | 20.983 | 11.751 | 10.905 | 8.607 | 6.855 | 6.120 | 5.596 |
| Cumulative Variance | 20.983 | 32.734 | 43.639 | 52.246 | 59.101 | 65.221 | 70.817 |
Table 2.
Factor Loadings: All Transcription Variables
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | Factor 7 | |
|---|---|---|---|---|---|---|---|
| IKBKB | 0.046 | −0.098 | 0.390 | 0.047 | −0.318 | 0.210 | 0.100 |
| IKBKG | 0.135 | −0.428 | 0.726 | 0.087 | 0.155 | 0.058 | 0.008 |
| NFKB1 | 0.172 | 0.003 | 0.017 | −0.005 | 0.212 | −0.044 | 0.093 |
| IKBA | −0.831 | 0.106 | −0.192 | 0.071 | −0.055 | −0.170 | −0.223 |
| RELB | −0.025 | 0.232 | 0.273 | −0.059 | 0.011 | −0.506 | −0.616 |
| REL | −0.196 | 0.358 | 0.130 | −0.050 | 0.699 | 0.112 | −0.305 |
| TLR3 | −0.004 | −0.104 | −0.061 | 0.058 | −0.111 | 0.007 | −0.026 |
| TLR4 | −0.205 | 0.295 | −0.206 | 0.728 | 0.180 | 0.151 | −0.086 |
| TNFRSF14 | 0.099 | −0.076 | 0.786 | 0.004 | −0.059 | −0.055 | −0.035 |
| TNFRSF1A | 0.355 | −0.336 | 0.042 | 0.654 | −0.352 | 0.111 | −0.161 |
| TNFSF6 | 0.203 | 0.085 | 0.043 | −0.022 | 0.082 | 0.864 | 0.021 |
| TNF | −0.921 | 0.030 | −0.007 | 0.067 | 0.019 | −0.093 | −0.138 |
| TRADDa | 0.161 | −0.818 | 0.162 | −0.082 | 0.059 | −0.111 | 0.201 |
| TRADDb | 0.141 | −0.162 | −0.022 | 0.146 | 0.772 | 0.058 | 0.066 |
| TRAF2 | −0.175 | −0.629 | 0.300 | −0.216 | −0.369 | 0.140 | −0.257 |
| TRAF5 | 0.136 | −0.376 | 0.193 | −0.418 | 0.063 | −0.017 | 0.592 |
| TRIFa | −0.115 | 0.144 | −0.024 | 0.015 | 0.047 | 0.040 | −0.194 |
| TRIFb | −0.333 | −0.111 | −0.016 | 0.079 | 0.107 | 0.023 | −0.813 |
| MYD88 | −0.140 | 0.085 | 0.329 | 0.746 | 0.138 | −0.257 | 0.009 |
Table 3.
Factors, Eigenvalues, and Variance Explained: TRIF-Dependent
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | |
|---|---|---|---|---|---|---|
| Eigenvalue | 3.835 | 2.161 | 1.801 | 1.488 | 1.263 | 1.053 |
| % Variance Explained | 21.918 | 12.351 | 10.293 | 8.504 | 7.218 | 6.017 |
| Cumulative Variance | 21.918 | 34.269 | 44.563 | 53.066 | 60.284 | 66.301 |
Table 6.
Factor Loadings: MyD88-Dependent
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | |
|---|---|---|---|---|---|---|
| IKBKB | 0.099 | −0.104 | −0.069 | 0.094 | −0.220 | 0.066 |
| IKBKG | 0.110 | −0.168 | −0.907 | 0.121 | 0.035 | 0.014 |
| NFKB1 | 0.142 | 0.065 | 0.031 | −0.023 | −0.942 | 0.030 |
| IKBA | −0.867 | 0.018 | 0.101 | −0.064 | 0.146 | −0.142 |
| RELB | −0.128 | −0.004 | −0.017 | −0.020 | 0.004 | −0.127 |
| REL | −0.100 | 0.100 | −0.058 | −0.162 | −0.015 | 0.042 |
| TLR3 | −0.018 | −0.024 | −0.022 | 0.046 | 0.078 | −0.009 |
| TLR4 | −0.123 | 0.133 | 0.073 | 0.134 | −0.016 | 0.020 |
| TNFRSF14 | 0.091 | −0.124 | −0.203 | 0.034 | −0.125 | −0.024 |
| TNFRSF1A | 0.150 | −0.063 | −0.114 | 0.930 | 0.022 | 0.047 |
| TNFSF6 | 0.140 | 0.004 | −0.010 | 0.040 | −0.028 | 0.974 |
| TNF | −0.939 | −0.084 | 0.037 | −0.111 | 0.051 | −0.065 |
| TRADDa | 0.132 | −0.216 | −0.205 | 0.098 | −0.041 | −0.052 |
| TRADDb | 0.074 | 0.121 | −0.048 | −0.045 | −0.112 | 0.068 |
| TRAF2 | −0.073 | −0.917 | −0.163 | 0.065 | 0.070 | −0.005 |
| TRAF5 | 0.180 | −0.045 | −0.133 | −0.111 | −0.128 | −0.002 |
| MYD88 | −0.099 | 0.072 | −0.090 | 0.132 | −0.028 | −0.068 |
3.3. Heritability and Linkage
In the ensuing, factors will have AT, TD, or MD as a suffix to indicate which set they were derived from. The heritability for each factor across the three variable sets are reported in Table 7. All of the factors were significantly heritable. The heritabilities ranged from a low of 19% to a high of 52%.
