Abstract
In situ biomonitoring has been used to assess the effects of pollution on aquatic species in heavily polluted waterways. In the current study, we used in situ biomonitoring in conjunction with molecular biomarker analysis to determine the effects of pollutant exposure in salmon caged in the Duwamish waterway, a Pacific Northwest Superfund site that has been subject to remediation. The Duwamish waterway is an important migratory route for Pacific salmon and has received historic inputs of polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs). Juvenile pre-smolt Chinook salmon (Oncorhynchus tshawytscha) caged for 8 days in the three contaminated sites in close proximity within the Duwamish were analyzed for steady state hepatic mRNA expression of 7 exposure biomarker genes encompassing several gene families and known to be responsive to pollutants, including cytochrome P4501A (CYP1A) and CYP2K1, glutathione S-transferase π class (GST pi), microsomal GST (mGST), glutamylcysteine ligase catalytic subunit (GCLC), UDP-glucuronyltransferase family 1 (UDPGT), and type 2 deiodinase (type 2 DI, or D2). Quantitation of gene expression was accomplished by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) in assays developed specifically for Chinook salmon genes. Gill PAH-DNA adducts were assessed as a chemical effects biomarker using 32P-postlabeling. The biomarkers in the field-caged fish were analyzed with respect to caged animals maintained at the hatchery receiving flow-through water. Chemical analysis of sediment samples from three field sampling sites revealed relatively high concentrations of total PAHs in one site (site B2, 6711 ng/g dry weight) and somewhat lower concentrations of PAHs in two adjacent sites (sites B3 and B4, 1482 and 1987 ng/g, respectively). In contrast, waterborne PAHs at all of the sampling sites were relatively low (<1 ng/L). Sediment PCBs at the sites ranged from a low of 421 ng/g at site B3, to 1160 ng/g at site B4, and there were no detectable waterborne PCBs at any of the sites (detection limit=10 ng/L). There were no significant differences (P<0.05) in biomarker gene expression in the Duwamish-caged fish relative to controls, although there was a pattern of gene expression suppression at site B3, the most heavily PAH-enriched site. The lack of a marked perturbation of mRNA biomarkers was consistent with relatively low levels of gill PAH-DNA adduct levels that did not differ among caged reference and field fish, and which were also consistent with relatively low waterborne concentrations of chemicals. The results of our study suggest a low bioavailability of sediment pollutants in caged juvenile Chinook potentially reflecting low waterborne exposures occurring at contaminated sites within the Duwamish waterway that have undergone partial remediation.
Keywords: biomarkers, Chinook salmon, quantitative RT-PCR, sediment pollutants, 32P-postlabeling
1. Introduction
Biomarkers of chemical exposure and effect have been widely used in field and in situ biomonitoring to assess the temporal and spatial effects of pollution in aquatic species (Barbee et al., 2008; Schlenk et al., 2008). These field sites are often contaminated with a host of organic compounds such as polychlorinated biphenyls (PCBs) and polycyclic aromatic hydrocarbons (PAHs), as well as trace metals (Dexter et al., 1985; PSAT, 2007a). Locally in the Pacific Northwest, fish from various trophic levels residing in urbanized waterways of the Puget Sound have received relatively high exposures to pollutants (Missildine et al., 2005; PSAT, 2007a). Pollutant exposures are of particular concern to sensitive species such as Pacific salmon, including Chinook (Oncorhynchus tschawytscha), sockeye (O. nerka), and coho (O. kisutch), whose populations are in decline. Although anthropogenic factors such as habitat loss play a key role in salmon population declines, exposures to environmental pollutants are also of importance. In particular, exposure to PCBs, heavy metals, and pesticides have been correlated with immunosupression leading to reduced salmon survival (Varanasi et al., 1993; Arkoosh et al., 1994; Arkoosh et al., 1998a; Arkoosh et al., 1998b; Arkoosh et al., 2001), whereas exposures to PAHs have been implicated in reproductive dysfunction and altered growth in salmon populations (Spromberg and Meador, 2005; Meador et al., 2006).
The scenarios under which Pacific salmon are most likely to receive exposure to anthropogenic chemicals include migratory passage through urbanized waterways or return to natal streams (Missildine et al., 2005). The impacted waterway selected for the present study has a history of pollution from point and non-point sources and is an important navigational passage for several species of local salmonids. In 2001, the USEPA placed this waterway on the national Superfund priority site listing, largely due to historically high concentrations of a number of pollutants including PAHs, PCBs and heavy metals (PSAT, 2007a). Exposure to these chemicals has been associated with increased incidences of liver lesions and other structural abnormalities in English sole and other benthic fish (Malins et al., 1984; Malins et al., 1985; Krahn et al., 1986; Krahn et al., 1987; McCain et al., 1990). In 2004, remediation efforts were initiated that included the removal of 66,000 cubic yards of contaminated sediment. To the best of our knowledge, however, there have been comparatively few studies to address the biological success of remediation in this area.
In the present study, we used a multidisciplinary approach involving chemical analysis in conjunction with molecular biomarkers in caged juvenile Chinook salmon (Onchorhynchus tshawytscha) to investigate physiological responses in this historically polluted waterway containing some areas that have undergone remediation. The presence of bulky PAH-DNA adducts in the gills as assessed by 32P-postlabeling was used as a biomarker of pollution effects at the tissue level. Quantitative gene expression assays were developed for a suite of seven hepatic genes that have shown promise as biomarkers of pollutant exposure. The criteria for selection of these genes of relevance to at least one of several processes, including biotransformation of environmental pollutants such as PAHs and PCBs, or physiological homeostatic processes sensitive to disruption by chemical exposures (i.e. thyroid hormone regulation, protection against oxidative stress), as well as those being previously identified in other studies to be modulated by exposure to waterborne pollutants in the field. The target genes included two cytochrome P450 isoforms (cytochrome P450S 1A and 2K1), two glutathione S-transferases (GST-π and microsomal GST), UDP-glucuronyltransferase family 1 (UDPGT-1), glutamylcysteine ligase catalytic subunit (GCLC), the rate limiting enzyme in glutathione biosynthesis, and type 2 deiodinase, which encodes an enzyme critical for thyroid hormone metabolism and whose expression has been shown to be altered in fish in polluted areas (Pickard-Aiken, 2009).
2. Materials and Methods
2.1 Chemicals
MS-222 (tricaine) was obtained from Argent Chemical Laboratories (Redmond, WA). RNeasy® mini kit was purchased from Qiagen (Valencia, CA). TRIzol® reagent and the first strand cDNA synthesis kit were purchased from Invitrogen (Carlsbad, CA). The PCR primers were obtained from MWG-Eurofins (Huntsville, AL). Finnzymes sybr green was purchased from New England Biolabs, Inc (Ipswich, MA). Protein assay reagent c was purchased from BioRad (Hercules, CA). Spleen phosphodiesterase, micrococcal endonuclease, nuclease P1, phenylmethylsulphonylfluoride (PMSF), bovine serum albumin, ethoxyresorufin (resorufin ethyl ether), resorufin and NADPH were purchased from Sigma-Aldrich (St. Louis, MO). Dibasic sodium phosphate was obtained from J.T. Baker (Phillipsburg, NJ). Proteinase K (recombinant, PCR grade, lyphilizate) was purchased from Roche Diagnostics (Indianapolis, IN). [γ-32P]-ATP was purchased from MP Biomedicals (Santa Ana, CA). All other solvents used for extraction and analysis of water and sediment samples were of analytical grade and purchased from standard sources.
