Abstract
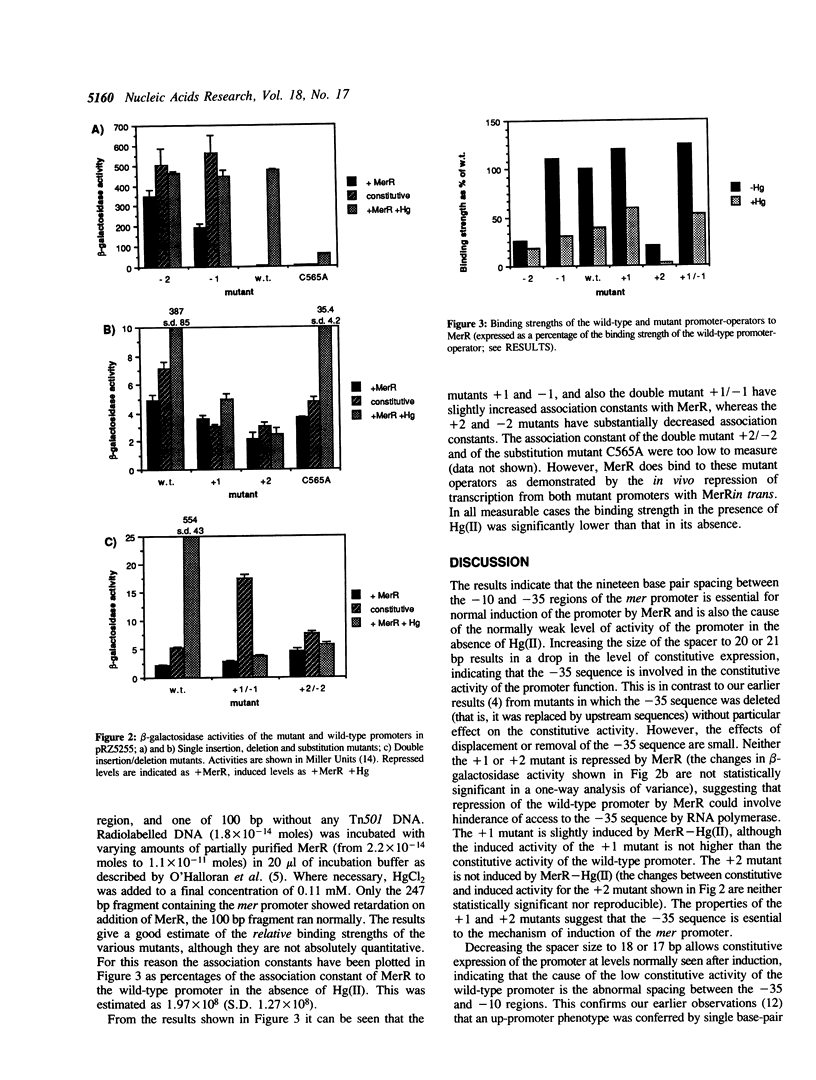
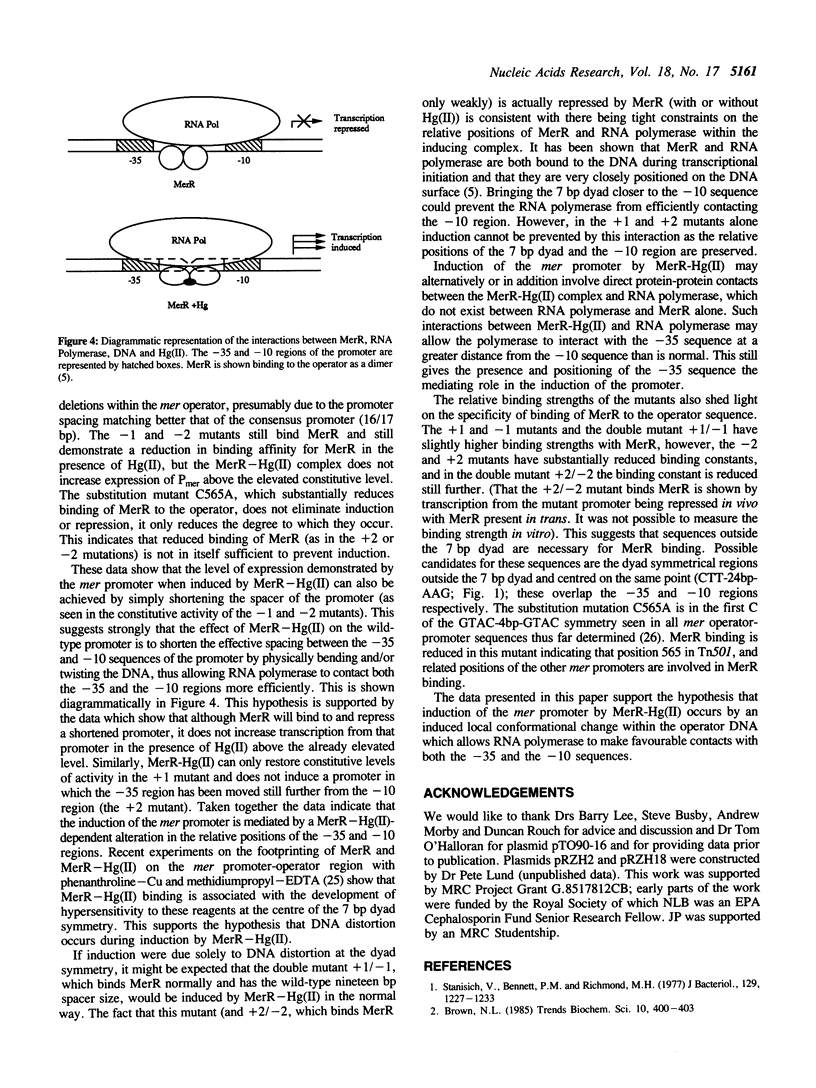
We have made site-specific mutations in the promoter-operator region of the mercury-resistance (mer) operon of transposon Tn501. Mutations were selected to alter the spacing between the -10 and -35 promoter elements without altering the sequence of a 7 bp dyad symmetrical sequence, which is the site of binding of the regulatory protein, MerR. (MerR acts as a repressor in the absence of mercuric salts and as an inducer in their presence, and binds to the same site in each case). Transcription from the mutant promoters was measured in vivo in the presence and absence of MerR and of mercuric salts; and the relative affinities of the mutant promoters for partially purified MerR were determined in vitro by gel-shift assay in the presence and absence of mercuric salts. The 19 bp spacer was found to be essential for correct induction and repression of the operon; a spacer size of 20 or 21 bp prevents induction, and a spacer size of 18 or 17 bp causes the promoter to be highly active under all conditions. Double mutations, which alter the position of the 7 bp dyad relative to the -10 and -35 sequences without altering their spacing prevent induction by the MerR-Hg(II) complex, demonstrating the tight constraints on the positions of MerR and RNA polymerase in the transcriptional complex. The data are compatible with a model for induction of the mer promoter involving a local conformational change in the DNA structure caused by the MerR-Hg(II) complex.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., O'Halloran T. V. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry. 1990 May 22;29(20):4747–4751. doi: 10.1021/bi00472a001. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Wang Y., Mahler I., Walsh C. T. Homologous metalloregulatory proteins from both gram-positive and gram-negative bacteria control transcription of mercury resistance operons. J Bacteriol. 1989 Jan;171(1):222–229. doi: 10.1128/jb.171.1.222-229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. S., Wulff D. L., Rosenberg M. Bacteriophage lambda protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature. 