Abstract
Ectopic accumulation of adipose in the skeletal muscle is associated with muscle wasting, insulin resistance and diabetes. However, the developmental origin of postnatal intramuscular adipose and its interaction with muscle tissue are unclear. We report here that compared to the fast EDL muscles, slow SOL muscles are more enriched with adipogenic progenitors and have higher propensity to form adipose. Using Cre/LoxP mediated lineage tracing in mice, we show that intramuscular adipose in both EDL and SOL muscles is exclusively derived from a Pax3− non-myogenic lineage. In contrast, inter-scapular brown adipose is derived from the Pax3+ lineage. To dissect the interaction between adipose and skeletal muscle tissues, we used Myf5-Cre and aP2-Cre mice in combination with ROSA26-iDTR mice to genetically ablate myogenic and adipogenic cell lineages, respectively. Whereas ablation of the myogenic cell lineage facilitated adipogenic differentiation, ablation of the adipogenic cell lineage surprisingly impaired the regeneration of acutely injured skeletal muscles. These results reveal striking heterogeneity of tissue-specific adipose and a previously unappreciated role of intramuscular adipose in skeletal muscle regeneration.
Keywords: White adipose tissue (WAT), brown adipose tissue (BAT), tissue stem cells, satellite cells, stem cell lineage, tissue regeneration
Introduction
Obesity due to metabolic dysfunction and excessive fat (adipose) accumulation has become a global health concern. Increased body fat content further leads to reduced insulin sensitivity and diabetic mellitus. Among the many fat depots, one that is not well appreciated is the fat in skeletal muscles(Vettor et al., 2009). It is estimated that muscle fat forms a depot in size equivalent to the visceral fat and its increases correlate to muscle insulin resistance(Gallagher et al., 2005). The muscle fat includes acellular lipid droplets within muscle cells (myofibers) known as intramyocellular triacylglycerides (IMTG) and adipocytes distributed in the muscle interstitium or surrounding muscle fascicles that are referred to as intramuscular adipose tissue (IMAT)(Vettor et al., 2009). Whereas IMTG can act as an energy source besides glucose to fuel muscle contraction and its content may or may not be linked to metabolic syndromes, increased IMAT has been shown to be directly linked to insulin resistance and diabetes(Schrauwen-Hinderling et al., 2006; Yim et al., 2007). Currently, little is known about the origin, or progenitor cell lineage that contributes to the development, of IMAT.
Increased muscle fat content is commonly associated with muscle wasting conditions such as ageing-related sarcopenia and disuse-related muscle atrophy. In these conditions, a loss of muscle tissue is always accompanied by fat and connective tissue infiltration, a condition known as myosteatosis (Taaffe et al., 2009). Muscular atrophy found with ageing and disuse is characterized by a transition in muscle contractile protein from slow to fast myosin heavy chain isoforms, accompanied by metabolic changes including a fuel transition toward glycolysis, decreased capacity for fat oxidation and energy substrate accumulation in the atrophied muscles (Stein and Wade, 2005). Similar irreversible fatty infiltration, impaired capacity of mitochondria to oxidize fatty acids and increased synthesis of fatty acids occur under disease conditions such as Duchenne Muscular Dystrophy (Lin et al., 1969; Petrof et al., 1993). These changes may lead to accumulation of fatty acids in the muscle interstitial preadipocytes and expansion of IMAT in dystrophic and atrophic muscles.
Adipose infiltration into skeletal muscle is also common if myogenic development or regeneration is impaired. Pax7 mutant muscles fail to regenerate due to the critical role of Pax7 in muscle stem cell (satellite cell) function (Kuang et al., 2006; Lepper et al., 2009). Instead, injured and degenerated Pax7 mutant muscle is largely replaced by fibrous and adipose tissues(Kuang et al., 2006). Likewise, adipose tissue infiltration into skeletal muscle is documented in Myogenin, Myf5, MyoD knockouts or Myf5/MyoD compound mutants that are characterized by defective differentiation or transient amplification of satellite cells(Braun et al., 1992; Gayraud-Morel et al., 2007; Hasty et al., 1993; Nabeshima et al., 1993; Rudnicki et al., 1992; Rudnicki et al., 1993). These observations lead to two assumptions: 1) Satellite cells may have taken an alternative differentiation path to contribute to IMAT upon blockage of their default myogenic pathway; 2) The infiltrated adipose may have further inhibited muscle differentiation. The first assumption seems to be supported by in vitro experiments showing that satellite cells are capable of multilineage, including myogenic, adipogenic and osteogenic differentiation(Asakura et al., 2001; Csete et al., 2001; Shefer et al., 2004).
However, recent studies demonstrate that adipocytes in the skeletal muscle may be derived from a mesenchymal progenitor population distinct from satellite cells. MyoD-Cre lineage tracing indicates that adipocytes derived from skeletal muscle cultures are from a MyoD− lineage(Starkey et al., 2011). Using Fluorescence Activated Cell Sorting technique, Uezumi et al. isolated non-myogenic PDGFRα+ mesenchymal progenitors from skeletal muscles and demonstrated that they efficiently differentiate into adipocytes in vitro and in vivo upon transplantation into injured muscles(Uezumi et al., 2010). Meanwhile, Joe et al. independently sorted CD34+/Sca1+ fibro/adipogenic progenitors (FAPs) from muscle tissue and showed that they do not have myogenic potential but are able to facilitate myogenesis in co-cultures (Joe et al., 2010). Furthermore, Myf5-Cre mediated lineage tracing indicate that FAPs are derived from a Myf5− lineage (Joe et al., 2010). Similarly, muscle resident progenitors that are capable of differentiating into brown adipocytes upon BMP7 treatment are shown to be derived from a Myf5− lineage (Schulz et al., 2011). These results suggest that a muscle resident Myf5− progenitor population is capable of adipogenic differentiation but do not rule out the possibility that Myf5+ lineage satellite cells can also adopt an adipogenic cell fate in vivo when local and systemic environment changes due to injury or ageing. It also remains unclear whether Myf5− satellite cells, accounting for about 10% of all Pax7+ satellite cells(Kuang et al., 2007), represent the muscle resident adipogenic progenitors.
Another unresolved question is whether there are distinct populations of progenitors in the muscle that give rise to white adipose tissue (WAT) and brown adipose tissue (BAT), respectively. The Myf5-independency of muscle resident adipogenic progenitors indicates that they are white preadipocyte, since BAT is derived from a Myf5+ lineage(Seale et al., 2008). Yet BAT is abundantly distributed in the skeletal muscle of the Sv129 strain mouse, and brown adipocyte progenitors can be purified from human skeletal muscles(Almind et al., 2007; Crisan et al., 2008). Therefore it is important to characterize whether IMAT represent brown or white adipose.
In this study, we first examined the abundance of adipogenic progenitors and the adipogenic potential of different muscles enriched with either oxidative slow fibers or glycolytic fast fibers. We then carried out genetic lineage tracing using Pax3-Cre allele to dissect the cell lineage origin of IMAT, and compared it to that of the BAT. Finally, we dissected the interactions between adipogenic and myogenic processes by selective ablation of one or the other cell lineages. Understanding the progenitor cell lineage origin of intramuscular adipose and their interaction with skeletal muscle tissues may lead to novel strategies to prevent or alleviate muscle wasting, insulin resistance and obesity.
Materials and Methods
Animals
All procedures on animals were performed in accordance with Purdue University’s Animal Care and Use Committee. Mice were housed in the animal facility with free access to water and standard rodent chow. The mice were harvested around 2-month-old unless specified. The Myf5-Cre mice were provided by Dr. Philip Soriano (Mount Sinai School of Medicine)(Tallquist et al., 2000). Other mice were from Jackson Laboratory mice (Pax3-Cre stock #005549, aP2-Cre stock #005069, mTmG stock #007576, ROSA26-iDTR stock #007900) and PCR genotyping was done using protocols described by the supplier.