Table 7.
Heritabilities for all factors
| Variable set or subset | Heritability, and standard error for: | ||||||
|---|---|---|---|---|---|---|---|
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | Factor 7 | |
| All Transcripts | 0.51 (0.05) | 0.34 (0.05) | 0.21 (0.06) | 0.49 (0.05) | 0.27 (0.06) | 0.41 (0.06) | 0.44 (0.05) |
| TRIF-Dependent | 0.48 (0.05) | 0.34 (0.05) | 0.30 (0.05) | 0.48 (0.05) | 0.29 (0.06) | 0.48 (0.05) | NA |
| MyD88-Dependent | 0.52 (0.05) | 0.24 (0.05) | 0.19 (0.05) | 0.40 (0.05) | 0.23 (0.05) | 0.30 (0.06) | NA |
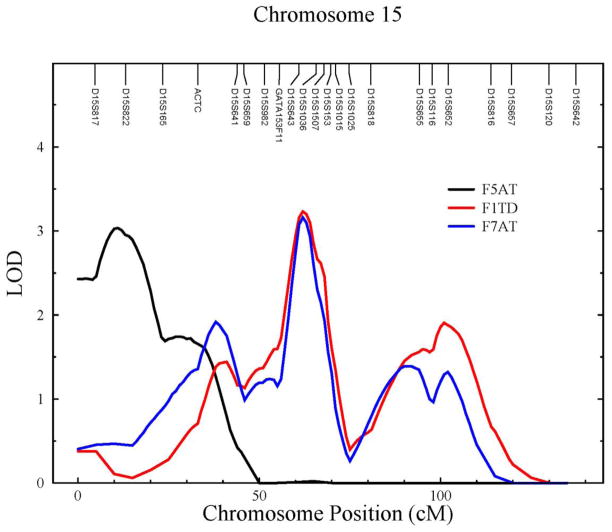
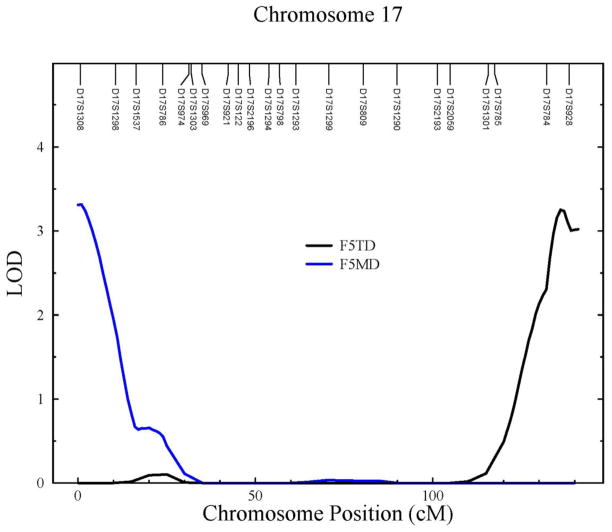
The genome-wide maximum LOD scores for all factors are reported in Table 8. Each variable set gave rise to at least one factor with a LOD score greater than 3, two on chromosome 15 at 15q12 and 15q22.2, and another two on chromosome 17 at 17p13.3, and 17q25.3 (Table 8; Figs. 2&3). We also found several suggestive signals (2 < LOD score < 3) at 1q32.1, 1q41, 2q34, 3q23, and 7p15.3 (Table 8). We do not believe that the three suggestive signals on chromosomes 15 and 17 are distinct from the nearby locations reported just above at 15q12, 15q22.2, and 17p13.3.
Table 8.
Standard linkage results: genome-wide maximum LOD scores for all factors
| Variable set or subset | LOD score, cytogenetic location: | ||||||
|---|---|---|---|---|---|---|---|
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | Factor 7 | |
| All Transcripts | 1.96 1q32.3 |
2.22 7p15.3 |
1.96 7q36.2 |
2.08 2q34 |
3.03 15q12 |
1.73 5p13.3 |
3.16 15q22.2 |
| TRIF-Dependent | 3.23 15q22.2 |
2.04 1q32.1 |
1.87 9p24.1-p23 |
2.48 15q14 |
3.25 17q25.3 |
2.12 17p12 |
NA |
| MyD88-Dependent | 2.64 1q41 |
1.50 4q34.3 |
1.97 18p11.21 |
2.61 15q22.3 |
3.31 17p13.3 |
2.10 3q23 |
NA |
Figure 2.
LOD plots on chromosome 15 for F5AT (black), F1TD (red), and F7AT (blue).
Figure 3.
LOD plots on chromosome 17 for F5TD (black), and F5MD (blue).