2.2 Animals and field exposures
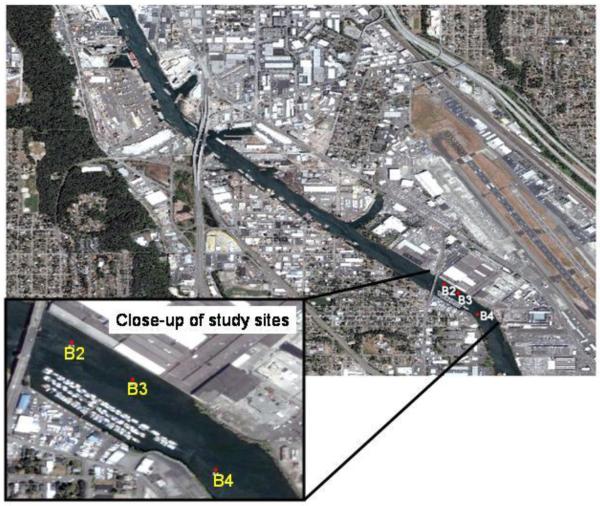
All animal care procedures were conducted in accordance with institutional animal care and use committees (IACUC) for the University of Washington and Texas A&M University. Juvenile pre-smolt Chinook salmon (mean length 9.53 cm, mean mass 8.1 g) were maintained at the NOAA Mukilteo fish hatchery in Washington State. In July 2007, the fish were transferred to 4 cages, each containing approximately 20 juvenile Chinook and deployed into each of 3 adjacent exposure sites in the historically polluted Duwamish waterway (Figure 1). Table 1 presents the typical physicochemical characteristics of each caging site. The GPS coordinates for each site are as follows B2: N47.52925 W122.31320, B3: N47.52848 W122.31193, B4: N47.52663 W122.31023. A control group of age-matched juvenile Chinook serving as reference animals were caged at the hatchery and received flow-through water. This approach was used due to the lack of an appropriate non-polluted reference site with similar physicochemical characteristics in the Duwamish area. In this regard, the Mukilteo hatchery draws upon Puget Sound seawater from an area considered non-polluted relative to the urbanized waterways of Seattle. The seawater that supplies the Mukilteo hatchery is drawn up from 20-30 m below the surface and processed through a bank of sand filters prior to sterilization with UV sterilizers and chilling as necessary. Accordingly, the water for control animals was of very high quality with a flow-through velocity in the holding tanks of several L/min. During the caging experiments, the control fish at Mukiliteo were held in this filtered seawater at 12°C at >90% (approximately 9 ppm) dissolved oxygen during the experiment (Table 1). These hatchery maintained reference Chinook were not fed because juvenile fish caged in the Duwamish system have limited access to prey in the water column and do not feed. This is supported by previous unpublished data from a 2006 study showing no stomach contents for all caged juvenile salmon in the Duwamish (J.P. Meador, NOAA NWFSC). We selected 8 days as a suitable caging period based on the fact that juvenile salmon migrating through this waterway can spend similar migratory time in this area and can bioaccumulate chemicals during this residence (Stein et al 1995). Also, it is our experience that caging juvenile salmon for longer periods may result in excessive stress and mortality (personal observations). Upon completion of the eight day caging period, all cages were retrieved by a USEPA dive team. The juvenile Chinook, including field site exposed and caged controls, were sacrificed by a lethal overdose of MS-222 followed by severing the spinal cord. The livers and gills were excised and rinsed in ice-cold PBS and snap frozen in liquid nitrogen and later transferred to a −80 °C freezer prior to isolation of subcellular fractions, and total tissue DNA and RNA.
Figure 1.
Map of the caging sites for juvenile Chinook salmon in the Lower Duwamish Waterway, Puget Sound Washington.
Table 1.
Typical physicochemical characteristics of the field sites and hatchery*
| Site | B2 | B3 | B4 | Reference controls |
|---|---|---|---|---|
| Depth of cage1 (m) | 12.2 | 0.4 | 2.2 | NA |
| Temperature (°C) | 18.5 | 19.1 | 18.8 | 12 |
| Salinity (pss2) | 9.3 | 11.9 | 3.4 | 28 |
| Dissolved oxygen (% sat) | 92 | 89 | 86 | >90 |
GPS coordinates for each site are listed in materials and methods. All field site measurements were taken at one time point on day of cage retrieval and do not the variations in temperature, salinity and DO that can occur at the LDW field sites.
reflects general depth below mean low water mark
pss= practical salinity scale
NA: not applicable, control fish were maintained in holding tanks with flow-through filtered seawater
2.3 Sediment and water collection and analysis
Sediment samples from each field site were collected from a boat using a petite ponar grab sampler (WILDCO, Buffalo, NY) on the day of cage deployment. Upon collection, the sediment samples were homogenized in stainless steel bowls and transferred to glass I-CHEM certified 1L sampling jars with Teflon lined lids (VWR, West Chester, PA). The samples were then shipped on ice overnight following chain-of-custody protocols to the laboratory at Texas A&M University, where they were stored at −20 °C. Prior to extraction, each sediment sample was oven-dried at 40 °C, homogenized, ground in a mortar and pestle and then passed through an 850 μm sieve. Approximately 10 g of dried sample was then extracted using dichloromethane in a Dionex (Dionex Corp., Sunnyvale, CA) Model 200 Accelerated Solvent Extractor (ASE) following USEPA Method 3545A (EPA, 1996a). Sediment extracts were analyzed for PAHs using USEPA method 8270C (EPA, 1996b) and for total PCBs and PCB homologs following USEPA method 680 (Alford-Stevens et al., 1985). Analysis was performed using an Agilent 5980 gas chromatograph with an Agilent 5972 mass selective detector in selected ion monitoring mode. A 60 m × 0.25 mm ID × 0.25 mm film thickness column (Agilent Technologies, Palo Alto, CA) was utilized. The injection port was maintained at 300 °C and the transfer line at 280 °C. The temperature program was as follows: 60 °C for 6 min, increased at 12 °C/min to 180 °C and then increased at 6 °C/min to 310 °C and held for 11 min for a total run time of 47 min.
Water samples were collected at each site using a Beta bottle sampling device (Wildlife Supply Co., Buffalo, NY) placed just above the sediment surface within the exposure zone of the deployed cages on the day of cage deployment. Samples were stored in 1 L I-CHEM certified amber bottles with Teflon lids (VWR, West Chester, PA) and were shipped on ice overnight following chain of custody protocols to the laboratory at Texas A&M University, where they were stored at 4 °C until extraction. Liquid:liquid extractions were performed with dichloromethane according to USEPA standard extraction method 3510C (EPA, 1996a). The surface water sample extracts were then analyzed for PAHs using USEPA method 8270C (EPA, 1996b) and for total PCBs and PCB homologues following USEPA method 680 (Alford-Stevens et al., 1985), as described above for sediment samples.
