Abstract
The glutathione S-transferases (GSTs) are a multifunctional family of phase II enzymes that detoxify a variety of environmental chemicals, reactive intermediates, and secondary products of oxidative damage. GST mRNA expression and catalytic activity have been used as biomarkers of exposure to environmental chemicals. However, factors such as species differences in induction, partial analyses of multiple GST isoforms, and lack of understanding of fish GST gene regulation, have confounded the use of GST as markers of pollutant exposure. In the present study, we examined the effect of exposure to cadmium (Cd), a prototypical environmental contaminant and inducer of mammalian GST, on GST mRNA expression in coho salmon (Oncorhynchus kisutch) liver, gill, and olfactory tissues. GST expression data were compared to those for metallothionein (MT), a prototypical biomarker of metal exposure. Data mining of genomic databases led to the development of quantitative real-time PCR (qPCR) assays for salmon GST isoforms encompassing 9 subfamilies, including alpha, mu, pi, theta, omega, kappa, rho, zeta and microsomal GST. In vivo acute (8-48 hr) exposures to low (3.7 ppb) and high (347 ppb) levels of Cd relevant to environmental scenarios elicited a variety of transient, albeit minor changes (<2.5-fold) in tissue GST profiles, including some reductions in GST mRNA expression. In general, olfactory GSTs were the earliest to respond to cadmium, whereas, more pronounced effects in olfactory and gill GST expression were observed at 48 hr relative to earlier time points. Although evaluation of GSTs reflected a cadmium-associated oxidative stress response, there was no clear GST isoform in any tissue that could serve as a reliable biomarker of acute cadmium exposure. By contrast, metallothionein (MT) mRNA was consistently and markedly induced in all three tissues by cadmium, and among the tissues examined, olfactory MT was the most sensitive marker of cadmium exposures. In summary, coho salmon exhibit a complex GST tissue profile consisting of at least 9 isoforms, all of which are present in the peripheral olfactory system. Short-term exposure to environmental levels of Cd causes transient changes in salmon GST consistent with oxidative stress, and in some cases, includes a loss of GST. In a biomarker context, however, monitoring of tissue MT mRNA expression, especially in the peripheral olfactory system, may be of greater utility for assessing short-term environmental exposures to cadmium.
Keywords: glutathione S-transferases, cloning, quantitative PCR, coho salmon, olfaction, cadmium, liver, gills
1.0 Introduction
The glutathione S-transferases (GSTs) are a phase II detoxification enzyme family that can mitigate the cellular toxicity of a number of endogenous and environmental chemicals by facilitating nucleophilic attack by reduced glutathione (GSH) (Hayes et al., 2005). Presently, 14 classes of mammalian GST have been identified based on primary amino acid sequences, and include alpha, mu, pi, sigma, theta, omega, kappa, and zeta GSTs (Hayes et al., 2005). In addition, membrane bound microsomal GSTs are a separate GST subfamily belonging to the MAPEG (Membrane Associated Proteins in Eicosanoid and Glutathione metabolism) pathway (Hayes et al., 2005). Although fish GSTs have not been well characterized relative to their mammalian counterparts, all fish species examined to date exhibit tissue GST catalytic activity and express multiple soluble hepatic GST isoforms, some of which share structural similarity to the rodent GSTs (Schlenk et al., 2008). Of the various isoforms, the predominant GST in cyprinids, salmonids and gadoids is a pi class GST, whereas the major isoform in flatfish, mullet, carp, sea bream and bass is a rho class GST that is expressed only in aquatic species (Konishi et al., 2005; Leaver et al., 1993; Schlenk et al., 2008).