4. Discussion
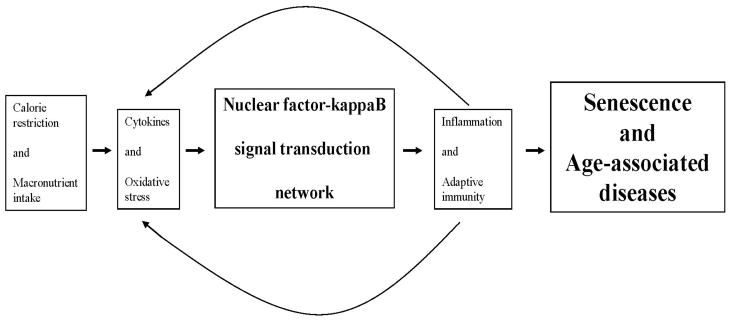
It has long been thought that NF-κB and its associated STN plays a major role in aging (Vellanoweth et al. 1994; Supakar et al., 1995; Roy et al. 1996, 2002; Roy 1997; Chung et al. 2000, 2001, 2002, 2006, 2009; Lavrovsky et al. 2000; Giardina and Hubbard, 2002; Gosselin and Abbadie, 2003; Herbein et al. 2006; Yu & Chung, 2006; Salminen et al. 2008a&b). We have dubbed this work the theory of NF-κB-dependent senescence. An attractive characteristic of the theory of NF-κB-dependent senescence is the elegance of mechanistically unifying it with at least four other prominent theories of aging, namely the oxidative stress, calorie restriction, inflammation, and immunological theories, and with a related theory on the etiology of the metabolic syndrome, namely the macronutrient intake theory of Dandona and colleagues (Dandona et al. 2004a,b,c, 2005).
The canonical and alternative signaling pathways of the NF-κB STN are set in train by extracellular signals and stimuli, such as proinflammatory cytokines and oxidative stress (Xiao 2004; Perkins 2007). Acting independently and/or in concert, the two pathways regulate the innate and adaptive arms of the immune response, including inflammation and cell-mediated pathways, respectively (Li and Verma 2002; Liou 2002; Bonizzi and Karin 2003; Liang et al. 2004; Hayden et al. 2006; Xiao et al. 2006; Vallabhapurapu and Karin 2009). In related work, it has been shown that by directly modulating oxidative stress, calorie restriction indirectly regulates NF-kappa B expression (Kim et al. 2002a&b, 2008; Jung et al. 2009). This is consistent with demonstrations that macronutrient intake promotes oxidative stress, which in turn upregulates the NF-κB STN (Dandona et al. 2004a,b,c, 2005). Thus, the NF-κB STN constitutes a critical nexus for linking calorie restriction, macronutrient intake, and oxidative stress to inflammatory and adaptive immunity pathways in relation to senescence (Figure 4).
Figure 4.
Schematic diagram of the nuclear factor kappa B signal transduction network in relation to other physiological processes important in senescence and age-associated disease.
To better understand the genetic regulation of the NF-κB STN, and, by extension the genetic architecture underlying NF-κB-dependent senescence, we used a comprehensive systems genetics approach. First, we constructed a gene expression network representative of the NF-κB STN. Then, with the goal of concentrating our efforts on the most important sources of variation, we used a PCFA approach on a subset of the NF-κB STN. Finally, we analyzed the factors so derived using a statistical genetic VC approach. We found that these factors are all significantly heritable, and, further, we found strong evidence of four QTLS at chromosomes 15q12, 15q22.2, 17p13.3, and 17q25.3, and suggestive evidence of potential QTLs on chromosomes 1, 2, 3, and 7.
To facilitate discussion of these QTLs in relation to the PCFA results, we abbreviate the factors by “F” followed by the factor number, and then by the AT, TD, or MD abbreviations as appropriate. For example, factor 5 under the AT set is abbreviated as F5AT. We also restrict our discussion to our strongest results (i.e. QTLs with a LOD score > 3). We found that five PCFA factors gave rise to the 4 main linkage signals on chromosomes 15 and 17. The composite variables F7AT and F1TD have the same three NF-κB STN variables loading onto the factor, namely RELB, TRAF5, and TRIF, and as expected they give rise to what appears to be the same linkage signals. Similarly, F5AT and F5TD also have the same NF-κB STN variables loading onto the factor, namely REL and TRADD, and their linkage signals on chromosomes 15 and 17 are similar. The maximum LOD scores for F5AT and F5TD are 3.03 at chromosome 15q12 and 3.25 at chromosome 17q25.3, respectively, and suggestive LODS scores for the same traits are 2.21 at chromosome 17q25.3 and 2.74 at chromosome 15q12, respectively. Given that MyD88 is the main difference between the AT and TD sets, it may be that the putative QTL on chromosome 15q12 is relatively more important in the MD TLR signaling pathway and that the putative QTL on chromosome 17q25.3 is relatively more important in the TD TLR signaling pathway. F5MD had only one NF-κB STN variable loading onto the factor, namely NFKB1. We note that none of the cytogenetic locations of the putative QTLs coincide with the cytogenetic locations of the constituent genes in the relevant factors. Thus, we have identified what appear to be trans-QTLs influencing three of the five main core proteins of the NF-κB STN, namely Rel, RelB and NFKB1, and three other important signaling proteins, namely TRADD, TRIF, and TRAF5.
As reported in Schadt et al. (2005) and discussed in Sieberts and Schadt (2007), trans-QTLs can play prominent roles as drivers of complex disease causation if they are centrally located in a network known to be important in complex disease. Their group was able to identify and functionally validate three candidate genes as causal for obesity, and these genes were first identified as trans-QTLs centrally located in a gene expression network known to be important in obesity. Regarding our situation, the work of Schadt and colleagues is encouraging because our trans-QTLs are centrally located within the NF-κB STN.