2.4 Quantitative gene expression assays
RNA was extracted from frozen liver tissue of a minimum of 5 individual fish per group using the TRIzol® method. The RNA samples were cleaned using Qiagen (Valencia, CA) RNeasy® mini kit prior to analysis on a Nanodrop spectrophotometer (Thermo Scientific, Wilmington DE) to ensure integrity of RNA prior to first strand cDNA synthesis, followed by assessment of 260/280 absorbance ratios, which fell between 1.9 and 2.1. Total first strand cDNA was synthesized from liver RNA using Invitrogen® (Carlsbad, CA) Superscript first stand synthesis system with oligo dT18 primers and DNAse treated RNA samples. To design PCR primers for type 2 deiodinase and GCLC, multiple sequence alignments of rainbow trout (Oncorhynchus mykiss), Atlantic salmon (Salmo salar), and zebrafish (Danio rerio) target sequences were examined for conserved sequences, prior to primer design using Oligo ® Software, Version 6.71 (Cascade, CO). For UDPGT, a previously published primer pair from Atlantic Salmon (S. salar) (Mortensen et al., 2007) was utilized. For ß-actin. CYP1A, and CYP2K1, previously published primer pairs from rainbow trout (O. mykiss) (Matsuo et al., 2008) were utilized. For GST-π, 18s RNA, and microsomal GST, previously published primer pairs from coho salmon were utilized (Gallagher et al., 2008). All primers were tested to optimize annealing temperature and ensure a single product formation (See Table 2 for primer sequences). Amplification products from Chinook liver cDNA were sequenced and verified at the UW Department of Biochemistry DNA sequencing facility. Once the cDNA sequences were obtained, the results were compared to other teleost DNA sequences using BLAST in the TGI (The Gene Index) database (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/Blast/index.cgi) to confirm the amplification of the target gene. After primer validation and optimization, SYBR green qRT-PCR assays were conducted using a BioRad® IQ5 thermocycler under the optimal PCR conditions for each primer set. For all comparative gene analysis, a standard curve generating using purified plasmid containing the target sequence was included in order to quantify the pg of cDNA.
Table 2.
Sequences and origins of gene specific PCR primers for Chinook salmon.
| Gene | Primer (5′ to 3′) | (Source Species) accession # / reference or position |
|---|---|---|
| 18s RNA | Forward: AAC GGC TAC CAC ATC CAA GGA Reverse: CGA GAT CCA ACT ACG AGC TTT TTA ACT |
(O. kisutch)AF030250 607-960 (Gallagher et al, 2008) |
| β-actin | Forward: GAC CCA CAC AGT GCC CAT CT Reverse: GTG CCC ATC TCC TGC TCA AA |
(O. mykiss)AF157514 528-767 (Matsuo et al, 2008) |
| CYP1A | Forward: AGT GCT GAT GGC ACA GAA CTC AA Reverse: AGC TGA CAG CGC TTG TGC TT |
(O. mykiss) AF059711 1441-1658 (Matsuo et al, 2008) |
| CYP2K1 | Forward: CTC ACA CCA CCA GCC GAG AT Reverse: CTT GAC AAA TCC TCC CTG CTC AT |
(O. mykiss) AF045053 1231-1372 (Matsuo et al, 2008) |
| Deiodinase (Type 2) |
Forward: CAG GTG ATG CGA GAG GTA GAG Reverse: CAA GTC TGG GCC GTC AAG |
(O. mykiss) AF207900 1881-2041 |
| GCLC | Forward: ACA TAG CCC ATC TCT TCA Reverse: GAA CTC AAC TCG CCA TC |
(O. mykiss) TC161597 476-709 |
| GST-π | Forward: CTC TGC TCC AGT TGC CTG GAT Reverse: GTT GCC ATT ATT GGG CAG TTT CT |
(O. nerka)AB026119 490-615 (Gallagher et al., 2008) |
| Microsomal GST |
Forward: GGG TGA GGC CTG GGA TGA Reverse: CAC AAG TAC GGA TGC CCA CAA |
(O. mykiss) CF752713 562-713 (Gallagher et al., 2008) |
| UDPGT | Forward: ATA AGG ACC GTC CCA TCG AG Reverse: ATC CAG TTG AGG TCG TGA GC |
(S. salar) DY802180 1528-1608 (Mortensen et al., 2007) |
2.5 Gill 32P-Postlabeling Assay
Gills from 4 fish at each caging site, as well as hatchery caged controls, were removed and composited for each tissue sample. Total DNA was isolated from the gill filaments by solvent extraction combined with enzymatic digestion of the protein and RNA (Randerath et al., 1986) for 32P-postlabeling. Gill PAH-DNA adducts were quantified using nuclease p1-enhanced bisphosphate 32P-postlabeling (Randerath et al., 1986). Briefly, DNA (10 μg) was enzymatically degraded to normal (Np) and modified (Xp) deoxyribonucleoside 3′-monophosphates with micrococcal nuclease and spleen phosphodiesterase at pH 6.0 and was incubated at 37 °C for 3.5 hours. After treatment of the mixture with nuclease P1 to convert normal nucleotides to nucleosides, modified nucleotides (Xp) were converted to 5′-32P-labeled deoxyribonucleoside 3′,5′-bisphosphates (pXp) by incubation with carrier-free [γ-32P]ATP and polynucleotide kinase. Radioactively labeled modified nucleotides were mapped by multidirectional anion-exchange thin-layer chromatography (TLC) on polyethyleneimine (PEI)-cellulose sheets (Mabon et al., 1996). After removal of orthophosphate and traces of radioactive impurities by one-dimensional development with 2.3 M sodium phosphate, pH 5.75 overnight (D1), bulky labeled DNA adducts retained in the lower (2.8 × 1.0 cm) part of the D1 chromatogram were contact-transferred to fresh thin-layer sheets and resolved by two-dimensional TLC after brief autoradiography on Cronex 4 X-ray film. The non-polar I-compounds were separated with solvents 3.82 M lithium formate, 6.75 M urea, pH 3.35 and 0.72 M sodium phosphate, 0.4 M Tris-HCl, 7.65 M urea, pH 8.2 in the first and second dimensions, respectively. 32P-labeled adducts were visualized by screen-enhanced autoradiography at −80 °C using Kodak XAR-5 film or with the aid of an InstantImager (Packard Instruments) (Zhou et al., 1999).
2.6 Microsomal ethoxyresorufin-O-deethylase (EROD) activities
Microsomal fractions from a pool of 16-20 juvenile Chinook livers from each of the exposure sites and the caged reference site were isolated according to standard methods (McKinney et al., 2006). The protein content of each sample was quantified using the method of Bradford (Bradford, 1976). The ethoxyresorufin-O-deethylase (EROD) catalytic activity assay was conducted using a 96-well fluorescent plate reader (Whyte et al., 2000) . This assay was selected to serve as a reference control and enzymatic activity corresponding to results of our CYP1A mRNA analyses. The assay incubations contained 0.1 M sodium phosphate buffer (pH 7.8) and 100 μM ethoxyresorufin, and microsomal fraction, and were initiated after a 3 minute incubation at 30 °C by the addition of 2 mM NADPH. The change in fluorescence was recorded at excitation and emission wavelengths of 550 and 585 nm, respectively, over 2 minutes. A resorufin standard curve was utilized to convert for arbitrary fluorescent units to ρmols of resorufin formed per unit time in the assay.
2.7 Statistical Analysis
Several approaches were employed to quantitate site-related effects in biomarker gene analysis. These included normalization to two different internal control genes (e.g. 18s RNA, β-actin), analysis of unnormalized biomarker gene mRNA data, as well as consideration of data transformations prior to and post data normalization. Because all gene expression data followed log normal distributions, we elected to log transform the data prior to undergoing statistical analyses using parametric methods. Following log transformations, biomarker gene expression for the individual genes was normalized to the individual expression of 18s RNA. Site differences in the normalized biomarker genes were then assessed using a one-way ANOVA. The effect of caging on the number of gill PAH-DNA adducts in control and field-exposed groups followed normal distributions and the data was directly compared by ANOVA using SigmaStat software (Aspire Software, Ashburn, VA). All other ANOVAs were performed with Statview (SAS 1998). Treatment differences were determined with Fisher’s Protected Least Significant Difference (PLSD) post-hoc test. We consider the p-values from the posthoc tests an important indicator of treatment effect. The strongest response was noted for those p-values below 0.05. P-values between 0.05 and 0.15 were also considered biologically important, especially in light of the relatively few replicates available for this experiment.