The majority of aquatic studies have been directed toward using GSTs as biomarkers of exposure to environmental chemicals (Schlenk et al., 2008). This is because an important aspect of GSTs is the inducibility of some isozymes upon exposure to pollutants. When enzymatic activity is used as an endpoint, a modest induction, or even repression (2-fold or less), of overall GST activity is typically observed in fish exposed to prototypical GST inducing agents in the laboratory (Schlenk et al., 2008). However, GST enzyme activities integrate multiple isoforms and the effect of chemical exposures on certain GST isoforms is not easily distinguished by enzymatic analysis. Accordingly, field GST biomarker studies have yielded inconsistent results ranging from either a clear induction of GST isoforms and catalytic activity in response to pollutants (Blahova et al., 2010; Crago et al., 2011; Kim et al., 2010; Nahrgang et al., 2009; Pathiratne and Hemachandra, 2010), no clear observable effects (Kim et al., 2010) or a depression of GST activity (Browne et al., 2010; Ferreira et al., 2010; Toni et al., 2010). Factors contributing to these variable GST responses include species-, tissue-, and isoform-associated differences in GST induction (Hayes et al., 2005; Hayes et al., 1991a; Hayes et al., 1991b), and also the fact that GST regulation is a complex phenomenon that can be influenced by hormones, exposure to natural toxins, phytochemicals and environmental chemicals (Hayes and Pulford, 1995). Some, but not all, GST genes contain an antioxidant response element (ARE, also known as electrophile response element (EpRE) (Hayes et al., 2005; Hayes et al., 1991b) in their promoters which can be activated by natural and synthetic chemicals. These chemical inducers trigger a protective cellular antioxidant response involving GSTs and other antioxidant enzymes (i.e. GSH biosynthetic genes, NADPH quinone reductase) via activation of a master transcriptional regulator nuclear factor-erythroid 2 p45-related factor 2 (Nrf2) (Casalino et al., 2007). Cadmium is a classic Nrf2 agonist that induces mammalian GSTs (Casalino et al., 2007; He et al., 2008; Simmons et al., 2011) and is of relevance to examine induction of GSTs in the context of aquatic pollution (Durou et al., 2007; Kim et al., 2010; Moreira and Guilhermino, 2005; Suzuki et al., 2005; Won et al., 2011).
In the present study, we used bioinformatic approaches to identify candidate coho salmon (Oncorhynchus kisutch) GST genes and develop quantitative real time PCR (qPCR) assays to discriminate the various GST mRNAs in response to cadmium. Coho salmon are experiencing population declines and receive chemical exposures during migration through urban and agricultural waterways containing metals, pesticides, herbicides, and persistent pollutants (Hoffman et al., 2000). In addition to their utility as an ecological indicator, coho are easily cultured in the laboratory and have a three-year lifecycle. Juvenile coho were exposed for 8, 24 and 48 hrs to two environmentally relevant concentrations of cadmium, including a low concentration (3.7 ppb) below federal drinking water guidelines of 5 ppb (USEPA; http://water.epa.gov/drink/contaminants/basicinformation/cadmium.cfm) and a higher concentration (347 ppb) within the upper range of environmental exposures associated with highly polluted waterways (Maceda-Veiga et al., 2011; Srinivasa Gowd and Govil, 2008) and below ppm cadmium levels often employed in toxicological studies. GST expression was analyzed in three target organ systems, including the liver, gills and peripheral olfactory system, to evaluate tissue differences in responses and identify particularly sensitive tissues for subsequent biomarker studies. The olfactory rosettes are the major component of the peripheral olfactory system, which is in direct contact with the surrounding water. Similar to the gills, the olfactory rosettes are highly vulnerable to the toxic effects of dissolved pollutants, but are relatively understudied with regards to antioxidant defenses. By contrast, the liver represents the major organ for detoxification and is a target of cadmium (Khoshnood et al, 2010; Vinodhini and Narayanan, 2008; Malik et al, 2009). We compared our results with those using the classic metal biomarker metallothionein (MT), which encodes a protein within a family of cysteine-rich, low molecular weight proteins that play an important role in metal transport and storage (Coyle et al., 2002; Vasak, 2005) and which has been shown to be highly sensitive to induction in liver tissues of fish exposed to Cd and to other metals (Chen et al., 2007; Man and Woo, 2008; Roberts and Oris, 2004; Woo et al., 2006).