The work reported herein is part of an ongoing investigation of the genetic regulation of the NF-κB STN by way of a systems genetics approach. While our findings regarding the heritable factors of the NF-κB STN and their associated trans-QTLs are important, we emphasize that together they constitute what is only a first step in the process of gene discovery. We are currently pursuing more fine-detail genetic analyses by examining the transcripts and single nucleotide polymorphisms within the 1-LOD intervals of the QTLs identified here.
Table 4.
Factor Loadings: TRIF-Dependent
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | |
|---|---|---|---|---|---|---|
| IKBKB | 0.169 | −0.093 | 0.613 | 0.165 | −0.249 | −0.030 |
| IKBKG | −0.028 | −0.392 | −0.184 | 0.122 | 0.229 | −0.120 |
| NFKB1 | 0.028 | −0.002 | 0.904 | −0.059 | 0.201 | −0.179 |
| IKBA | −0.175 | 0.131 | −0.167 | −0.053 | −0.078 | 0.844 |
| RELB | −0.792 | 0.097 | 0.023 | −0.122 | −0.086 | 0.015 |
| REL | −0.387 | 0.335 | −0.060 | −0.350 | 0.541 | 0.182 |
| TLR3 | −0.003 | −0.066 | −0.100 | 0.064 | −0.148 | 0.016 |
| TLR4 | 0.006 | 0.579 | 0.053 | 0.508 | 0.302 | 0.319 |
| TNFRSF14 | −0.069 | −0.084 | 0.255 | 0.001 | −0.022 | −0.103 |
| TNFRSF1A | −0.027 | −0.101 | −0.004 | 0.882 | −0.095 | −0.202 |
| TNFSF6 | 0.102 | 0.042 | 0.048 | 0.020 | 0.054 | −0.202 |
| TNF | −0.119 | 0.036 | −0.057 | −0.118 | −0.015 | 0.913 |
| TRADDa | 0.179 | −0.812 | 0.081 | 0.115 | 0.164 | −0.159 |
| TRADDb | 0.069 | −0.004 | 0.122 | 0.004 | 0.849 | −0.100 |
| TRAF2 | −0.143 | −0.714 | −0.042 | 0.111 | −0.328 | 0.202 |
| TRAF5 | 0.518 | −0.482 | 0.201 | −0.353 | 0.041 | −0.236 |
| TRIFa | −0.187 | 0.149 | −0.045 | −0.046 | 0.073 | 0.118 |
| TRIFb | −0.728 | −0.101 | −0.114 | 0.139 | 0.091 | 0.421 |
Table 5.
Factors, Eigenvalues, and Variance Explained: MyD88-Dependent
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | |
|---|---|---|---|---|---|---|
| Eigenvalue | 3.496 | 2.055 | 1.933 | 1.576 | 1.245 | 1.016 |
| % Variance Explained | 21.163 | 12.443 | 11.699 | 9.538 | 7.539 | 6.153 |
| Cumulative Variance | 21.163 | 33.606 | 45.305 | 54.843 | 62.382 | 68.535 |
Highlights.
Gene expression network of the nuclear factor kappa B (NF-κB) signaling network.
Principal components factor analysis used to derive composite traits.
Identified and localized quantitative trait loci (QTL) underlying the NF-κB signaling network.
Acknowledgments
We thank the Mexican American families of San Antonio who participated in the SAFHS. This research was funded by National Institutes of Health (NIH) grants P01 HL45522 and MH 59490 and was conducted in facilities constructed with support from NIH Research Facilities Improvement Program grants C06 RR013556 and C06 RR017515 and from SBC Communications (now AT&T).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Joanne E. Curran, Email: jcurran@txbiomedgenetics.org.
Jac Charlesworth, Email: Jac.Charlesworth@utas.edu.au.
Juan M. Peralta, Email: jperalta@txbiomedgenetics.org.
V. Saroja Voruganti, Email: saroja@txbiomedgenetics.org.
Shelley A. Cole, Email: scole@txbiomedgenetics.org.
Thomas D. Dyer, Email: tdyer@txbiomedgenetics.org.
Matthew P. Johnson, Email: mjohnson@txbiomedgenetics.org.
Eric K. Moses, Email: texozmoses@txbiomedgenetics.org.
Harald H. H. Göring, Email: hgoring@txbiomedgenetics.org.
Jeff T. Williams, Email: jtw@txbiomedgenetics.org.
Anthony G. Comuzzie, Email: tony@txbiomedgenetics.org.
Laura Almasy, Email: almasy@txbiomedgenetics.org.
John Blangero, Email: john@txbiomedgenetics.org.
Sarah Williams-Blangero, Email: sarah@txbiomedgenetics.org.