3. Results
3.1 Water quality conditions at the field sites
The results of typical physical and chemical water quality parameters such as salinity, dissolved oxygen content, temperature, and depth of cage are presented in Table 1. As observed, the water quality parameters at the field sites can be highly variable, and the salinity and temperature recordings from the hydrolab at the time of cage retrieval differed from those at those hatchery maintaining control fish. It is important to note that the hydrolab recordings represent a snapshot taken at one particular sampling and thus are not representative of mean recordings during the period of caging. Variations in temperature and salinity associated with water quality are common at the sites and fluctuate at the surface, as well as at the depths that the salmon were caged. Because of the salt wedge in the river at the field sites, the salinity will increase with depth the bottom water will be closer to full salt water (28-30 ppt) and cooler temperatures the more resemble those at the hatchery. Despite the potential variability in salinity and temperature in water associated with the field sites relative to the hatchery, fish retrieved from all three exposure sites appeared healthy and active upon cage retrieval. However, due to the necessity for rapid handling of tissues to maintain the integrity of mRNA and gill DNA adducts as well as for analytical chemistry analysis, no additional quantitative indicators of fish health were measured.
Sediment PCBs, as well as sediment and water PAH concentrations from each site are summarized in Table 3. Despite their close geographic proximities, the sediment and water carcinogenic PAH concentrations varied markedly between sites. The total PAH sediment load varied by a factor of 5 between sites B3, B4 (1482, 1967 ng/g respectively) and site B2 (6711 ng/g). Interestingly, the sediment PAH levels for all three exposed sites were within the lower range of sediment PAH concentrations measured in the 1980s prior to remediation (range: 1000-49000 ppb sediment) (Malins et al., 1984) (Table 4).
Table 3.
Water and sediment PAH concentrations at the three sites in the Lower Duwamish Waterway. Sediment PCB concentrations are also included. All concentrations of PAHs are surrogate corrected.
| B2 | B3 | B4 | |
|---|---|---|---|
| Water PAH concentrations | ppt (ng/L) | ppt (ng/L) | ppt (ng/L) |
| Carcinogenic PAHs | 12 | 7 | nd |
| Low Molecular Weight PAHs | nd | nd | nd |
| High Molecular Weight PAHs | 16.7 | 8.3 | nd |
| Total PAHs | 18.3 | 10.4 | nd |
| Sediment PAH concentrations | ppb (ng/g dry) | ppb (ng/g dry) | ppb (ng/g dry) |
|
| |||
| Carcinogenic PAHs | 1056 | 425 | 541 |
| Low Molecular Weight PAHs | 1219 | 70 | 159 |
| High Molecular Weight PAHs | 3069 | 666 | 985 |
| Total PAHs | 6711 | 1482 | 1967 |
| Sediment PCB concentrations | ppb (ng/g dry) | ppb (ng/g dry) | ppb (ng/g dry) |
|
| |||
| Total PCBs | 1030 | 421 | 1160 |
nd*: not detected , <1 ng/L
Table 4.
Historical comparisons of PAH and PCB concentrations in sediment, water and fish from polluted areas of the Puget Sound, WA
| Year Sampled |
Site | Total PAH (unit)/ (Environmental media) |
Total PCB (unts)/ (Environmental media) |
Total PCB (units) /(fish species) |
Total PAHs (units)/ (fish species) |
Reference |
|---|---|---|---|---|---|---|
| 1979-1982 | Puget Sound, WA | 1 000- 49 000 ppb/ sediment |
0-770 ppb/ sediment |
47 ±25 ppm dry weight/ English Sole |
<0.05 ppm/ English sole liver |
Malins et al., 1984 |
|
| ||||||
| 1984-1988 | Puget Sound, WA | 5900 ppb/ sediment |
500 ppb/ sediment |
11,000 ppb (dry weight)/ English sole |
Myers et al., 1994 | |
|
| ||||||
| 1989-1990 | Puget Sound, WA including Duwamish River |
300± 40 ppb (wet weight) (mean ± SEM)/ Juvenile Chinook salmon stomach contents |
LMW# PAHs= 1,800- 360 000 ppb (wet weight ), HMW % PAHs= 21 000 ±15 000 ppb (wet weight)/ juvenile Chinook salmon stomach contents |
Stein et al., 1995 | ||
|
| ||||||
| 1989-1990 | Puget Sound, WA including Duwamish River |
200-450 ppb (wet weight)/ Juvenile Chinook salmon liver |
Stein et al., 1995 | |||
|
| ||||||
| 1992-1996 | Puget Sound WA including Duwamish River, Elliot Bay Seattle WA |
55 ppb (wet weight)/ Chinook salmon filets |
PSAT, 2007b | |||
|
| ||||||
| 1994 | Puget Sound WA: Duwamish waterway |
263- 21 828 ppb (dry weight)/ sediment |
35-26 000 ppb (dry weight)/ sediment |
LDWG, 2001 | ||
|
| ||||||
| 1996 | Puget Sound WA: Duwamish waterway |
1430- 6115 ppb (dry weight)/ sediment |
84-450 ppb (dry weight)/ sediment |
LDWG, 2001 | ||
|
| ||||||
| 1998 | Puget Sound, WA: Duwamish Waterway |
50-70 870 ppb (dry weight)/ sediment |
20- 12 000 ppb (dry weight)/ sediment |
LDWG, 2001 | ||
|
| ||||||
| 1998-2002 | Puget Sound, WA including Duwamish River, Elliot Bay Seattle WA |
12 ppb (wet weight)/ Coho Salmon |
PSAT, 2007b | |||
|
| ||||||
| 1999 | Puget Sound, WA including Duwamish River, Elliot Bay Seattle WA |
0.18-3.1 (ppb)/ water |
PSAT, 2007b | |||
|
| ||||||
| 1999-2004 | Puget Sound, WA including Duwamish River, Elliot Bay Seattle WA |
150 ppb (wet weight)/ Pacific Herring |
PSAT, 2007b | |||
|
| ||||||
| 2003 | Puget Sound, WA | 14-170 ppb (wet weight)/ Chinook salmon |
Missildine et al., 2005 | |||
| 2000 – 2004 |
Puget Sound, WA Duwamish Waterway |
4- 25,000 ppb (dry weight)/ sediment |
4-1,100 ppb (wet weight)/ Chinook salmon |
Meador et al. 2009 | ||
LMW= Low molecular weight, defined as two to three benzene rings
HMW= High molecular weight, defined as containing four to six benzene rings
Although, water PAH levels were relatively low, our data suggests the potential for bioaccumulation of PAHs in the water column. As observed in Table 3, carcinogenic PAHs accounted for more than 50% of the total PAHs detected in the water column. Carcinogenic PAHs are defined by the USEPAs priority list of pollutants and included benzo[a]pyrene. Overall water column PAH levels were low, ranging from non-detectable (<1 ng/L) to 18 ng/L. Sediment PCB levels varied two fold between exposure sites that are located within 100 meters of each other (sites B2 and B3). Specifically, site B3 had total PCB concentrations of 421 ng/g dry weight, while site B2 had total PCB concentrations of 1030 ng/g dry weight.. This disparity in sediment PAH and PCB concentrations is suggestive of the fact that the riverbed does not contain a homogeneous pollutant mixture in the sediments (Meador et al., 2009).