2.0 Materials and Methods
2.1 Chemicals
MS-222 (Tricaine methanesulfonate) was obtained from Argent Chemical Laboratories (Redmond, WA) whereas analytical grade cadmium chloride was purchased from Mallinckrodt Baker (Phillipsburg, NJ). RNeasy® mini kit was purchased from Qiagen (Valencia, CA). TRIzol® reagent and the SuperScript® First-Strand Synthesis System were purchased from Invitrogen (Carlsbad, CA). Finnzymes® DyNAmo® SYBR Green 2-Step qPCR Kit was purchased from New England Biolabs, Inc. (Ipswich, MA). The qPCR primers were obtained from Eurofins MWG Operon (Huntsville, AL). All other chemicals and solvents were of analytical grade and purchased from standard sources.
2.2 Animals and exposures
Juvenile coho salmon (1 yr of age, 15.0 g ± 5.7 gm) were provided by the National Marine Fisheries Service Northwest Fisheries Science Center, Seattle, Washington. Fish were raised in cylindrical tanks in recirculating freshwater at 10-12°C in dechlorinated municipal water under a natural photoperiod. The fish were fed Bio Vita Fry Feed (Bio-Oregon Inc, OR) and water quality conditions were typically 80-120 mg/L total hardness as calcium carbonate, pH 7.4 ± 0.2, and 8.1 mg/L dissolved oxygen content. Twenty four hours prior to initiation of exposures, the juvenile coho were transferred to 120 L aquaria receiving filtered city water. Water quality conditions were monitored during experiments and were pH 7.4 ± 0.1, 12 ± 1 °C, alkalinity 50 ppm, hardness 80 ppm, dissolved oxygen 8.6 ± 0.6 mg/L. For the exposures, 8-10 juvenile coho per treatment were exposed to the intended concentrations of 3.1 and 310 ppb cadmium for 8, 24, and 48 hrs in 120 L aquaria with an equal number of control animals receiving carrier (water). Total cadmium analysis and water samples were conducted pre- and post-exposure in all treatment groups with sample collections following a 24 hr static renewal protocol (Frontier GeoSciences Inc. Seattle, WA) and are presented in the supplementary material (Appendix 1). Measured waterborne cadmium concentrations in the tanks at the initiation of exposures were 3.7 and 347 ppb in the low and high dose exposure tanks, respectively. Hereafter, measured cadmium concentrations are used in all text, tables and figures. The aquaria were contained within a large chilled re-circulating water bath and received aeration during the static exposures. Exposures were staggered for each of the three experimental periods (8, 24 and 48 hrs) so that equal numbers of fish were continually exposed for the treatments and to avoid ammonia buildup. Nitrate and nitrite levels were below detection for all treatment groups, and ammonia levels were <0.25 ppm. A 90% static renewal approach was implemented with water containing cadmium replaced after a 24 hour period.
2.3 Tissue processing, RNA isolation and first strand cDNA synthesis
Following exposures, fish were anesthetized with tricaine methanesulfonate (MS-222) prior to cervical dislocation. The olfactory rosettes, gill arches, and liver tissues were then quickly excised and rinsed in PBS, transferred to TRIzol® reagent, frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated and the samples were cleaned using the RNeasy® Mini Kit from Qiagen (Valencia, CA) prior to analysis on a Nanodrop spectrophotometer (Thermo Scientific, Wilmington DE) to ensure integrity of RNA. Total first strand cDNA was synthesized from DNAse treated RNA samples using the SuperScript® First-Strand Synthesis System (Invitrogen®, Carlsbad, CA).