References
- Akira S, Takeda K, Kaisho T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Contemporary model-free methods for linkage analysis. Genetic Dissection of Complex Traits. In: Rao DC, Gu CC, editors. Advances in Genetics. 2. Vol. 60. 2008. pp. 175–193. [DOI] [PubMed] [Google Scholar]
- Alter O. Genomic signal processing: From matrix algebra to genetic networks. In: Korenberg MJ, editor. Microarray Data Analysis: Methods and Applications, Methods in Molecular Biology. Vol. 377. 2007. pp. 17–59. [DOI] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Nat Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, Blangero J, Williams K, Almasy L, Dyer TD, Leach RJ, O’Connell P, Stern MP, Duggirala R. Factors of insulin resistance syndrome-related phenotypes are linked to genetic locations on chromosome 6 and 7 in nondiabetic Mexican-Americans. Diabetes. 2002;51:841–847. doi: 10.2337/diabetes.51.3.841. [DOI] [PubMed] [Google Scholar]
- Atkinson TJ. Toll-like receptors, transduction-effector pathways, and disease diversity: Evidence of an immunobiological paradigm explaining all human illness? Int Rev Immunol. 2008;27:255–281. doi: 10.1080/08830180801959072. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RRH, Mackay TFC. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard J-P, Maachi M, Lagathu C, Kim JM, Caron M, Vidal H, Capeau H, Feve B. Recent advances in the relationship between obesity, inflammation, and insulin resistance. Eur Cyt Netw. 2006;17:4–12. [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Edu Behav Stat. 2000;25:60–83. [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- Blangero J. Statistical genetic approaches to human adaptability. Hum Biol. 1993;65:941–966. [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Quantitative trait locus mapping using human pedigrees. Hum Biol. 2000;72:35–62. [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Variance component methods for detecting complex trait loci, Genetic Dissection of Complex Traits. In: Rao DC, Province MA, editors. Advances in Genetics. Vol. 42. 2001. pp. 151–181. [DOI] [PubMed] [Google Scholar]
- Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, Krainc D. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc Nat Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüünsgaard H, Pedersen M, Pedersen BK. Aging and proinflammatory cytokines. Curr Opin Hematol. 2001;8:131–136. doi: 10.1097/00062752-200105000-00001. [DOI] [PubMed] [Google Scholar]
- Brüünsgaard H, Pedersen BK. Age-related inflammatory cytokines and disease. Immunol Allergy Clin N Am. 2003;23:15–39. doi: 10.1016/s0889-8561(02)00056-5. [DOI] [PubMed] [Google Scholar]
- Cai G, Cole SA, Freeland-Graves JH, MacCluer JW, Blangero J, Comuzzie AG. Principal component for metabolic syndrome risk maps to chromosome 4p in Mexican Americans: The San Antonio Family Heart Study. Hum Biol. 2004;76:651–665. doi: 10.1353/hub.2005.0001. [DOI] [PubMed] [Google Scholar]
- Chung HY, Kim HJ, Jung KJ, Yoon JS, Yoo MA, Kim KW, Yu BP. The inflammatory process in aging. Rev Clin Gerontol. 2000;10:207–222. [Google Scholar]
- Chung HY, Kim HJ, Kim JW, Yu BP. The inflammation hypothesis of aging: Molecular modulation by calorie restriction. Ann NY Acad Sci. 2001;928:327–335. [PubMed] [Google Scholar]
- Chung HY, Kim HJ, Kim KW, Choi JS, Yu BP. Molecular inflammation hypothesis of aging based on the anti-aging mechanism of calorie restriction. Microsc Res Techniq. 2002;59:264–272. doi: 10.1002/jemt.10203. [DOI] [PubMed] [Google Scholar]
- Chung HY, Sung B, Jung KJ, Zou Y, Yu BP. The molecular inflammatory process in aging. Antiox Redox Signal. 2006;8:572–581. doi: 10.1089/ars.2006.8.572. [DOI] [PubMed] [Google Scholar]
- Chung HY, Cesari M, Anton S, Marzetti E, Giovannini S, Seo AY, Carter C, Yu BP, Leeuwenburgh C. Molecular inflammation: Underpinnings of aging and age-related diseases. Ageing Res Rev. 2009;8:18–30. doi: 10.1016/j.arr.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandona P, Aljada A, Bandyopadhyay A. Inflammation: The linke between insulin resistance, obesity and diabetes. Trends Immunol. 2004a;25:4–7. doi: 10.1016/j.it.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Dandona P, Aljada A, Chaudhuri A, Mohanty P. Endothelial dysfunction, inflammation and diabetes. Rev Endocr Metab Disord. 2004b;5:189–197. doi: 10.1023/B:REMD.0000032407.88070.0a. [DOI] [PubMed] [Google Scholar]
- Dandona P, Aljada A, Chaudhuri A, Mohanty P, Garg R. A novel view of metabolic syndrome. Metab Syndr Rel Disord. 2004c;2:2–8. doi: 10.1089/met.2004.2.2. [DOI] [PubMed] [Google Scholar]
- Dandona P, Aljada A, Chaudhuri A, Mohanty P, Garg R. Metabolic syndrome: A comprehensive perspective based on interactions between obesity, diabetes, and inflammation. Circulation. 2005;111:1448–1454. doi: 10.1161/01.CIR.0000158483.13093.9D. [DOI] [PubMed] [Google Scholar]
- de Winther MPJ, Kanters E, Kraal G, Hofker MH. Nuclear factor κB signaling in atherosclerosis. Arterioscler Thromb Vasc Biol. 2005;25:904–914. doi: 10.1161/01.ATV.0000160340.72641.87. [DOI] [PubMed] [Google Scholar]