3.2 Biomarker responses
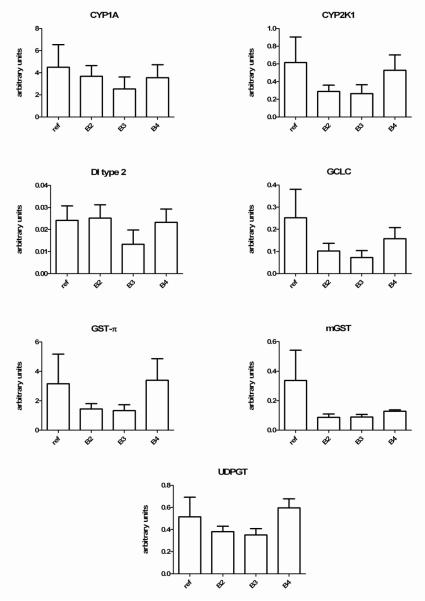
Fish caged at the field sites, as well as the reference controls, appeared healthy upon retrieval. Initial optimization of quantitative PCR assays was conducted after ensuring that selected PCR primer pairs were amplifying the correct gene sequence. All primers used in this study produced a single band PCR product and the BLAST query resulted in the correct gene hit with a BLAST e-value < 10 −15. Furthermore, we also compared similar sequences with lesser e-values before assigning identities to the target genes. Consistent with other aquatic toxicology studies (Arukwe, 2006), we observed statistically significant differences in the expression of two so-called “housekeeping genes” tested, including 18s and β-actin, among the field sites. However, careful inspection of the data revealed that the expression of all genes, including the biomarker genes as well as internal controls, followed log normal distributions, and the data were transformed prior to statistical analysis. Essentially, this approach provided similar results observed with those of the individual biomarker genes unnormalized to control housekeeping gene (data not shown). Figure 2 presents the results of qRT-PCR gene expression assays in juvenile pre-smolt Chinook hepatic cDNA normalized to the housekeeping gene 18s RNA.
Figure 2.
Comparative hepatic gene expression of 7 genes measured in the livers of Chinook salmon caged for 9 days in a polluted waterway. All data for the individual genes was normalized to the expression of 18s RNA and then multiplied by 1000 to provide the RNA expression units on the y-axis. As discussed in the methods, control fish (ref) were maintained in cages at the fish hatchery and were not fed. All data represent the mean ± SEM of n=5 individuals. There were no site related differences in mRNA expression at p<0.05. Individual p values for reference fish vs site B3 animals are provided in table 5.
There were no statistically significant differences in biomarker gene expression among field sampled fish relative to controls (Fig. 2). However, a careful inspection of the data revealed a trend toward repression of all of the biomarker genes in site B3 fish. The p values for these comparisons ranged from p=0.08 (type 2DI) through p=0.21 (GST pi, Table 5). There was also a trend in suppression of several biomarker genes, including CYP2K1, GCLC, GST pi, mGST, type 2 DI, and UDPGT in the site B2 fish relative to controls (Figure 2).
Table 5.
Statistical ( p) values for biomarker gene expression for Site B3 relative to control animals caged at the hatchery.
| Gene | Distribution |
p values for control v. site B3 |
|---|---|---|
| CYP1A | lognormal | 0.20 |
| CYP2K1 | lognormal | 0.19 |
| GCLC | lognormal | 0.13 |
| GST pi | lognormal | 0.21 |
| mGST | lognormal | 0.06 |
| UDPGT | lognormal | 0.12 |
| type 2 DI | lognormal | 0.08 |
Values represent gene expression normalized to 18s RNA of n=5 animals in each group. All data were log transformed prior to statistical analysis to follow parametric distributions (One-way ANOVA and Fisher’s PLSD test)
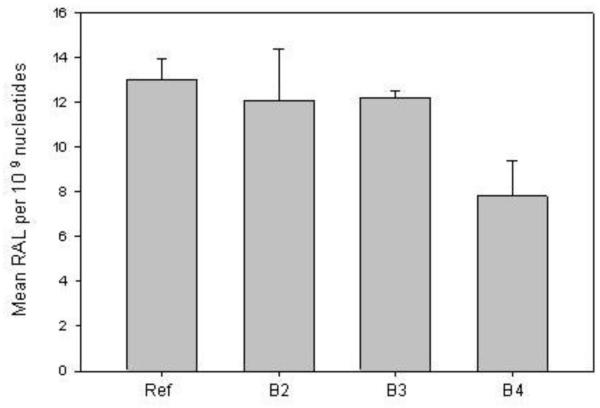
The CYP1A-associated microsomal EROD catalytic activities for each site-exposed group were 98(±7), 44(±3), 65(±3) ρmol/ minute/ mg protein (sites B2, B3 and B4 respectively), while control fish had initial rate EROD activity of 150 (±12.2) ρmol/ minute/ mg protein. Because of the small size of the fish livers, the EROD assay was conducted in triplicate on microsomal fractions made from a pool of 16-20 juvenile Chinook livers from each exposure site and therefore statistical differences among sites and controls could not be tested. As observed, however, the CYP1A biochemical assay was in good agreement with the CYP1A mRNA levels. Similar to the results of hepatic gene expression and biochemical analyses, there were no statistically significant differences among the control hatchery-maintained and field exposed Chinook with respect to the presence of bulky PAH-DNA adducts in the gills (Figure 3).
Figure 3.
Comparison of gill PAH-DNA adducts in Chinook salmon caged for 8 days in a polluted waterway. As described in methods, control fish were maintained in cages at the fish hatchery (indicated as reference, ref). All data represent the mean ± SD of PAH-DNA adducts in the gills per 109 nucleotides from n=4 individuals. There were no site related differences in DNA adducts at p<0.05.
4. Discussion
The expression of genes involved in cellular process such as protecting against oxidative stress and cell injury, or mediating chemical biotransformation have been used for biomarkers of exposure to chemical pollutants (for a review see Schlenk et.al. 2008). For example, cytochrome P4501A (CYP1A) is a classic biomarker gene that is inducible by a host of xenobiotics, and thus has been used in many studies as a sensitive indicator of pollutant exposure (Rice et al., 2000; EPA, 2007), including the Duwamish waterway (Stein et al., 1995). CYP1A has very low basal expression and is induced by a number of compounds including PAHs (Jönsson et al., 2006). Cytochrome P450 2K1 is an important phase 1 metabolism enzyme that is more highly expressed in salmonids and also functions in the metabolism of steroids, xenobiotics, and fatty acids (Schlenk et al., 2008). In addition to the phase 1 enzyme studies, phase II enzymes such as isoforms of cytosolic and microsomal GSTs as well as UDPGT, collectively conjugate a host of environmental compound and their metabolites and have also been used in biomonitoring studies involving several fish species (Machala et al., 1998; Bello et al., 2001; Schlenk et al., 2008). The expression of mRNAs encoding proteins involved in glutathione biosynthesis (i.e. glutathione synthetase, GCLC) have been less studied as biomarkers. However, GCLC is a highly inducible enzyme in rodents exposed to electrophilic (reactive) chemical compounds (Iles and Liu, 2005; Li et al., 2007; Han et al., 2008; Thimmulappa et al., 2008). GCLC induction has also been observed in fish exposed to xenobiotics (Hughes and Gallagher, 2004), suggesting that it is under similar gene regulatory pathways in rodents and fish. The iodothyronine deiodinases represent a family of enzymes responsible for converting the pro hormone T4 to biologically active T3, as well as for inactivating thyroid hormones. In addition to their roles in thyroid hormone homeostasis and the initiation of ovarian maturation in fish, these enzymes have been investigated for use as potential biomarkers of exposure to environmental contaminants in fish (Pickard-Aitken, 2007). Of the three isoforms, type 2 DI, in particular, has been shown in walleye (Sander vitreus) liver to be sensitive to waterborne pollutants in fish sampled from the Ottawa River ( Pickard-Aitken et al., 2007).