2.4 Data mining and development of qPCR assays
Candidate GSTs from genomic databases were identified by analysis of conserved GST classes using multiple sequence alignments of rainbow trout (Oncorhynchus mykiss), Atlantic salmon (Salmo salar), and zebrafish (Danio rerio) target sequences. qPCR primer design was accomplished using Oligo ® Software, Version 6.71 (Cascade, CO). All primers were tested to optimize annealing temperatures and ensure single product formation. Amplification products from coho olfactory, liver and gill cDNA were sequenced and verified at the UW Department of Biochemistry Sequencing Facility, and the sequences were compared to other teleost DNA sequences using BLAST in The Gene Index database (TGI, http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/Blast/index.cgi) to confirm amplification of the target gene. The BLAST queries resulted in correct gene hits with BLAST e-values < 10−15. As part of our cloning strategy, we also compared similar sequences with lesser e-values before assigning identities to the target GST genes. To analyze for metal–inducible MT mRNA expression, PCR primer design was accomplished using the corresponding sequenced data from Atlantic salmon (Salmo salar) MT-1A gene (Table 1).
Table 1.
Sequences of PCR primers used in this study.
| Gene | Primer (5′ to 3′) | (Source Species) accession # / reference |
|---|---|---|
| β-actin | Forward: GAC CCA CAC AGT GCC CAT CT | (O. mykiss) AF157514/(Matsuo et al., 2008) |
| Reverse: GTG CCC ATC TCC TGC TCA AA | ||
| GST alpha | Forward: GCT CAC AAG AAG TTT CCG ACA |
(O. mykiss) EZ790503.1; (S. salar) BT046898.1 |
| Reverse: GTC CCC AGG CAC CTT TTA TAC TA | ||
| GST kappa | Forward: CCC AAA CAA GTT CCT GTA TAT GA |
(O. mykiss) BT074032.1; (S. salar) BT043662.1 |
| Reverse: CGT GAC ACC TGC TCT ACC TG | ||
| GST mu | Forward: GAT GAG AAG CAC AAA CTG GG |
(O. mykiss) EZ764534.1 |
| Reverse: AAT CAC AAA GCC GTT CCG GAA G | ||
| GST omega | Forward: TTG GTG GAA ATG CAA TCA CA | (O. mykiss) BT073404.1; (S. salar) BT049799.1 |
| Reverse: GTA AAA TGC CTT GTA GGT CTC CG | ||
| GST pi | Forward: TAT TGT GGG CTA ATG TGT AAG AT |
(O. nerka) AB026119/ (Gallagher et al., 2008) |
| Reverse: CCC TGA AGA GCT TTG TCG | ||
| GST rho | Forward: CTG CTG TGG GGC TCC GGC | (O. mykiss) BT073173.1; (S. salar) BT057296.1 |
| Reverse: ATC TCA AAC ATG CGC TGG | ||
| GST theta | Forward: GGT GAG AAA GGT TCC GGT CA |
(S. salar) BT045588.1 |
| Reverse: GTG TTG CCA AGA CAG GTACTCGT | ||
| GST zeta | Forward: ACA AAG CCC ATT CTT CAT GG | (O. mykiss) EZ805030.1 |
| Reverse: GGC ACT TGT TGC ATA GGG TT | ||
| microsomal GST |
Forward: TAC AGA CAA TTT GCG AAC ATT AT |
(O. mykiss) BT074213; (S. salar) BT048458.2 |
| Reverse: CAC TGA CTA GGC GAG GAC T | ||
| Metallothionein (MT-1A) |
Forward: CAAGTGCTCCAACTGTGCAT | (S. salar) BT059876 |
| Reverse: TACACCAGGCCTCACTGACA | ||
| ribosomal protein L9 |
Forward: AAAAAGCTGCGTGTGGATAAAT | (S. salar) HM4_2302 |
| Reverse: GATCGCATCTTATAGCGGAAAC | ||
| GAPDH | Forward: TCTGTGTTGGAATCAACGGA | (S. salar) BT050071.1; (O. mykiss) AF360980.1 |
| Reverse: TGAAGAAGACTCCGGTGGAC |
After PCR product validation and assay optimization, qPCR assays were conducted in a 96-well format with SYBR Green using the relative standard curve method (Tilton et al., 2008) on a BioRad® IQ5 Real-Time PCR Detection System (Hercules, CA) under the optimal PCR conditions for each primer set. PCR amplifications were performed for 35 cycles with denaturation at 94°C for 10s, annealing at optimum temperature for primers (55-58°C) for 20s, and extension at 72°C for 30s followed by melt curve analysis. Standards for mRNA quantification were created from gel-purified qPCR products using QIAX II kit (Qiagen Inc.) and quantified before serial dilution from 0.001-100 pg DNA. Several salmon housekeeping genes were investigated and optimized in qPCR assays, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ß -actin and ribosomal protein L9 (rpl9). Stability analysis by geNorm (version 3.5) software (http://medgen.ugent.be/~jvdesomp/genorm/) indicated that ß-actin showed the greatest stability among tissues and treatments (data not shown). Accordingly, GST and MT gene expression in all samples was normalized against β-actin quantities.