- Dolcet X, Llobet D, Pallares J, Matias-Guiu X. NF-κB in development and progression of human cancer. Virchows Arch. 2005;446:475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- Drake TA, Schadt EE, Lusis AJ. Integrating genetic and gene expression data: Application to cardiovascular and metabolic traits in mice. Mamm Genome. 2006;17:466–479. doi: 10.1007/s00335-005-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunteman GH. Principal Components Analysis. SAGE Publications; Newbury Park, CA: 1989. [Google Scholar]
- Edwards KL, Austin MA, Newman B, Mayer E, Krauss RM, Selby JV. Multivariate analysis of the insulin resistance syndrome in women. Arterioscler Thromb. 1994;24:1940–1945. doi: 10.1161/01.atv.14.12.1940. [DOI] [PubMed] [Google Scholar]
- Edwards KL, Hutter CM, Wan JY, Kim H, Monks SA. Genome-wide Linkage scan for the metabolic syndrome: The GENNID Study. Obesity. 2008;16:1596–1601. doi: 10.1038/oby.2008.236. [DOI] [PubMed] [Google Scholar]
- Galitski T. Molecular networks in model systems. Annu Rev Genomics Hum Genet. 2004;5:177–187. doi: 10.1146/annurev.genom.5.061903.180053. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Giardina C, Hubbard AK. Growing old with nuclear factor-κB. Cell Stress Chaperones. 2002;7:207–212. doi: 10.1379/1466-1268(2002)007<0207:gownfb>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Aguilera CM, Gil-Campos M, Cañete R. Altered signaling and gene expression associated with the immune system and the inflammatory response in obesity. Br J Nutr. 2007;98:S121–S126. doi: 10.1017/S0007114507838050. [DOI] [PubMed] [Google Scholar]
- Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Prog Neuro-Psychopharmacol Biol Psychiatry. 2004;28:559–576. doi: 10.1016/j.pnpbp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Göring HHH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JBM, Abraham LJ, Rainwater DL, Comuzzie AG, Mahaney MC, Almasy L, MacCluer JW, Collier GR, Moses EK, Blangero J. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Gosselin K, Abbadie C. Involvement of Rel/NF-κB transcription factors in senescence. Exp Gerontol. 2003;38:1271–1283. doi: 10.1016/j.exger.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Han JDJ. Understanding biological functions through molecular networks. Cell Res. 2008;18:224–237. doi: 10.1038/cr.2008.16. [DOI] [PubMed] [Google Scholar]
- Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-κB and the immune response. Oncogene. 2005;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- Heath SM. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine H, Lien E. Toll-like receptors and their function in innate and adaptive immunity. Int Arch Allergy Immunol. 2003;130:180–192. doi: 10.1159/000069517. [DOI] [PubMed] [Google Scholar]
- Herbein G, Varin A, Fulop T. NF-κB, AP-1, Zinc-deficiency and aging. Biogerontol. 2006;7:409–419. doi: 10.1007/s10522-006-9056-4. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D. Circuitry of nuclear factor κB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Barkaii N. Defining transcription modules using large-scale gene expression data. Bioinformatics. 2004;20:1993–2003. doi: 10.1093/bioinformatics/bth166. [DOI] [PubMed] [Google Scholar]
- Inoue JI, Gohda J, Akiyama T, Semba K. NF-κB activation in development and progression of cancer. Cancer Sci. 2007;98:268–274. doi: 10.1111/j.1349-7006.2007.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT. Principal Component Analysis. 2. Springer; New York: 2002. [Google Scholar]
- Jung KJ, Lee EK, Kim JY, Zou Y, Sung B, Heo HS, Kim MK, Lee J, Kim ND, Yu BP, Chung HY. Effect of short term calorie restriction on pro-inflammatory NF-κB and AP-1 in aged rat kidney. Inflamm Res. 2009;58:143–150. doi: 10.1007/s00011-008-7227-2. [DOI] [PubMed] [Google Scholar]
- Kachigan S. Multivariate Statistical Analysis: A Conceptual Introduction. 2. Radius Press; New York: 1991. [Google Scholar]
- Kaltschmidt B, Widera D, Kaltschmidt C. Signaling via NF-κB in the nervous system. Biochim Biophys Acta. 2005;1745:287–299. doi: 10.1016/j.bbamcr.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Kardarmideen HN, von Rohr P, Janss LLG. From genetical genomics to systems genetics: Potential applications in quantitative genomics and animal breeding. Mamm Genome. 2006;17:548–564. doi: 10.1007/s00335-005-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kim DH, Kim JY, Yu BP, Chung HY. The activation of NF-κB through Akt-induced FOXO1 phosphorylation during aging and its modulation by calorie restriction. Biogerontology. 2008;9:33–47. doi: 10.1007/s10522-007-9114-6. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Yu B-P, Chung HY. Molecular exploration of age-related NF-κB/IKK downregulation by calorie restriction in rat kidney. Free Rad Biol Med. 2002;32:991–1005. doi: 10.1016/s0891-5849(02)00798-0. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Jung KJ, Yu BP, Cho CG, Choi JS, Chung HY. Modulation of redox-sensitive transcription factors by calorie restriction during aging. Mech Ageing Dev. 2002;123:1589–1595. doi: 10.1016/s0047-6374(02)00094-5. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Krabbe KS, Pedersen M, Brüünsgaard H. Inflammatory mediators in the elderly. Exp Gerontol. 2004;39:687–699. doi: 10.1016/j.exger.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Kutalik Z, Beckmann JS, Bergmann S. A modular approach for integrative analysis of large-scale gene-expression and drug-response data. Nat Biotechnol. 2008;26:531–539. doi: 10.1038/nbt1397. [DOI] [PubMed] [Google Scholar]