In a study sharing several similarities to ours, caged coho salmon and molecular biomarker analysis of genes encoding CYP1A and two antioxidant defense genes (superoxide dismutase and glutathione peroxidase) were used to detect environmental contamination effects in the Prince William Sound, Alaska. The authors also compared site-specific biomarker data to those in control animals maintained in hatchery water (Roberts et al., 2006). Interestingly, the authors reported site-related differences in the expression of these genes in the gill tissue (Roberts et al., 2006), but not in the livers. These data suggested that pharmacokinetic or tissue specific-responses in gene expression related to chemical exposures may also be a factor in modulation of gene expression. Although we did not measure gill biomarker mRNA expression in the present study, the fact that our gill DNA adduct levels in field sampled fish were not elevated was consistent with the lack of modulation of hepatic biomarker mRNA expression.
The Duwamish waterway has been the subject of numerous studies that have linked exposure to sediment-associated chemicals to physiological abnormalities in several fish species (Malins et al., 1984; Varanasi et al., 1989; McCain et al., 1990; Stein et al., 1995). Of relevance to the present study was a previous analysis of field sampled Chinook salmon (sampled in 1989 and 1990) returning to urban and non-urban natal streams in Seattle, including Chinook salmon migrating through field sites in our study. In that earlier study, the authors reported increased levels of gill PAH-DNA adducts in Chinook returning to urban natal streams (Stein et al., 1995). These data contrast those observed in our study in that despite the presence of sediment PAH and PCB levels, we did not detect increased levels of gill PAH DNA adducts.
The differences in results of the two studies may have been due to the fact that the caged salmon in our study likely did not bioaccumulate sediment associated chemicals due to restriction of feeding in the cages. In this regard, Duwamish caged fish feed poorly once caged in the waterway. The lack of active feeding not only limits exposure to contaminated prey, but may compromise their ability to undergo gene expression induction responses as a result of a poor nutritional status (unpublished observations). Many of the copepods and amphipods that the native juvenile Chinook consume have bioaccumulated high levels of PCBs and PAHs and thus dietary consumption may be a large contributing factor to PAH and PCB levels in Chinook from the Puget Sound (Stein et al., 1995). While dietary uptake is important for bioaccumulation, exposure to dissolved contaminants via gill ventilation is likely the only route of uptake for caged fish. Uptake from gill ventilation may be reduced in caged fish due to reduced activity and the lack of feeding. Studies have shown a reduction in metabolism (as a function of oxygen uptake and gill ventilation) in starved salmonids (Brett 1995). A more important variable may be the level of activity, which can have a large impact on gill ventilation rates and contaminant uptake (Meador et al. 2008). Based on these factors, caged fish were likely below their exposure potential compared to wild fish. Other factors that may lead to variability in hepatic mRNA response are temperature (Heise et al., 2003), dissolved oxygen content (Cooper et al., 2002) and salinity (Martinez-Alvarez et al., 2002), all of which can affect antioxidant parameters which may disrupt the expression of genes involved in protecting against oxidative stress (Di Giulio and Hinton, 2008).
Also of consideration is the potential for a reduction of waterborne chemical exposures due to the success of recent remediation efforts. However, analysis of sediment PAH and PCB levels in the current study did reveal contamination at the sites and in some cases were comparable to concentrations detected in the late 1970s and early 1980s (Malins et al., 1984; Myers et al., 1994). Accordingly, the fact that water PAH levels were either low or non-detectable in the present study was probably a major factor in the lack of responses. In essence, despite the continued presence of sediment pollution, it is likely that the caged salmon experienced reduced exposures compared to uncaged fish due to the lack of dietary uptake. In another study of caged juvenile coho salmon in a polluted waterway in the Puget Sound, mean hepatic PAH-DNA adducts as measured by 32P-postlabeling were 39 ±6.5 RAL × 109 nucleotides (Barbee et al., 2008), or approximately threefold higher than observed in the present study. Furthermore, the field site utilized in the aforementioned study had waterborne PAH concentrations as high as 1018 ng/L (Barbee et al., 2008), which greatly exceeded the water PAH levels measured in our field study. Collectively, these studies indicate relatively low water column PAH levels that were not sufficient to induce CYP1A-bioactivation pathways and facilitate the formation of gill PAH-DNA adducts.
5. Conclusion
In conclusion, we have developed a suite of 7 biomarker assays that are specific for the analysis of genes that encode proteins from several families of important toxicological function in Chinook salmon. Chinook constitute an ecologically sensitive species in the Western United States whose populations are in decline. Caging of juvenile Chinook in an urbanized waterway that has undergone remediation, but which still contains sediments polluted with PAHs and PCBs, revealed a relatively low waterborne bioavailability of sediment-associated chemicals that did not appear to be of sufficient magnitude to perturb liver mRNA expression markers, induce the formation of gill PAH-DNA adducts, or induce CYP1A-associated EROD activity. However, examination of biomarker responses in the present study revealed a subtle, but visible pattern of overall gene suppression in the most heavily contaminated subsampling area. Of interest would be a comparative analysis of biomarker mRNA responses in fish that preferentially feed on benthic organisms exposed to sediment associated PAH is and PCBs in the Duwamish waterway. In addition, laboratory experiments involving dose-response exposures of pure compounds and their mixtures is needed to further characterize molecular biomarkers of chemical exposures and effects in Pacific salmon.
Acknowledgements
We will miss the friendship, collegiality, and conversations with our good friend, Dr. K.C. Donnelly, who passed away in July 2009. This work is a collaboration among two NIEHS Superfund Research Program (SRP) program project awards to the University of Washington and Texas A&M University. Accordingly, this work was funded in part by P42 ES-004696 and P42 ES04917. Additional funding for the study was provided in part from the National Oceanic and Atmospheric Administration, Oceans and Human Health Program (NA05NS4781253), and the Region 10 office of the USEPA (USEPA Region 10). The technical comments of Dr. Karen Peck at NOAA fisheries are appreciated. Disclaimer: the views and opinions expressed in this manuscript do not necessarily reflect those of the US Environmental Protection Agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alford-Stevens A, Bellar TA, Eichelberger JW, Budde WL. Determination of pesticides and PCBs in water and soil/sediment by GC/MS (CD ROM) United States Environmental Protection Agency Office of Research and Development; Cincinnati, Ohio: Nov, 1985. [Google Scholar]
- Alford-Stevens A, Bellar TA, Eichelberger JW, Budde WL. Development, U.S.E.P.A.O.o.R.a., editor. Determination of pesticides and PCBs in water and soil/sediment by GC/MS (CD ROM) 1985.
- Arkoosh M, Casillas E, Clemons E, Kagley A, Olson R, Reno P, Stein J. Effect of pollution on fish diseases: Potential impacts on salmonid populations. Journal of Aquatic Animal Health. 1998a;10:182–190. [Google Scholar]
- Arkoosh M, Casillas E, Huffman P, Clemons E, Evered J, Stein J, Varanasi U. Increased susceptibility of juvenile chinook salmon from a contaminated estuary to Vibrio anguillarum. Transactions of the American fisheries Society. 1998b;127:360–374. [Google Scholar]
- Arkoosh M, Clemons E, Huffman P, Kagley A. Increased susceptibility of juvenile Chinook salmon to vibriosis after exposure to chlorinated and aromatic compounds found in contaminated urban estuaries. Journal of Aquatic Animal Health. 2001;13:257–268. [Google Scholar]
- Arkoosh M, Clemons E, Myers M, Casillas E. Suppression of B-cell mediated immunity in juvenile chinook salmon (Oncorhynchus tshawytscha) after exposure to either a polycyclic aromatic hydrocarbon or to polychlorinated biphenyls. Immunopharmacol Immunotoxicol. 1994;16:293–314. doi: 10.3109/08923979409007096. [DOI] [PubMed] [Google Scholar]
- Arukwe A. Toxicological housekeeping genes: do they really keep the house? Environmental Sci. and Tech. 2006;40:7944–7949. doi: 10.1021/es0615223. [DOI] [PubMed] [Google Scholar]
- Barbee GC, Barich J, Duncan B, Bickham JW, Matson CW, Hintze CJ, Autenrieth RL, Zhou GD, McDonald TJ, Cizmas L, Norton D, Donnelly KC. In situ biomonitoring of PAH-contaminated sediments using juvenile coho salmon (Oncorhynchus kisutch) Ecotoxicol Environ Saf. 2008;71:464–464. doi: 10.1016/j.ecoenv.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: in vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicol Sci. 2001;60:77–91. doi: 10.1093/toxsci/60.1.77. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brett JR. Energetics. In: Groot C, Margolis L, Clarke WC, editors. Physiological ecology of Pacific salmon. UBC Press; Vancouver, Canada: 1995. pp. 3–68. [Google Scholar]
- Cooper RU, Clough LM, Farwell MA, West TL. Hypoxia-induced metabolic and antioxidant enzymatic activities in the estuarine fish Leiostomus xanthurus. Journal of Experimental Marine Biology and Ecology. 2002;279:1–20. [Google Scholar]
- Dexter R, Goldstein L, Chapman P, Quinlan E. Administration, N.O., Atmospheric, editor. Temporal trends in selected environmental parameters monitored in Puget Sound. 1985. NOAA technical memorandum OMA-19.