Culture of skeletal muscle derived primary cells
EDL and SOL muscles were collected, minced, and digested in 3-5 volumes of collagenase/dispase (Roche, 10ug/ml collagenase B, 2.4U/ml dispase in PBS) for 24 minutes. The digestion was stopped by Dulbecco’s Modified Eagle Medium (DMEM) with 2%FBS and 1% Hepes, and was spun down at 450x g for 3 minutes. The pellet was resuspended in Ham’s complete medium containing 20% FBS, 2% penicillin/streptomycin (P/S) and 4ng/ml bFGF in Ham’s F-10 medium and cultured in collagen-coated plates at 37°C with 5% CO2. Debris and non-adherent cells were removed by medium change at day 3 and cells were fed with fresh medium every 2 days. Upon confluence, cells were differentiated in myogenic differentiation medium containing DMEM with 5% FBS and P/S, or differentiated in adipogenic medium containing 200nM insulin (20mg/ml in acetic acid, PH2-3, Sigma) and 10nM Triiodothyronine (T3, Sigma).
BAT primary adipocyte cultures
Interscapular BAT depots were collected and trimmed free of WAT, minced and digested with BAT isolation buffer for 30 minutes at 37°C with on a shaker. The isolation buffer contains 123mM NaCl, 5mM KCl, 1.3mM CaCl2, 5mM Glucose, 100mM Hepes, 4% BSA, 1%P/S and 1.5mg/ml Collagenase I. The digestion was stopped with DMEM containing 2%FBS and 1% Hepes, filtered through 100μm filters, and cells were pelleted at 450x g for 5 minutes. The cells were cultured in growth medium containing DMEM, 20% FBS, 2% Hepes and 1% P/S at 37°C with 5% CO2 for 3 days, and then fresh media was changed every two days. Upon confluence, cells were exposed to induction medium for 4 days and then differentiation medium for several days until adipocytes mature. The induction medium contains DMEM, 10% FBS, 2.85μM insulin, 0.3μM dexamethasone (DEXA) and 0.63mM 3-isobutyl-1-methylxanthine (IBMX) (Sigma), and the differentiation medium contains DMEM, 10% FBS, 200nM insulin and 10nM T3.
Fluorescent activated cell sorting (FACS)
Primary myoblasts were cultured from the hindlimb muscles of aP2-Cre/ROSA26-mTmG mice. Early passage (P3-P5) myoblasts were harvested by trypsin (0.25% W/V) and resuspended with Ham’s complete medium into 5×106 cells/ml. Cells were filtered through 50μm sterile nylon mesh right before the sorting. Cells are selected on the basis of fluorescence characteristics. Myf5-Cre/mTmG myoblasts were used as a control for gating GFP+ cells and mTmG myoblasts were used as a control for gating RFP+ cells. After sorting, cells were collected in Ham’s medium and cultured in CO2 incubator at 37 °C. Cells were differentiated in differentiation medium containing DMEM and 5% HS for 6 days before analysis.
Gene expression analysis
Quantitative Realtime polymerase chain reaction (qPCR) analysis was performed using a Roche Lightcycler 480. RNA was extracted and purified from muscle tissue or different cell cultures with NucleoSpin RNA XS kit (MN) and converted into cDNA using MMLV reverse transcriptase and random hexamer primers. The primers used in the qPCR analysis are listed in Table S1. Each PCR reaction contains 0.2μl of each primer (20μM), 4μl cDNA (2.5ng/μl), 5μl SBBR Green Mastermix (Roche) and 0.6μl H2O. qPCRs were run for 40 cycles and the fold changes for genes were calculated by 2−ΔΔct method using RPLP38 gene as housekeeping control.
Oil Red O staining and image analysis
Cultured cells were rinsed with PBS and fixed with 10% formaldehyde for 15 min at room temperature. Tissue sections were fixed with 3.7% formaldehyde for 1 hour at room temperature and washed out with deionized water. Cells and tissue sections were stained in working solution for 30-60 min at room temperature. The working solution, freshly made and filtered prior to staining, contained 6ml Oil Red O stock (5g/L in isopropanol, Sigma) and 4ml ddH20 for cultured cells; or 12ml Oil Red O stock (5g/L in 60% triethylphosphate) and 8ml ddH20 for tissue sections. After staining, plates and slides were washed with distilled water for 10 minutes and mounted in glycerin mounting medium. The signal density of Oil Red O staining was quantified with Image-J software. Color images were first converted to gray scales and threshold was adjusted consistently to remove background coloration. The Area, Mean Gray Value and Integrated Density were measured and calculated for relative signal intensity.
Cardiotoxin (CTX) and diphtheria toxin (DT) injections
Muscle regeneration was induced by injecting 50μL CTX (10μM, Sigma) into the mid-belly of TA muscles. DT powder (Calbiochem) was dissolved in PBS to make 1mg/ml stock solution. 10 μl DT working solutions (20μg/ml in saline) were mixed with CTX before injecting into TA muscles. Muscles were harvested at different time points (3-15 days) after injection and fixed in 4% paraformaldehyde, sunk in 30% sucrose and embedded in OCT compound (Sakura Finetek) and quickly frozen in dry ice cooled isopentane. Muscles were cut into 10μm thickness using a Leica CM 1850 cryostat and the sections were placed on superfrost plus glass slides. For cell ablation in culture, a final DT concentration of 200ng/ml was added to culture medium for 48 hours.
Immunostaining and image capture
Fixed cells or tissue sections were blocked for 30 minutes in blocking buffer containing 5% horse serum, 2% BSA, 0.2% triton X-100 and 0.1% sodium azide in PBS. Samples were then incubated with primary antibodies diluted in blocking buffer for 1 hour at room temperature, followed incubating with secondary antibodies and Hoechst for 30 minutes at room temperature, and mounted with Dako fluorescent mounting media (Glostrup, Denmark). Fluorescent images were captured with a Coolsnap HQ CCD camera (Photometrics, USA) driven by IP Lab software (Scanalytics Inc, USA) using Leica DMI 6000B fluorescent microscope (Mannheim, Germany).
Hematoxylin and Eosin (H&E) staining
Cryosections from fresh-frozen tissues were washed with PBS for 3 times and incubated in hematoxylin for 5 minutes, rinsed with deionized water followed by washing with tap water for at least 5 minutes to allow the stain to develop. Slides were then incubated in eosin for 45 seconds. The staining was dehydrated sequentially with 75%, 95% and 100% ethanol for 2-5 minutes each. Slides were finally placed in xylene for 10 minutes and mounted with cytoseal.
Statistical analysis
Data presented in this study were mean ± standard error of the mean. Numbers of repeats represent biological repeats unless indicated. P-values were calculated using two-tailed student’s t-test.
Results
The fast EDL and slow SOL muscles have different adipogenic potentials
Fast and slow muscles have distinct insulin sensitivity and respond differentially to muscle wasting (Lillioja et al., 1987). We thus investigated the adipogenic potentials of different muscles known to be primarily composed of fast or slow fiber types. The extensor digitorum longus (EDL) and soleus (SOL) were chosen as the EDL contains 80% type IIx and IIb (glycolytic fibers) and the SOL has 95% type I and IIa (oxidative) fibers (Waddell et al., 2010). Such dramatic differences in fiber type composition and metabolic properties make them ideal representatives for investigating fiber type specific regulation of adipogenesis.