- Lavrovsky Y, Chatterjee B, Clark RA, Roy AK. Role of redox-regulated transcription factors in inflammation, aging and age-related diseases. Exp Gerontol. 2000;35:521–532. doi: 10.1016/s0531-5565(00)00118-2. [DOI] [PubMed] [Google Scholar]
- Liang Y, Zhou Y, Shen P. NF-κB and its regulation on the immune system. Cell Mol Immunol. 2004;1:343–350. [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-κB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Lin HF, Boden-Albala B, Juo SH, Park N, Rundek T, Sacco RL. Heritabilities of the metabolic syndrome and its components in the Northern Manhattan Family Study. Diabetologia. 2005;48:2006–2012. doi: 10.1007/s00125-005-1892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HC. Regulation of the immune system by NF-κB and IκB. J Biochem Mol Biol. 2002;35:537–546. doi: 10.5483/bmbrep.2002.35.6.537. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Hanley AJG, Paterson AD. Genetic analysis of common factors underlying cardiovascular disease-related traits. BMC Genet. 2003;4:S56. doi: 10.1186/1471-2156-4-S1-S56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCluer JW, Stern MP, Almasy L, Atwood LA, Blangero J, Comuzzie AG, Dyke B, Haffner SM, Henkel RD, Hixson JE, Kammerer CM, Mahaney MC, Mitchell BD, Rainwater DL, Samollow PB, Sharp RM, VandeBerg JL, Williams JT. Genetics of atherosclerosis risk factors in Mexican Americans. Nutr Rev. 1999;57:S59–S65. doi: 10.1111/j.1753-4887.1999.tb01790.x. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: Challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Maeda S, Omata M. Inflammation and cancer: Role of nuclear factor-kappaB activation. Cancer Sci. 2008;99:836–842. doi: 10.1111/j.1349-7006.2008.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly BFJ. Multivariate Statistical Methods: A Primer. 2. Chapman & Hall/CRC; Boca Raton, FL: 1994. [Google Scholar]
- Mattson MP. NF-κB in the survival and plasticity of neurons. Neurochem Res. 2005;30:883–893. doi: 10.1007/s11064-005-6961-x. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Meffert MK. Roles for NF-κB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- Meigs JB. Insulin resistance syndrome? Syndrome X? Multiple metabolic syndrome? A syndrome at all? Factor analysis reveals patterns in the fabric of correlated metabolic risk factors. Am J Epidemiol. 2000;152:908–911. doi: 10.1093/aje/152.10.908. [DOI] [PubMed] [Google Scholar]
- Mémet S. NF-κB functions in the nervous system: From development to disease. Biochem Pharmacol. 2006;72:1180–1195. doi: 10.1016/j.bcp.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Ghosh S, Schneider JL, Birznieks G, Blangero J. Power of variance component linkage analysis to detect epistasis. Genet Epidemiol. 1997;14:1017–1022. doi: 10.1002/(SICI)1098-2272(1997)14:6<1017::AID-GEPI76>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- North KE, Almasy L, Göring HHH, Cole SA, Diego VP, Laston S, Cantu T, Williams JT, Howard BV, Lee ET, Best LG, Fabsitz RR, MacCluer JW. Linkage analysis of factors underlying insulin resistance: Strong Heart Family Study. Obes Res. 2005;13:1877–1884. doi: 10.1038/oby.2005.230. [DOI] [PubMed] [Google Scholar]
- Ott J. Analysis of Human Genetic Linkage. 3. Johns Hopkins University Press; Baltimore, MD: 1999. [Google Scholar]
- Perkins ND. Integrating cell-signaling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Purcell S, Sham PC. Epistasis in quantitative trait locus linkage analysis: Interaction or main effect? Behavior Genet. 2004;34:143–152. doi: 10.1023/B:BEGE.0000013728.96408.f9. [DOI] [PubMed] [Google Scholar]
- Ringnér M. What is principal component analysis? Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- Roden JC, King BW, Trout D, Mortazavi A, Wold BJ, Hart CE. Mining gene expression data by interpreting principal components. BMC Bioinformatics. 2006;7:194. doi: 10.1186/1471-2105-7-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AK. Transcription factors and aging. Mol Med. 1997;3:496–504. [PMC free article] [PubMed] [Google Scholar]
- Roy AK, Vellanoweth RL, Chen S, Supakar PC, Jung MH, Song CS, Chatterjee B. The evolutionary tangle of aging, sex, and reproduction and an experimental approach to its molecular dissection. Exp Gerontol. 1996;31:83–94. doi: 10.1016/0531-5565(95)00020-8. [DOI] [PubMed] [Google Scholar]
- Roy AK, Oh T, Rivera O, Mubiru J, Song CS, Chatterjee B. Impacts of transcriptional regulation on aging and senescence. Ageing Res Rev. 2002;1:367–380. doi: 10.1016/s1568-1637(02)00006-5. [DOI] [PubMed] [Google Scholar]