1983 Aug 25;304(5928):703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Nordeen S. K., Vogt K., Edgell M. H. A complete library of point substitution mutations in the glucocorticoid response element of mouse mammary tumor virus. Proc Natl Acad Sci U S A. 1986 Feb;83(3):710–714. doi: 10.1073/pnas.83.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Spassky A., Chapon C., Blazy B., Buc H. On the different binding affinities of CRP at the lac, gal and malT promoter regions. Nucleic Acids Res. 1983 Nov 25;11(22):7833–7852. doi: 10.1093/nar/11.22.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert P. F., Reznikoff W. S. Use of transcriptional repressors to stabilize plasmid copy number of transcriptional fusion vectors. J Bacteriol. 1985 Apr;162(1):441–444. doi: 10.1128/jb.162.1.441-444.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P. A., Brown N. L. Regulation of transcription in Escherichia coli from the mer and merR promoters in the transposon Tn501. J Mol Biol. 1989 Jan 20;205(2):343–353. doi: 10.1016/0022-2836(89)90345-8. [DOI] [PubMed] [Google Scholar]
- Lund P. A., Ford S. J., Brown N. L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986 Feb;132(2):465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- Lund P., Brown N. Up-promoter mutations in the positively-regulated mer promoter of Tn501. Nucleic Acids Res. 1989 Jul 25;17(14):5517–5527. doi: 10.1093/nar/17.14.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- O'Halloran T., Walsh C. Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization. Science. 1987 Jan 9;235(4785):211–214. doi: 10.1126/science.3798107. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B., Rupp W. D., Little J. W., Mount D. W. LexA protein inhibits transcription of the E. coli uvrA gene in vitro. Nature. 1982 Jul 1;298(5869):96–98. doi: 10.1038/298096a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M., Richmond M. H. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J Bacteriol. 1977 Mar;129(3):1227–1233. doi: 10.1128/jb.129.3.1227-1233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanssens P., Opsomer C., McKeown Y. M., Kramer W., Zabeau M., Fritz H. J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989 Jun 26;17(12):4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]