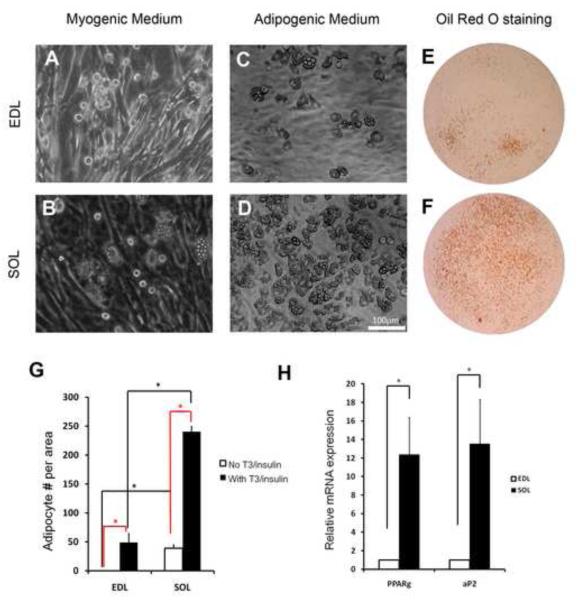
Primary cells from equal amount of EDL and SOL muscles were induced to differentiate under myogenic culture conditions. Within 3-4 days, differentiated myotubes were readily detectable in both cultures (Fig. 1A-B). Interestingly, multilocular adipocytes appeared in SOL-, but not EDL-, cultures at 6-10 days after induction of differentiation (Fig. 1A-B). Addition of adipogenic inducing agents, insulin and T3, greatly enhanced the efficiency of adipogenesis and resulted in adipocyte formation in EDL culture as well (Fig. 1C-D). Oil red O staining demonstrated that the multilocular cells seen in the culture are lipid-filled adipocytes, and the signal is more robust in SOL-compared to EDL-derived cell cultures (Fig. 1E-F). Quantitative analysis confirmed that there are more adipocytes in the SOL than EDL cultures, with or without Insulin/T3 induction (Fig. 1G). Consistent to the marked differences in cell numbers, the relative mRNA levels of the adipocyte-specific Pparγ and aP2 genes were more than 10-fold higher in the SOL than those in the EDL cultures (Fig. 1H). Together, these results demonstrate the existence of IMAT progenitors capable of adipogenic differentiation in both fast and slow muscles, and reveal distinct intrinsic adipogenic capacity in the fast EDL and slow SOL muscles.
Figure 1.
- A-B: EDL (A) and SOL (B) muscle derived primary cells differentiated under myogenic culture conditions. Darker tubular cells are myotubes and multilocular cells (in B) are adipocytes.
- C-D: EDL (C) and SOL (D) muscle derived primary cells differentiated under adipogenic culture conditions (with insulin and T3 induction).
- E-F: Oil Red O staining showing lipid-filled cells in EDL (E) and SOL (F) muscle derived primary cultures differentiated under adipogenic medium.
- G: Relative numbers of adipocytes formed in EDL and EDL muscle derived primary cultures with or without induction by insulin and T3. Asterisks denote p < 0.05 (t-test). Error bars represent standard error of the mean, n=3.
- H: Relative level of adipogenic gene expressions in EDL and SOL muscle derived primary cultures upon differentiation in adipogenic medium, n=4.
Different abundance and properties of IMAT progenitors residing in the slow SOL and fast EDL muscles
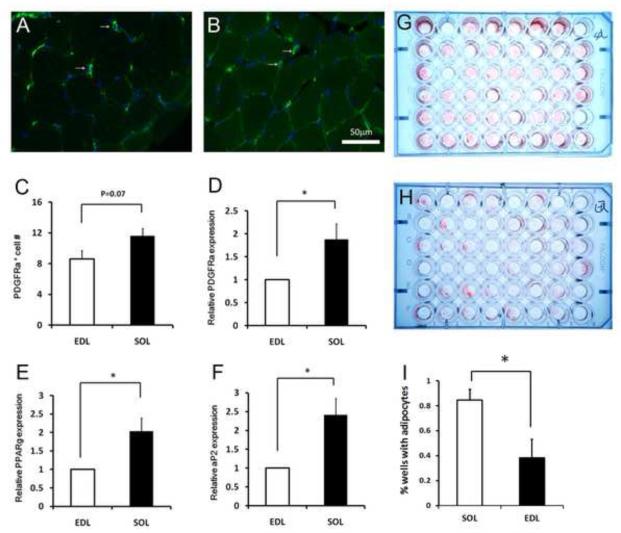
The differential adipogenic potentials of EDL versus SOL muscles prompted us to investigate if they are due to intrinsic difference in progenitor cell properties or extrinsic regulation of the progenitors. EDL and SOL conditioned media and co-culture experiments (Figs. S1&S2) suggest that muscle specific extrinsic factors do not play a major role in the adipogenic potential of IMAT progenitors. We therefore sought to determine the intrinsic difference (such as relative abundance) of progenitor cells in EDL and SOL muscles. We assessed the relative abundance of IMAT progenitors in these muscles using several techniques. First, we labeled EDL and SOL sections with an antibody to platelet-derived growth factor receptor alpha (PDGFRα) that labels a population of muscle resident adipogenic progenitors (Uezumi et al., 2010). PDGFRα+ cells can be identified in both EDL (Fig. 2A) and SOL (Fig. 2B) muscle interstitium. The PDGFRα+ cell number per unit square area was higher in the SOL compared to EDL muscles (Fig. 2C). This observation is further supported by qPCR results showing that PDGFRα mRNA was expressed more abundantly in SOL-than EDL-derived adherent cells (Fig. 2D). It is important to mention, however, that not all PDGFRα positive cells are preadipocytes (Hu et al., 1998). Thus, we examined the expression of two additional adipogenic markers, Pparγ and aP2, known to be expressed by preadipocytes (Tang et al., 2008; Tchoukalova et al., 2004). Both Pparγ and aP2 mRNAs were about 2 times more abundant in the SOL than EDL muscles (Fig. 2E-F). Furthermore, we conducted limiting dilution culture analysis of all cells derived from a single EDL or SOL muscle, and estimated that 1:100 dilution reliably gave rise to adipocyte colonies in 20-90% of the cultures (Fig. 2G-H). Under this dilution factor, adipocyte colonies were found in 85% of the SOL cultures, but only in less than 40% of the cultures (Fig. 2I; p<0.05, n=3). Together, these data strongly suggest that a higher abundance (roughly 2 times) of IMAT progenitors resides in the slow SOL than fast EDL muscles.
Figure 2.
- A-B: Representative images of PDGFRα immunofluorescence (labeled in Green as a marker for adipogenic progenitors) in EDL (A) and SOL (B) muscle sections. Nuclei are counterstained with DAPI.
- C: PDGFRα+ cells/unit area in EDL and SOL muscles. Error bars represent SEM, n=5. P value is based on two tail t-test.
- D: Relative levels of PDGFRα mRNA expression in EDL (normalized to 1) and SOL muscle derived adherent cells (never been passaged). Error bars represent SEM, n=4.
- E: Relative levels of Pparγ mRNA expression in EDL (normalized to 1) and SOL muscles. Error bars represent SEM, n=4.
- F: Relative levels of aP2 mRNA expression in EDL (normalized to 1) and SOL muscles. Error bars represent SEM, n=4.
- G-H: Limiting dilution cultures (1:100 in each well) of adipocytes (stained with Oil Red O) differentiated from a SOL (G) and an EDL (H) muscle.
- I: Percentage of wells that exhibited Oil Red O signal calculated from limiting dilution cultures. Error bars represent SEM, n=3.
The estimated two fold difference in progenitor cell number cannot fully explain the 10-fold difference in the Oil Red O signaling and adipogenic gene expression in adipocytes differentiated from SOL and EDL muscles (Fig. 1). Additional intrinsic differences such as in the survival, growth and differentiation must also exist between fast and slow muscle IMAT progenitors. In our limiting dilution culture analysis, the average progenitor cell density is less than one per well at 1:100 dilution (Fig. 2G-H), therefore the differentiated adipocytes in each well should in theory be derived from one progenitor (one clone). Inspection of Oil Red O signaling intensity in individual wells indicate that almost all SOL adipocyte colonies fully covered the whole area of the wells, but most EDL adipocytes only partially covered the wells (Fig. 2G-H). This indicates that the average SOL adipocyte clone size is much bigger than that of EDL colonies, and further suggests that SOL and EDL muscle resident adipogenic progenitors have intrinsically different growth and differentiation properties.
Muscle resident adipogenic progenitors are from a Pax3−/Myf5− lineage and give rise to white adipocytes in culture
To explore the developmental origin of skeletal muscle resident adipogenic progenitors and their relationship to satellite cells, we performed Cre/LoxP mediated genetic cell lineage tracing and lineage ablation. We used Myf5-Cre and Pax3-Cre knockin mice(Engleka et al., 2005; Tallquist et al., 2000) as the lineage driver and Cre-inducible mTmG transgenic mouse (Muzumdar et al., 2007) as the lineage reporter. In the absence of Cre, mTmG mouse ubiquitously expresses membrane-targeted tandem dimer Tomato (mT, a RFP), but switches to express membrane-targeted GFP (mG) in the presence of Cre. Lineage ablation is achieved using ROSA26-iDTR mice that specifically express the diphtheria toxin receptor (DTR) in Cre-induced cell lineages, which can then be ablated by administration of diphtheria toxin (DT).