- Di Giulio RT, Hinton D. The Toxicology of Fishes. Taylor and Francis; 2008. [Google Scholar]
- EPA, U. SW-846, Test methods for evaluating solid waste, physical/chemical methods. United States Environmental Protection Agency; Washington, D.C.: 1996a. Method 3510C: Separatory funnel liquid-liquid extraction. [Google Scholar]
- EPA, U. SW-846, Test methods for evaluating solid waste, physical/chemical methods. United States Environmental Protection Agency; Washington, D.C.: 1996b. Method 8270C: Semivolatile organic compounds by gas chromatography/mass spectrometry (GC/MS) [Google Scholar]
- Gallagher EP, LaVire HM, Bammler TK, Stapleton PL, Beyer RP, Farin FM. Hepatic expression profiling in smolting and adult coho salmon (Onchorhynchus kisutch) Environ Res. 2008;106:365–378. doi: 10.1016/j.envres.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher EP, Sheehy KM. Effects of phenytoin on glutathione status and oxidative stress biomarker gene mRNA levels in cultured precision human liver slices. Toxicol Sci. 2001;59:118–126. doi: 10.1093/toxsci/59.1.118. [DOI] [PubMed] [Google Scholar]
- Han E, Muller F, Pérez V, Qi W, Liang H, Xi L, Fu C, Doyle E, Hickey M, Cornell J, Epstein C, Roberts L, Van Remmen H, Richardson A. The in vivo gene expression signature of oxidative stress. Physiol Genomics. 2008;34:112–126. doi: 10.1152/physiolgenomics.00239.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise K, Puntarulo S, Pörtner HO, Abele D. Production of reactive oxygen species by isolated mitochondria of the Antarctic bivalve Laternula elliptica (King and Broderip) under heat stress. Comp Biochem Physiol C Toxicol Pharmacol. 2003;134:79–90. doi: 10.1016/s1532-0456(02)00212-0. [DOI] [PubMed] [Google Scholar]
- Hughes EM, Gallagher EP. Effects of beta-naphthoflavone on hepatic biotransformation and glutathione biosynthesis in largemouth bass (Micropterus salmoides) Mar Environ Res. 2004;58:675–679. doi: 10.1016/j.marenvres.2004.03.061. [DOI] [PubMed] [Google Scholar]
- Iles K, Liu R. Mechanisms of glutamate cysteine ligase (GCL) induction by 4-hydroxynonenal. Free Radic Biol Med. 2005;38:547–556. doi: 10.1016/j.freeradbiomed.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Jönsson E, Abrahamson A, Brunström B, Brandt I. Cytochrome P4501A induction in rainbow trout gills and liver following exposure to waterborne indigo, benzo[a]pyrene and 3,3′,4,4′,5-pentachlorobiphenyl. Aquat Toxicol. 2006;79:226–232. doi: 10.1016/j.aquatox.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Krahn MM, Burrows DG, MacLeod WD, Jr., Malins DC. Determination of individual metabolites of aromatic compounds in hydrolyzed bile of English sole (Parophrys vetulus) from polluted sites in Puget Sound, Washington. Arch Environ Contam Toxicol. 1987;16:511–522. doi: 10.1007/BF01055807. [DOI] [PubMed] [Google Scholar]
- Krahn MM, Rhodes LD, Myers MS, Moore LK, MacLeod WD, Jr., Malins DC. Associations between metabolites of aromatic compounds in bile and the occurrence of hepatic lesions in English sole (Parophrys vetulus) from Puget Sound, Washington. Arch Environ Contam Toxicol. 1986;15:61–67. doi: 10.1007/BF01055249. [DOI] [PubMed] [Google Scholar]
- LDWG, (Lower Duwamish Waterway Group) Sediment Summary Techical Document. Seattle, WA: 2001. Duwamish Waterway REmedial Investigation. [Google Scholar]
- Li M, Jang J, Na H, Cha Y, Surh Y. Carbon monoxide produced by heme oxygenase-1 in response to nitrosative stress induces expression of glutamate-cysteine ligase in PC12 cells via activation of phosphatidylinositol 3-kinase and Nrf2 signaling. J Biol Chem. 2007;282:28577–28586. doi: 10.1074/jbc.M701916200. [DOI] [PubMed] [Google Scholar]
- Mabon N, Moorthy B, Randerath E, Randerath K. Monophosphate 32P-postlabeling assay of DNA adducts from 1,2:3,4-diepoxybutane, the most genotoxic metabolite of 1,3-butadiene: in vitro methodological studies and in vivo dosimetry. Mutat Res. 1996;371:87–104. doi: 10.1016/s0165-1218(96)90098-1. [DOI] [PubMed] [Google Scholar]
- Machala M, Drabek P, Neca J, Kolarova J, Svobodova Z. Biochemical markers for differentiation of exposures to nonplanar polychlorinated biphenyls, organochlorine pesticides, or 2,3,7, 8-tetrachlorodibenzo-p-dioxin in trout liver. Ecotoxicol Environ Saf. 1998;41:107–111. doi: 10.1006/eesa.1998.1675. [DOI] [PubMed] [Google Scholar]
- Malins D, McCain B, Brown D, Chan S, Myers M, Landahl J, Prohaska P, Friedman A, Rhodes L, Burrows D, Gronlund W, Hodgins H. Chemical-Pollutants in Sediments and Diseases of Bottom-dwelling Fish in Puget Sound, Washington. Environmental Science & Technology. 1984;18:705–713. [Google Scholar]
- Malins DC, Krahn MM, Brown DW, Rhodes LD, Myers MS, McCain BB, Chan SL. Toxic chemicals in marine sediment and biota from Mukilteo, Washington: relationships with hepatic neoplasms and other hepatic lesions in English sole (Parophrys vetulus) J Natl Cancer Inst. 1985;74:487–494. [PubMed] [Google Scholar]
- Martinez-Alvarez RM, Hidalgo MC, Domezain A, Morales AE, Garcia-Gallego M, Sanz A. Physiological changes of sturgeon Acipenser naccarii caused by increasing environmental salinity. J Exp Biol. 2002;205:3699–3706. doi: 10.1242/jeb.205.23.3699. [DOI] [PubMed] [Google Scholar]
- Matsuo A, Gallagher E, Trute M, Stapleton P, Levado R, Schlenk D. Characterization of Phase I biotransformation enzymes in coho salmon (Oncorhynchus kisutch) Comp Biochem Physiol C Toxicol Pharmacol. 2008;147:78–84. doi: 10.1016/j.cbpc.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCain BB, Malins DC, Krahn MM, Brown DW, Gronlund WD, Moore LK, Chan SL. Uptake of aromatic and chlorinated hydrocarbons by juvenile chinook salmon (Oncorhynchus tshawytscha) in an urban estuary. Arch Environ Contam Toxicol. 1990;19:10–16. doi: 10.1007/BF01059807. [DOI] [PubMed] [Google Scholar]