- Salminen A, Ojala J, Huuskonen J, Kauppinen A, Suuronen T, Kaarniranta K. Interaction of aging-associated signaling cascades: Inhibition of NF-κB signaling by longevity factors FoxOs and SIRT1. Cell Mol Life Sci. 2008a;65:1049–1058. doi: 10.1007/s00018-008-7461-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Huuskonen J, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Activation of innate immunity system during aging: NF-κB signaling is the molecular culprit of inflamm-aging. Ageing Res Rev. 2008b;7:83–105. doi: 10.1016/j.arr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Sarkar SK. Some results on false discovery rate in stepwise multiple testing procedures. Ann Stat. 2002;30:239–257. [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, GuhaThakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EA, Reaven PD. Molecular and signaling mechanisms of atherosclerosis in insulin resistance. Endocrinol Metab Clin N Am. 2006;35:525–549. doi: 10.1016/j.ecl.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Selaru FM, Yin J, Olaru A, Mori Y, Xu Y, Epstein SH, Sato F, Deacu E, Wang S, Sterian A, Fulton A, Abraham JM, Shibata D, Baquet C, Stass SA, Meltzer SJ. An unsupervised approach to identify molecular phenotypic components influencing breast cancer features. Cancer Res. 2004;64:1584–1588. doi: 10.1158/0008-5472.can-03-3208. [DOI] [PubMed] [Google Scholar]
- Self SG, Liang KY. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc. 1987;82:605–610. [Google Scholar]
- Sieberts SK, Schadt EE. Moving towards a system genetics view of disease. Mamm Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GE, Krakower GR, Kissebah AH. A novel pathway to the manifestations of metabolic syndrome. Obes Res. 2004;12:180–186. doi: 10.1038/oby.2004.24. [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. Applied Multivariate Statistics for the Social Sciences. 4. Lawrence Erlbaum Associates, Publishers; Mahwah, New Jersey: [Google Scholar]
- Supakar PC, Jung MH, Song CS, Chatterjee B, Roy AK. Nuclear factor κB functions as a negative regulator for the rat androgen receptor gene and NF-κB activity increases during the age-dependent desensitization of the liver. J Biol Chem. 1995;270:837–842. doi: 10.1074/jbc.270.2.837. [DOI] [PubMed] [Google Scholar]
- Tedgui A, Mallat Z. Cytokines in atherosclerosis: Pathogenic and regulatory pathways. Physiol Rev. 2006;86:515–581. doi: 10.1152/physrev.00024.2005. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Nossoya N, Yager T, Tsuang MM, Guo SC, Shyu KG, Glatt SJ, Liew CC. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Vellanoweth RL, Supakar PC, Roy AK. Biology of disease: Transcription factors in development, growth, and aging. Lab Invest. 1994;70:784–799. [PubMed] [Google Scholar]
- Wang A, Gehan EA. Gene selection for microarray data analysis using principal component analysis. Statist Med. 2005;24:2069–2087. doi: 10.1002/sim.2082. [DOI] [PubMed] [Google Scholar]
- Wang X, Dalkic E, Wu M, Chan C. Gene module level analysis: Identification to networks and dynamics. Curr Op Biotechnol. 2008;19:482–491. doi: 10.1016/j.copbio.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fang Y, Wang S. Clustering and principal-components approach based on heritability for mapping multiple gene expressions. BMC Proc. 2007;1:S121. doi: 10.1186/1753-6561-1-s1-s121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T. Regulatory networks: Linking microarray data to systems biology. Mech Ageing Dev. 2007;128:168–172. doi: 10.1016/j.mad.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Xanthoulea S, Curfs DMJ, Hofker MH, de Winther MPJ. Nuclear factor kappaB signaling in macrophage function and atherogenesis. Curr Opin Lipidol. 2005;16:536–542. doi: 10.1097/01.mol.0000180167.15820.ae. [DOI] [PubMed] [Google Scholar]
- Xia K, Xue H, Dong D, Zhu S, Wang J, Zhang Q, Hou L, Chen H, Tao R, Huang Z, Fu Z, Chen YG, Han JDJ. Identification of the proliferation/differentiation switch in the cellular network of multicellular organisms. PLoS Comput Biol. 2006;2:e145. doi: 10.1371/journal.pcbi.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Rabson AB, Young W, Qing G, Qu Z. Alternative pathways of NF-κB activation: A double-edged sword in health and disease. Cytokine Growth Factor Rev. 2006;17:281–293. doi: 10.1016/j.cytogfr.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Xiao W. Advances in NF-κB signaling transduction and transcription. Cell Mol Immunol. 2004;1:425–435. [PubMed] [Google Scholar]
- Xue H, Xian B, Dong D, Xia K, Zhu S, Zhang Z, Hou L, Zhang Q, Zhang Y, Han J-DJ. A modular network model of aging. Mol Sys Biol. 2007;3(147) doi: 10.1038/msb4100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BP, Chung HY. The inflammatory process in aging. Rev Clin Gerontol. 2006;16:179–187. [Google Scholar]
- Zhan M. Deciphering modular and dynamic behaviors of transcriptional networks. Genomic Med. 2007;2007:19–28. doi: 10.1007/s11568-007-9004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]