It has been recently shown during the course of our study that muscle resident Sca-1+CD34+CD31−CD45− fibro/adipogenic progenitors (FAPs) are derived from a Myf5− lineage (Joe et al., 2010). Consistent with this observation, we found that all adipocytes differentiated from Myf5-Cre/mTmG EDL and SOL muscles displayed mT (Fig. S3), suggesting that IMAT is exclusively derived from a non-Myf5 lineage. As Myf5 lineage cells only account for 90% of the satellite cell population, this result however does not exclude the possibility that some IMAT is derived from satellite cells.
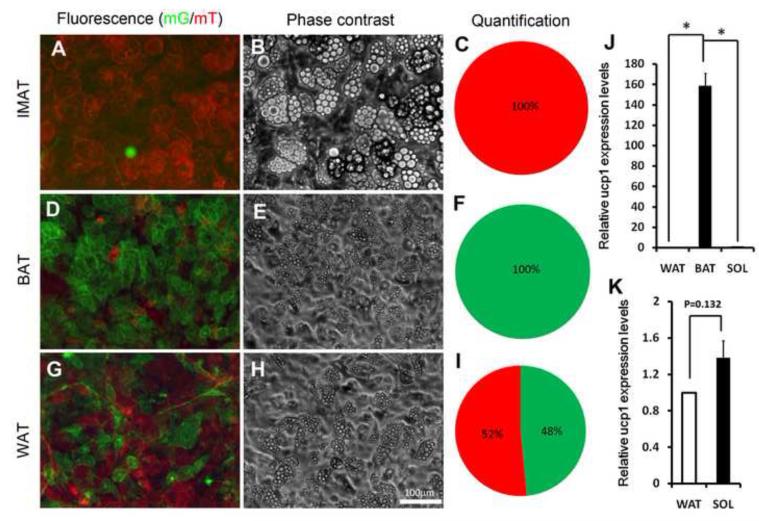
During muscle development, Pax3 is known to activate Myf5 (Bajard et al., 2006; Sato et al., 2010; Tajbakhsh et al., 1997) and all satellite cells have been shown to be derived from the Pax3 lineage(Schienda et al., 2006). We further investigated the Pax3 lineage origin of IMAT using Pax3-Cre/mTmG mice. All skeletal muscle derived adipocytes in culture were found to be mT+, suggesting that they are exclusively derived from a non-Pax3 lineage (Fig. 3A-C). By contrast, all BAT-derived adipocytes were mG+ in culture (Fig. 3D-F), indicating that they originated from the Pax3 lineage. Interestingly, adipocytes formed in cultures of anterior subcutaneous WAT were a mixture of roughly equal proportions of mG+ and mT+ cells (Fig. 3G-I). To determine if the intramuscular adipocytes originate from the Pax3 lineage in vivo, we examined Pax3-Cre/mTmG regenerating muscles (healthy resting muscle has few intramuscular adipocytes). Oil Red O staining revealed massive accumulation of adipose in the TA muscles after CTX induced regeneration, with smaller lipid droplets at day 3 (Fig. S4A) and larger ones at day 7(Fig. S4B). As expected, all muscle fibers were mG+, demonstrating their Pax3 lineage origin. By contrast, none of the Oil Red O labeled cells exhibited mG fluorescence (Fig. S4). These results provide compelling in vitro and in vivo evidence that IMAT is derived from a Pax3− non-satellite cell lineage.
Figure 3.
- A-C: Skeletal muscle derived adipocytes in culture. (A) mT and mG fluorescence, (B) Phase-contrast images, (C) The relative percentage composition of mT and mG positive adipocytes (n=3).
- D-F: BAT derived adipocytes in culture. (D) mT and mG fluorescence, (E) Phase-contrast image, (F) The relative percentage composition of mT and mG positive adipocytes (n=3).
- G-I: Anterior subcutaneous WAT derived adipocytes in culture. (G) mT and mG fluorescence, (H) Phase-contrast image, (I) The relative percentage composition of mT and mG positive adipocytes (n=3).
- J: Relative level of Ucp1 expression in IMAT (SOL), BAT, WAT (normalized to 1) derived adipocytes. Error bars represent SEM, n=2.
- K: Relative level of Ucp1 expression in IMAT (SOL) and WAT derived adipocytes. Error bars represent SEM, n=2.
To determine if muscle-derived adipocytes are brown or white adipocytes, we conducted qPCR to examine brown adipose-specific UCP1 gene expression. The UCP1 expression in BAT cultures were about 160-fold higher than those expressed in the muscle- or WAT-derived cultures (Fig. 3J). While no significant difference in UCP1 expression was found in the muscle- and WAT-derived adipocytes (Fig. 3K). We conclude that muscle-derived adipocytes in culture are white rather than brown adipocytes.
Ablation of myogenic cell lineage facilitates adipogenic differentiation
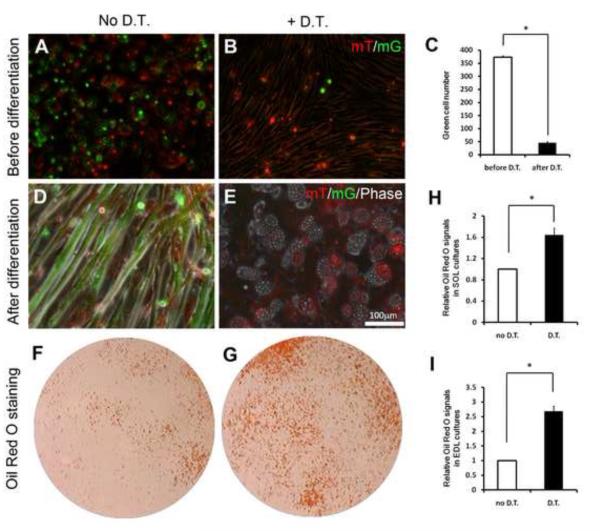
The coexistence of adipogenic and myogenic progenitors in the skeletal muscle suggests that they may interact with each other to maintain tissue homeostasis. We used genetic cell ablation techniques to dissect such interactions. To achieve this, Myf5-Cre/ROSA26-iDTR and aP2-Cre/ROSA26-iDTR mice were established for ablation of muscle resident myogenic and adipogenic cells, respectively. We further created Myf5-Cre/ROSA26-iDTR/mTmG mice in order to monitor the efficiency of cell ablation. Skeletal muscle derived cells under growth conditions contained both mG+ (myogenic) and mT+ (adipogenic and fibroblastic) cells (Fig. 4A). DT treatment nearly eliminated all the mG+cells (Fig. 4B-C), the few remaining mG+ cells were mostly floating and unable to fuse into myotubes under differentiation conditions. In contrast, non-DT treated mG+ cells readily fused into myotubes upon induced differentiation (Fig. 4D); whereas DT treatment efficiently eliminated these mG+ cells resulting in only mT+ adpocytes (Fig. 4E). These data demonstrate that the iDTR mouse model provides an effective tool for lineage-specific cell ablation.
Figure 4.
- A-B: Skeletal muscle derived cells under growth conditions (before differentiation). (A) mT and mG fluorescence can be detected without DT treatment, (B) DT treatment selectively ablates mG positive Myf5-lineage cells.
- C-D: Skeletal muscle derived cells after differentiation in adipogenic medium. (C) Myotubes are mG positive and non-myogenic interstiatial cells are mT positive prior to DT treatment, (D) DT treatment selectively ablates mG positive myotubes but not mT positive adipocytes.
- E-F: Representative Oil Red O staining of skeletal muscle derived adipocytes in culture with (F) or without (E) DT treatment.
- G: Efficiency of DT mediated cell ablation indicated by quantification of mG positive cells before and after DT treatment. Error bars represent SEM, n=3.
- H-I: Relative Oil Red O signal intensity of SOL (H) and EDL (I) muscle derived primary cultures before and after DT mediated ablation of myogenic cells. Error bars represent SEM, n=3.