- McKinney MA, De Guise S, Martineau D, Beland P, Arukwe A, Letcher RJ. Biotransformation of polybrominated diphenyl ethers and polychlorinated biphenyls in beluga whale (Delphinapterus leucas) and rat mammalian model using an in vitro hepatic microsomal assay. Aquat Toxicol. 2006;77:87–97. doi: 10.1016/j.aquatox.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Meador JP, Ylitalo GM, Sommers FC, Boyd DT. Bioaccumulation of polychlorinated biphenyls (PCBs) in juvenile chinook salmon (Oncorhynchus tshawytscha) outmigrating through a contaminated urban estuary. Dynamics and application. Ecotox. 2009 doi: 10.1007/s10646-009-0399-x. In press. [DOI] [PubMed] [Google Scholar]
- Meador JP, Buzitis J, Bravo C. Using fluorescent aromatic compounds (FACs) in bile from juvenile salmonids to determine exposure to polycyclic aromatic hydrocarbons. Env Tox Chem. 2008;27:845–853. doi: 10.1897/07-434.1. [DOI] [PubMed] [Google Scholar]
- Meador JP, Sommers FC, Ylitalo GM, Sloan CA. Altered growth and related physiological responses in juvenile chinook salmon (Oncorhynchus tshawytscha) from dietary exposure to polycyclic aromatic hydrocarbons (PAHs) Can J Fish Aquat Sci. 2006;63:2364–2376. [Google Scholar]
- Missildine BR, Peters RJ, Chin-Leo G, Houck D. Polychlorinated biphenyl concentrations in adult chinook salmon (Oncorhynchus tshawytscha) returning to coastal and Puget Sound hatcheries of Washington State. Environ Sci Technol. 2005;39:6944–6951. doi: 10.1021/es0506408. [DOI] [PubMed] [Google Scholar]
- Mortensen AS, Braathen M, Sandvik M, Arukwe A. Effects of hydroxy-polychlorinated biphenyl (OH-PCB) congeners on the xenobiotic biotransformation gene expression patterns in primary culture of Atlantic salmon (Salmo salar) hepatocytes. Ecotoxicol Environ Saf. 2007 doi: 10.1016/j.ecoenv.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Myers M, Stehr C, Olson O, Johnson L, McCain B, Chan S, Varanasi U. Relationships between toxicopathic hepatic lesions and exposure to chemical contaminants in English sole (Pleuronectes vetulus), starry flounder (Platichthys stellatus), and white croaker (Genyonemus lineatus) from selected marine sites on the Pacific Coast, USA. Environ Health Perspect. 1994;102:200–215. doi: 10.1289/ehp.94102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard-Aitken M, Fournier H, Pariseau R, Marcoglies D, Cyr D. Thyroid disruption in walleye (Sander vitreous) exposed to environmental contaminants: cloning and use of iodthyronine okay deiodinases as molecular biomarkers. Aquatic toxicol. 2007;83:200–211. doi: 10.1016/j.aquatox.2007.04.004. [DOI] [PubMed] [Google Scholar]
- PSAT . In: 2007 Puget Sound Update: Ninth Report of the Puget Sound Assessment and Monitoring Program. Team, P.S.A., editor. Puget Sound Action Team; Olympia, WA: 2007a. p. 260. [Google Scholar]
- PSAT, P.S.A.T. In: 2007 Puget Sound Update: Ninth Report of the Puget Sound Assessment and Monitoring Program. Team, P.S.A., editor. Olympia, WA: 2007b. p. 260. [Google Scholar]
- Randerath K, Reddy MV, Disher RM. Age- and tissue-related DNA modifications in untreated rats: detection by 32P-postlabeling assay and possible significance for spontaneous tumor induction and aging. Carcinogenesis. 1986;7:1615–1617. doi: 10.1093/carcin/7.9.1615. [DOI] [PubMed] [Google Scholar]
- Rice CA, Myers MS, Willis ML, French BL, Casillas E. From sediment bioassay to fish biomarker--connecting the dots using simple trophic relationships. Mar Environ Res. 2000;50:527–533. doi: 10.1016/s0141-1136(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Oris JT, Stubblefield WA. Gene expression in caged juvenile Coho Salmon (Oncorhynchys kisutch) exposed to the waters of Prince William Sound, Alaska. Mar Pollut Bull. 2006;52:1527–1532. doi: 10.1016/j.marpolbul.2006.05.016. [DOI] [PubMed] [Google Scholar]
- SAS . Statview Statistical software. SAS Institute Inc SAS Campus Drive; Cary, N.C.: 1998. SAS Institute, Inc. [Google Scholar]
- Schlenk D, Handy R, Steinert S, Depledge M, Benson W. Biomarkers. In: Di giulio RT, Hinton DE, editors. The Toxicology of Fishes. CRC Press; Boca Raton, FL: 2008. pp. 683–715. [Google Scholar]
- Spromberg JA, Meador JP. Relating results of chronic toxicity responses to population-level effects: modeling effects on wild Chinook salmon populations. Integr Environ Assess Management. 2005;1:9–21. doi: 10.1897/ieam_2004a-005.1. [DOI] [PubMed] [Google Scholar]
- Stein J, Hom T, Collier T, Brown D, Varanasi U. Contaminant exposure and biochemical effects in outmigrant juvenile Chinook salmon from urban and nonurban estuararies of Puget Sound, Washington. Environmental Toxicology and Chemistry. 1995;14:1019–1029. [Google Scholar]
- Thimmulappa R, Rangasamy T, Alam J, Biswal S. Dibenzoylmethane activates Nrf2-dependent detoxification pathway and inhibits benzo(a)pyrene induced DNA adducts in lungs. Med Chem. 2008;4:473–481. doi: 10.2174/157340608785700199. [DOI] [PubMed] [Google Scholar]
- Varanasi U, Casillas E, Arkoosh MR, Hom T, Misitano DA, Brown DW, Chan S-L, Collier TK, McCain BB, Stein JE. Contaminant Exposure and Associated Biological Effects in Juvenile Chinook Salmon (Oncorhynchus tshawytscha) from Urban and Nonurban Estuaries of Puget Sound. National Marine Fisheries Service; Seattle WA: 1993. [Google Scholar]
- Varanasi U, Reichert W, Stein J. 32P-postlabeling analysis of DNA adducts in liver of wild English sole (Parophrys vetulus) and winter flounder (Pseudopleuronectes americanus) Cancer Res. 1989;49:1171–1177. [PubMed] [Google Scholar]
- Whyte JJ, Jung RE, Schmitt CJ, Tillitt DE. Ethoxyresorufin-O-deethylase (EROD) activity in fish as a biomarker of chemical exposure. Crit Rev Toxicol. 2000;30:347–570. doi: 10.1080/10408440091159239. [DOI] [PubMed] [Google Scholar]
- Zhou G, Hernandez N, Randerath E, Randerath K. Acute elevation by short-term dietary restriction or food deprivation of type I I-compound levels in rat liver DNA. Nutr Cancer. 1999;35:87–95. doi: 10.1207/S1532791487-95. [DOI] [PubMed] [Google Scholar]