Notably, DT induced ablation of myogenic Myf5-lineage robustly increased the number of adipocytes in SOL cultures (Fig. 4F-G), as indicated by the significant increase in Oil Red O signal intensity (Fig. 4H). Similar enhancement of adipogenesis was observed in EDL cultures after ablation of myogenic cells (Fig. 4I). These results suggest that myogenesis would normally inhibit adipogenesis in the skeletal muscle and failures of myogenesis, commonly seen in muscular dystrophy and atrophy, lead to enhanced adipogenesis and fat infiltration.
aP2 lineage interstitial cells rapidly increase in regenerating muscles but rarely directly fuse into myofibers
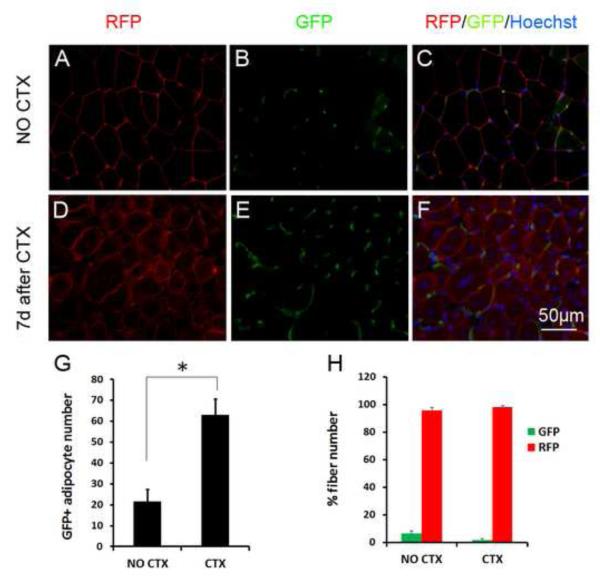
To investigate if adipogenic cell lineage plays a role in skeletal muscle homeostasis, we first examined the dynamics of aP2 lineage cells in both resting and regenerating muscles, as well in muscle cell cultures. We examined aP2-Cre/mTmG muscle sections and found that mG is predominantly expressed by muscle interstitial cells (Fig. S5A-B; Fig. 5A-C). Importantly, the number of aP2 labeled (mG+) interstitial cells increased rapidly during muscle regeneration (Fig. 5D-F). The relative number of mG+ cells at day 7 after CTX induced injury was about 3 times of that in non-injured muscle (Fig. 5G). Interestingly, we found that about 8% of myofibers also express mG+ in non-CTX treated resting muscles, and this frequency dropped to below 5% at day 7 regenerating muscles (Fig. 5H). As the mG+ interstitial cells increase during regeneration (Fig. 5G), a reduction in mG+ regenerating fibers suggest that the mG+ cells rarely fuse into regenerating myotubes.
Figure 5.
- A-C: GFP and RFP expression in non-CTX treated resting TA muscle section. Nuclei are counterstained by DAPI. Myofibers are the large red round cells.
- D-F: Regenerating muscle at 7 day after CTX treatment, showing increased abundance of interstitial GFP+ cells.
- G: GFP+ interstitial number per unit area in non-CTX and CTX treated muscles, n =4
- H: Percentage of myofibers that are labeled with GFP or RFP in non-CTX and CTX treated muscles, n=4. * P<0.05, ** P<0.01.
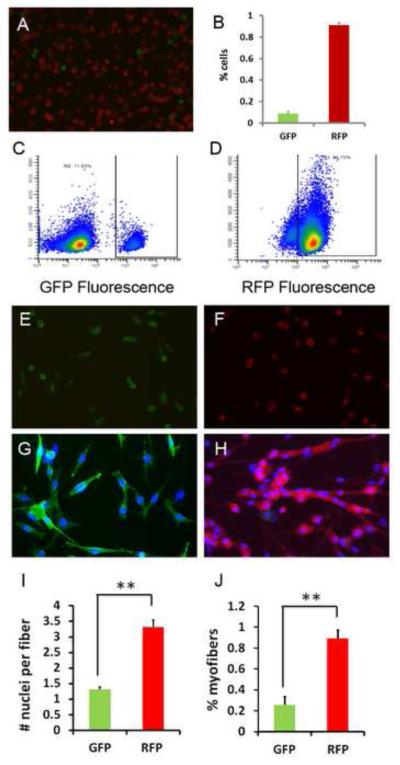
The above results suggest a low frequency of aP2 activation in satellite cells or post differentiation myotubes. Indeed, cultures of satellite cell derived primary myoblasts from aP2-Cre/mTmG mice contained a small fraction of mG+ cells (Fig. 6A). FACS analysis indicates that there are roughly 10% of mG+ and 90% mT+ cells (Fig. 6B-D); this ratio of myoblast composition is in relative consistency with the ratio of mG+ to mT+ muscle fibers in vivo. We further sorted the mG+ and mT+ cells by FACS and placed them in culture (Fig. 6E-F). Proliferating mG+ and mT+ cells both had round myoblastic shape and similar size (Fig. 6E-F). When induced to differentiate, the mT+ cells robustly formed mutinuclear myotubes, whereas the mG+ cells mostly exhibited an elongated shape but barely fused into multinuclear cells (Fig. 6G-H). At day 4 after induced to differentiate, each mG+ cell contains 1.3 ± 0.1 nuclei, compared to 3.3 ± 0.2 nuclei per mT+ cell (Fig. 6I). Using a different criterion, only 25% of the mG+ nuclei were found to have fused into multinuclear cells, compared to 90% of mT+ nuclei that have fused into multinuclear cells (Fig. 6J). These results indicate that although 10% of myoblast-like cells have activated aP2 gene expression, these cells don’t fuse into myotubes efficiently.
Figure 6.
- A: Early passage (p3) myoblasts derived from aP2-Cre/mTmG muscle and enriched by preplating to removed fibroblastic cells. Notice that most myoblasts are RFP+ (non-aP2 lineage), with only few GFP+ (aP2 lineage) cells.
- B-D: Percentage composition of aP2 and non-aP2 lineage cells (B) determined by fluorescence activated cell sorting (FACS) based on GFP (C) and RFP (D) fluorescence.
- E-F: FACS sorted mG+ (E) and mT+ (F) cells under growth conditions.
- G-H: FACS sorted mG+ (G) and mT+ (H) cells 4 days after induced to differentiate.
- I-J: Extremely poor fusion ability of aP2 lineage cells (GFP) compared to non-aP2 lineage cells (RFP), based on the average number of nuclei per cell (I), or the percentage of nuclei that have been fused (J) at day 4 after induced differentiation, n=3. * P<0.05, ** P<0.01.
Ablation of aP2+ adipogenic cell lineage inhibits muscle regeneration
To directly examine the role of adipogenic cells in skeletal muscle homeostasis, we generated aP2-Cre/ROSA26-iDTR mice (called aP2-iDTR), which allowed us to ablate aP2 cell lineage by DT administration. As the aP2 lineage cells are predominantly non-myogenic (Fig. 6), this cell ablation strategy should preserve the myogenic cell lineage. IP injection of DT (200 ng/25g mouse) had no effect on wildtype, aP2-Cre or ROSA26-iDTR mice, but killed all aP2-iDTR mice (data not shown), presumably due to ablation of some neuronal cells and macrophages that are known to express aP2 (Furuhashi et al., 2008; Urs et al., 2006). The leaky expression of aP2 outside the adipogenic lineage represents a caveat of the aP2-Cre model. We therefore focally injected 20 ng of DT into the middle belly of TA muscles of aP2-iDTR mice to circumvent lethality. Compared to CTX only injection, DT+CTX injection of TA muscles resulted in robust reduction of PDGFRa+ cells at 2 days post injury (dpi) and 5 dpi (Fig. S6A-D). Quantitative Realtime PCR analysis indicates that transcripts of Dlk1 and Pdfrα, markers of preadipocytes, were reduced by over 80% at 2dpi (Fig. S6E) and by 40% - 80% at 5dpi (Fig. S6F), and returned to normal at 10 dpi (Fig. S6G). These results demonstrate the efficient ablation of adipogenic cells within 5 days after DT treatment. The later recovery of Dlk1 and Pdfrα expression at 10 dpi suggests a replenishment of adipogenic cells by progenitors from non-injured region or from the circulation.
Whereas CTX injection into aP2-iDTR TA muscles resulted in widespread regeneration, DT alone injection caused little or no damage to myofibers with only few visible regenerating myofibers at 15 dpi (Fig. 7A-F) that likely represent the 10% aP2 activated myofibers (Fig. S5A-B). The only noticeable changes in muscle morphology after DT injection are slightly increased interstitial space at 5 and 10 dpi (Fig. 7B, D), presumably due to ablation of interstitial aP2 lineage cells, which returned to normal at 15 dpi (Fig. 7F). These results suggest that adipogenic lineage cells are not essential for short-term muscle maintenance. However, their long-term effect on muscle homeostasis is still unclear but can be assessed after repeated bouts of DT injection to prevent replenishment of the adipogenic cells.
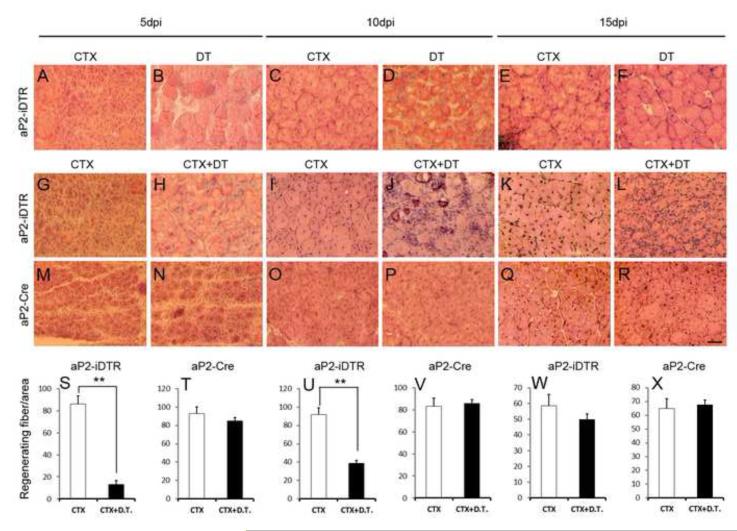
Figure 7.
- A-F: aP2-iDTR TA muscles contralaterally injected with CTX or DT, and harvested at different time points for H&E staining. Notice widespread muscle regeneration (characterized by central nuclei in regenerating muscle fibers) in CTX but not DT treated muscles.
- G-L: aP2-iDTR TA muscles contralaterally injected with CTX or CTX+DT, and harvested at different time points for H&E staining. CTX+DT treatment impaired muscle regeneration demonstrated by the lack of regenerating fibers with central nuclei and excessive accumulation of interstitial nuclei.
- M-R: aP2-Cre TA muscles treated identically as described in G-L. Muscle regeneration is morphologically indistinguishable between the CTX and CTX+DT treatment.
- S-X: Number of regenerating fibers per unit area showing reduced fiber number in CTX+DT treated aP2-iDTR muscles but not in the control aP2-Cre muscles, n=3. * P<0.05, ** P<0.01.
Next, we injected CTX alone or CTX+DT into the TA muscles of aP2-iDTR mice and inspected muscle morphology based on H&E staining of muscle sections harvested at 5, 10 and 15 dpi (Fig. 7G-L). Strikingly, the muscle regeneration was severely inhibited in the aP2-iDTR mice whose adipogenic cells were ablated by DT. Specifically, CTX+DT injected aP2-iDTR muscles poorly regenerated as compared to CTX alone injection at all three time points examined (Fig. 7G-L). In sharp contrast, the control aP2-Cre or ROSA26-iDTR (data not shown) skeletal muscles were regenerated equally well whether they were injured by CTX alone or CTX+DT injection (Fig. 7M-R). Enumerating newly regenerated myofibers with central nuclei confirms the regenerative defect caused by the ablation of aP2 cell lineage (Fig. 7S-X). In the control aP2-Cre mice, the regenerating myofiber number per unit area muscle was not affected by CTX alone or CTX+DT treatment at all time points (Fig. 7T, V, X). By contrast, the number of regenerating myofibers was dramatically reduced by DT in the aP2-iDTR mice at 5 and 10 dpi (Fig. 7S, U). Although the number of regenerating myofiber was only slightly reduced at 15 dpi (Fig. 7W), these myofibers were obviously smaller in the CTX+DT compared to CTX only treatment (Fig. 7K, L). Furthermore, substantial interstitial cell nuclei infiltration was apparent in the CTX+DT treated TA muscles at 10 and 15 dpi (Fig. 7J, L). These observations indicate elevated inflammatory responses in regenerating muscles devoid of aP2 lineage cells. Immunohistochemistry labeling revealed similar defective muscle regeneration upon ablation of aP2 lineage cells (Fig. S5C-E). These data show that aP2+ adipogenic lineage cells play a positive role in mediating skeletal muscle regeneration upon acute injuries.
Reduced myoblast differentiation after ablation of aP2 lineage adipogenic cells
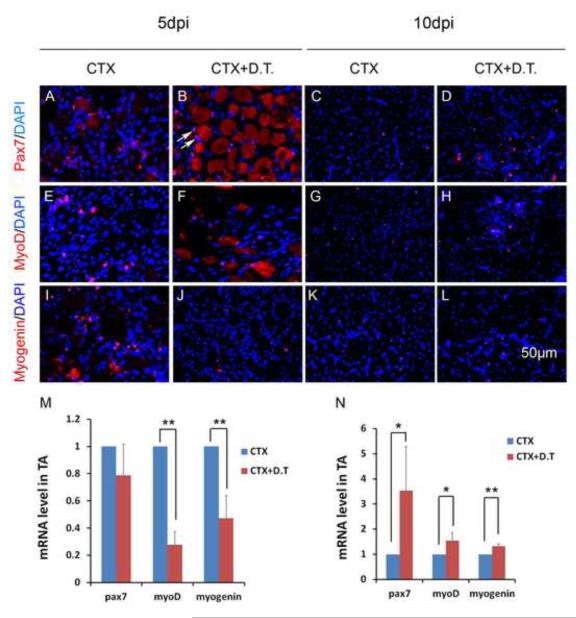
To assess d the effect of aP2 cell lineage ablation on satellite cells, we examined Pax7 (a myogenic progenitor marker), MyoD and Myogenin (markers for early and late myogenic differentiation) expression in regenerating aP2-iDTR muscles with or without DT treatment. Pax7 immunofluorescence signal was similar in both CTX and CTX+DT treated TA sections at 5 dpi (Fig. 8A-B), but an increased Pax7+ cells were found in CTX+DT than CTX treated sections at 10 dpi (Fig. 8C-D). In contrast, there was a decrease in MyoD+ and Myogenin+ cells in CTX+DT treated sections at 5 dpi, but not at 10 dpi (Fig. 8E-L). Quantitative PCR analysis indicate that Pax7 transcripts were not changed, whereas MyoD and Myogenin were both decreased by more than 50% in CTX+DT treated muscles at 5 dpi (Fig. 8M). Interestingly, there was a 3-fold increase in Pax7, and marginal increases in MyoD and Myogenin gene transcription at 10 dpi in the CTX+DT treated muscles (Fig. 8N). The robust increase in Pax7 at 10 dpi may be induced a recovery of aP2 cells after initial cell ablation, combined with the residual effect of blocked myogenic differentiation (thus increase in progenitor cell number). These results indicate that ablation of aP2 cell lineage leads to an inhibition of myogenic differentiation early during muscle regeneration, leading to regenerative defects upon acute muscle injury.
Figure 8.
- A-L: Representative immunofluorescence labeling of Pax7 (A-L), MyoD (E-H) and Myogenin (I-L) in TA muscle sections treated with CTX or CTX+DT at 5 and 10 days post injection (dpi). Pax7, MyoD and Myogenin signals are shown in Red. Nuclei are counterstained by DAPI in blue. Myogenic cell nuclei are labeled by Pax7/MyoD/Myogenin and DAPI and therefore are purple (for example, the two Pax7+ nuclei indicated by arrows). Notice that degenerated/necrotic fibers (large round structure) are none specifically labeled in red.
- M-N: Relative levels of Pax7, MyoD and Myogenin at 5 dpi (M) and 10 dpi (N) quantified by Realtime qPCR. The control (CTX alone treatment) levels are normalized to 1, n=3. * P<0.05, ** P<0.01.
Decreased myogenesis is accompanied by elevated infiltration of IMAT in old mice
To establish the interaction between myogenesis and adipogenesis under physiological conditions, we examined 18-month old mice. TA muscles were collected from the old mice and IMAT was labeled by Oil red O staining. We found substantial fat infiltration in the old, compared to the young (2-month-old), muscle sections (Fig. S7A-B). Upon CTX injection into TA muscles, the injured young muscles robustly regenerated, manifested by newly regenerated fibers with central nuclei and lack of fat infiltration (Fig. S7C). In contrast, muscle regeneration in the old muscles was extremely poor and adipocyte infiltration was very common (Fig. S7D). Compared with old mice, there were about 5-times more newly regenerated muscle fibers in the young mice (Fig. S7E). Importantly, lineage tracing using Myf5-Cre/mTmG mice indicates that none of the adipocytes emerging in regenerating old muscles exhibited mG. Thus, decreased myogenic potential in the old mice is accompanied by elevated adipogenesis mediated by a population of Myf5-independent progenitors.
Discussion
Our results support a conclusion that compared to the fast EDL muscle, the slow SOL muscle contains higher density of adipogenic progenitors and thus have higher adipogenic activities. Intriguingly, satellite cell density is also higher in the SOL compared to EDL muscle. Similar correlations in the muscle type and adipogenic potential were reported in rat muscles(Yada et al., 2006). Slow SOL muscle contains over 90% of oxidative (type I and IIa) myofibers, therefore they are more enriched with mitochondria and have a higher demand for oxygen supply. In contrast, the EDL muscle contains predominantly glycolytic (Type IIx and IIb) myofibers and has a lower demand for oxygen supply. In response to the higher demand of oxygen supply, slow muscles develop higher densities of vasculature(Lillioja et al., 1987). As adipogenic progenitors are known to be mostly associated with blood vessels, this higher density of vasculature in the slow muscles explains why adipose progenitors are more abundant in the SOL than EDL muscles.
Because reduced myogenesis in the skeletal muscle is often associated with ectopic accumulation of IMAT, it has long been assumed that IMAT is derived from myogenic satellite cells, which may take alternative cell fates under disease conditions (Mora, 1989). Satellite cells were thought to be the progenitors for both adipocytes and skeletal muscles, and in vitro experiments showed that myogenic cells can be induced to form adipocytes (Asakura et al., 2001; Csete et al., 2001; Shefer et al., 2004; Yeow et al., 2001). Our lineage tracing experiments using Myf5Cre/mTmG transgenic mice clearly demonstrated that IMAT formed in the skeletal muscle both in vivo and in the culture were not from the Myf5-lineage. This is consistent with recent lineage tracing studies (Joe et al., 2010; Schulz et al., 2011; Uezumi et al., 2010), and further indicates that the adipocytes ectopically formed in skeletal muscle are not from myogenic satellite cells that express Myf5. However, it is unclear whether the Myf5-independent satellite cells (which account for 90% of all satellite cells(Kuang et al., 2007) contributed to the intramuscular adipocytes. To answer that question, Pax3-Cre/mTmG cell lineage tracing was performed. As all satellite cells are derived from embryonic Pax3-progenitors (Kassar-Duchossoy et al., 2005; Relaix et al., 2005; Schienda et al., 2006), if they ever differentiate into adipocytes then these adipocytes should be labeled by mG. Again, we found that none of the IMAT was labeled with mG. These data suggest that intramuscular adipocytes are not derived from satellite cells or Pax3-expressing mesoderm. It is likely the IMAT originate from a mesenchymal or haematopoietic progenitor population associated with blood vessels (Majka et al., 2010; Sera et al., 2009).
Whether IMAT represents a population of brown or white adipocyte, or a mixture of both, is not established. Previous studies have shown that brown adipocytes are present in the skeletal muscles (Almind et al., 2007; Crisan et al., 2008). In addition, brown adipocyte progenitors can be identified and isolated from human skeletal muscles (Crisan et al., 2008). Furthermore, skeletal muscle resident progenitors (including satellite cell derived myoblasts and other non-myogenic progenitors) can give rise to brown adipocytes if induced with Prdm16 or BMP7 under culture conditions(Schulz et al., 2011; Seale et al., 2008). It is unclear if there is a progenitor cell population in the skeletal muscle that can differentiate into brown adipocytes without Prdm16 or BMP7 induction. Our qPCR analysis indicates that the UCP1 expression level in the skeletal muscle derived adipocytes is similar to that of the WAT progenitor-derived adipocytes, both are about 160 times lower than that of BAT progenitor-derived adipocytes. These results demonstrate that muscle resident adipogenic progenitors predominantly give rise to white adipocytes in culture. However, it is possible that Myf5-lineage brown adipocytes do present in the muscle in vivo under certain physiological conditions and formation of ectopic white adipocytes only occurs during aging or under pathological conditions when normal myogenesis is inhibited.
Our adipogenic and myogenic lineage ablation results suggest a model in which skeletal muscle and IMAT dynamically interact. In young healthy muscles, little adipocyte infiltration is evident due to the inhibitory effect of myogenesis on adipogenesis. As muscle degenerates, adipogenesis is enhanced in muscle (Wagatsuma, 2007), which in turn facilitates muscle regeneration probably mediated by secretion of promyogenic cytokines. Subsequent muscle regeneration then inhibits adipocytes and results in few ectopic adipocytes upon completion of muscle regeneration. Under disease conditions or in mutant mice when myogenesis is defective, ectopic adipocytes and fibroblastic cells gradually expand and replace muscle cells (Mora, 1989). In our old mice studies, we also observed that elevated adipogenesis accompanying decreased myogenensis. This dynamic interaction of adipogenesis and myogenesis, regulated by local environmental and systemic signals, underlies the kinetics of muscle regeneration and the pathological fat infiltration in aged muscles (Gopinath and Rando, 2008; Wagatsuma, 2007).
Obesity as a major health concern in modern era is associated with other health problems like type II diabetes and cardiovascular disease (Kopelman, 2000). In addition to the obesity metabolic risks, adipose tissue development in skeletal muscle is widely recognized as one of the hallmarks of muscle wasting conditions such as sarcopenia (Song et al., 2004). The effect of adipose tissue in skeletal muscle is not only important in muscle diseases but also has been of interest to animal agriculture, as IMAT contributes to the juicy and tender taste in meat quality assessment (Karlsson et al., 1999) (Kim et al., 2009). Understanding of the origin of IMAT and molecular mechanisms regulating muscle-adipose interaction will contribute to our ability to influence relative fat and lean deposition to improve meat quality in animal production and achieve optimal body composition in human health.
Supplementary Material
Highlights.
Slow soleus muscles have more adipo-progenitors than fast EDL muscles.
Muscle resident adipose, BAT and WAT are distinct in Pax3/Myf5-lineage origin.
Myogenic cells inhibit ectopic adipogenesis in the muscle.
Muscle resident adipose is required for efficient regeneration of damaged muscles.
Thus, various tissue-specific adipose are heterogeneous and dynamically interact with skeletal muscles.
ACKNOWLEDGEMENTS
We thank Xin Yang and Jun Wu for assistance with cell culture and mouse colonies, and other members of the Kuang laboratory for comments. The project is partially supported by funding from MDA, NIH and USDA to SK.
Footnotes
The authors declare no conflict of interests.
Author Contribution: Weiyi Liu: Conception and design of experiments, collection and assembly of data, data analysis and interpretation, final approval of manuscript.
Yaqin Liu: Conception and design of experiments, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Xinsheng Lai: Collection and assembly of data, final approval of manuscript.
Shihuan Kuang: Conception and design of experiments, financial support, administrative support, provision of study materials, data analysis and interpretation, manuscript writing, final approval of manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almind K, Manieri M, Sivitz WI, Cinti S, Kahn CR. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2366–2371. doi: 10.1073/pnas.0610416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura A, Komaki M, Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation; research in biological diversity. 2001;68:245–253. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham ME. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Rudnicki MA, Arnold HH, Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- Crisan M, Casteilla L, Lehr L, Carmona M, Paoloni-Giacobino A, Yap S, Sun B, Leger B, Logar A, Penicaud L, Schrauwen P, Cameron-Smith D, Russell AP, Peault B, Giacobino JP. A reservoir of brown adipocyte progenitors in human skeletal muscle. Stem cells (Dayton, Ohio) 2008;26:2425–2433. doi: 10.1634/stemcells.2008-0325. [DOI] [PubMed] [Google Scholar]
- Csete M, Walikonis J, Slawny N, Wei Y, Korsnes S, Doyle JC, Wold B. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. Journal of cellular physiology. 2001;189:189–196. doi: 10.1002/jcp.10016. [DOI] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Developmental biology. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Furuhashi M, Fucho R, Gorgun CZ, Tuncman G, Cao H, Hotamisligil GS. Adipocyte/macrophage fatty acid-binding proteins contribute to metabolic deterioration through actions in both macrophages and adipocytes in mice. The Journal of clinical investigation. 2008;118:2640–2650. doi: 10.1172/JCI34750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher D, Kuznia P, Heshka S, Albu J, Heymsfield SB, Goodpaster B, Visser M, Harris TB. Adipose tissue in muscle: a novel depot similar in size to visceral adipose tissue. The American journal of clinical nutrition. 2005;81:903–910. doi: 10.1093/ajcn/81.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayraud-Morel B, Chretien F, Flamant P, Gomes D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Developmental biology. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Gopinath SD, Rando TA. Stem cell review series: aging of the skeletal muscle stem cell niche. Aging cell. 2008;7:590–598. doi: 10.1111/j.1474-9726.2008.00399.x. [DOI] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Hu Y, Bock G, Wick G, Xu Q. Activation of PDGF receptor alpha in vascular smooth muscle cells by mechanical stress. FASEB J. 1998;12:1135–1142. doi: 10.1096/fasebj.12.12.1135. [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson AH, Klont RE, Fernandez X. Skeletal muscle fibres as factors for pork quality. Livestock Production Science. 1999;60:255–269. [Google Scholar]
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Choi BD, Kim BC, Park SS, Hong KC. Associations of the variation in the porcine myogenin gene with muscle fibre characteristics, lean meat production and meat quality traits. J Anim Breed Genet. 2009;126:134–141. doi: 10.1111/j.1439-0388.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. The Journal of cell biology. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillioja S, Young AA, Culter CL, Ivy JL, Abbott WG, Zawadzki JK, Yki-Jarvinen H, Christin L, Secomb TW, Bogardus C. Skeletal muscle capillary density and fiber type are possible determinants of in vivo insulin resistance in man. The Journal of clinical investigation. 1987;80:415–424. doi: 10.1172/JCI113088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Hudson AJ, Strickland KP. Fatty acid metabolism in dystrophic muscle in vitro. Life Sci. 1969;8:21–26. doi: 10.1016/0024-3205(69)90112-x. [DOI] [PubMed] [Google Scholar]
- Majka SM, Fox KE, Psilas JC, Helm KM, Childs CR, Acosta AS, Janssen RC, Friedman JE, Woessner BT, Shade TR, Varella-Garcia M, Klemm DJ. De novo generation of white adipocytes from the myeloid lineage via mesenchymal intermediates is age, adipose depot, and gender specific. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14781–14786. doi: 10.1073/pnas.1003512107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M. Fibrous-adipose replacement in skeletal muscle biopsy. Eur Heart J. 1989;10(Suppl D):103–104. doi: 10.1093/eurheartj/10.suppl_d.103. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sato T, Rocancourt D, Marques L, Thorsteinsdottir S, Buckingham M. A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 2010;6:e1000897. doi: 10.1371/journal.pgen.1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schienda J, Engleka KA, Jun S, Hansen MS, Epstein JA, Tabin CJ, Kunkel LM, Kardon G. Somitic origin of limb muscle satellite and side population cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen-Hinderling VB, Hesselink MK, Schrauwen P, Kooi ME. Intramyocellular lipid content in human skeletal muscle. Obesity. 2006;14:357–367. doi: 10.1038/oby.2006.47. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, Schrier D, Falb D, Kirkland JL, Wagers AJ, Tseng YH. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sera Y, LaRue AC, Moussa O, Mehrotra M, Duncan JD, Williams CR, Nishimoto E, Schulte BA, Watson PM, Watson DK, Ogawa M. Hematopoietic stem cell origin of adipocytes. Exp Hematol. 2009;37:1108–1120. 1120, e1101–1104. doi: 10.1016/j.exphem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Wleklinski-Lee M, Yablonka-Reuveni Z. Skeletal muscle satellite cells can spontaneously enter an alternative mesenchymal pathway. Journal of cell science. 2004;117:5393–5404. doi: 10.1242/jcs.01419. [DOI] [PubMed] [Google Scholar]
- Song MY, Ruts E, Kim J, Janumala I, Heymsfield S, Gallagher D. Sarcopenia and increased adipose tissue infiltration of muscle in elderly African American women. The American journal of clinical nutrition. 2004;79:874–880. doi: 10.1093/ajcn/79.5.874. [DOI] [PubMed] [Google Scholar]
- Starkey JD, Yamamoto M, Yamamoto S, Goldhamer DJ. Skeletal muscle satellite cells are committed to myogenesis and do not spontaneously adopt nonmyogenic fates. J Histochem Cytochem. 2011;59:33–46. doi: 10.1369/jhc.2010.956995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein TP, Wade CE. Metabolic consequences of muscle disuse atrophy. J Nutr. 2005;135:1824S–1828S. doi: 10.1093/jn/135.7.1824S. [DOI] [PubMed] [Google Scholar]
- Taaffe DR, Henwood TR, Nalls MA, Walker DG, Lang TF, Harris TB. Alterations in muscle attenuation following detraining and retraining in resistance-trained older adults. Gerontology. 2009;55:217–223. doi: 10.1159/000182084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development (Cambridge, England) 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science (New York, N.Y. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchoukalova YD, Sarr MG, Jensen MD. Measuring committed preadipocytes in human adipose tissue from severely obese patients by using adipocyte fatty acid binding protein. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1132–1140. doi: 10.1152/ajpregu.00337.2004. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- Urs S, Harrington A, Liaw L, Small D. Selective expression of an aP2/Fatty Acid Binding Protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res. 2006;15:647–653. doi: 10.1007/s11248-006-9000-z. [DOI] [PubMed] [Google Scholar]
- Vettor R, Milan G, Franzin C, Sanna M, De Coppi P, Rizzuto R, Federspil G. The Origin of Intermuscular Adipose Tissue and Its Pathophysiological Implications. American journal of physiology. 2009 doi: 10.1152/ajpendo.00229.2009. [DOI] [PubMed] [Google Scholar]
- Waddell JN, Zhang P, Wen Y, Gupta SK, Yevtodiyenko A, Schmidt JV, Bidwell CA, Kumar A, Kuang S. Dlk1 is necessary for proper skeletal muscle development and regeneration. PLoS One. 2010;5:e15055. doi: 10.1371/journal.pone.0015055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagatsuma A. Adipogenic potential can be activated during muscle regeneration. Molecular and cellular biochemistry. 2007;304:25–33. doi: 10.1007/s11010-007-9482-x. [DOI] [PubMed] [Google Scholar]
- Yada E, Yamanouchi K, Nishihara M. Adipogenic potential of satellite cells from distinct skeletal muscle origins in the rat. J Vet Med Sci. 2006;68:479–486. doi: 10.1292/jvms.68.479. [DOI] [PubMed] [Google Scholar]
- Yeow K, Phillips B, Dani C, Cabane C, Amri EZ, Derijard B. Inhibition of myogenesis enables adipogenic trans-differentiation in the C2C12 myogenic cell line. FEBS Lett. 2001;506:157–162. doi: 10.1016/s0014-5793(01)02900-3. [DOI] [PubMed] [Google Scholar]
- Yim JE, Heshka S, Albu J, Heymsfield S, Kuznia P, Harris T, Gallagher D. Intermuscular adipose tissue rivals visceral adipose tissue in independent associations with cardiovascular risk. International journal of obesity. 2007;31:1400–1405. doi: 10.1038/sj.ijo.0803621. (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.