2.5 Statistical Analysis
All gene expression data presented represents the mean + standard error (SEM) of triplicate reactions for n=8 individuals for each experimental group unless otherwise indicated. Data were inspected for homogeneity of variances using the Bartlett’s test. Data sets conforming to normal distributions were assessed for significance using a one-way ANOVA followed by a Dunnett’s test. For non-parametric distributions, the effects of cadmium exposure on gene expression were assessed for significance using the Kruskal-Wallis nonparametric one-way analysis of variance test followed by a Dunn’s test. All differences in gene expression were considered statistically significant at p < 0.05. All statistical analyses were conducted using GraphPad Prism Ver 5.0 (Graph Pad Software Inc, San Diego, CA, USA).
3.0 Results
3.1 Quantitative real time PCR analysis of constitutive GST isozyme and MT expression in coho salmon tissues
Data mining for salmonid MT and GST-like DNA sequences revealed the presence of a number of candidate sequences related to alpha, kappa, mu, pi, theta, omega, rho, zeta and microsomal GST classes as well as for MT isoform 1A. Multiple sequence alignments of the candidate sequences allowed for the development of PCR primers that were specific to coho MT and GST genes (Table 1). All products from initial qPCR reactions were sequenced and confirmed to be similar to the corresponding MT and GST class-specific genes (Table 1).
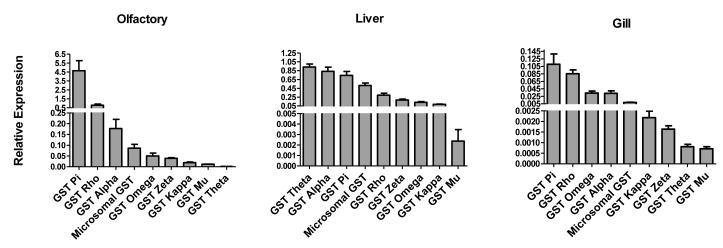
The results of constitutive GST isoform expression analysis in coho liver, olfactory rosettes and gills are shown in Figure 1. Notable differences in constitutive GST expression patterns were observed among the three tissues. For example, pi and rho class GSTs dominated in the olfactory rosettes and gills, whereas theta and alpha class GSTs were the more predominant isoforms in the liver (Figure 1). Among the isoforms, the theta class GST showed the most striking tissue-specific expression. Specifically, GST theta was the dominant GST isoform in liver, but was the least expressed isoform in the gills, and was among the two minor GSTs in the olfactory rosettes (Figure 1).
Figure 1. Tissue-specific constitutive GST isoform expression in juvenile coho salmon.
All gene expression data for the individual GST isoforms was normalized to the expression of β-actin mRNA and data represents the mean ± SEM of olfactory rosettes, liver, and gill tissues from 5 juvenile coho salmon.
3.2 Waterborne cadmium concentrations and fish health during exposures
Waterborne cadmium concentrations in tanks containing juvenile coho were relatively similar at 24 hr of exposure when compared to those at the initiation of exposures (time 0, Appendix 1). No overt cadmium-related effects on fish behavior as assessed by visual inspection of swimming activity, abnormal opercular movements, or failure to maintain buoyancy were noted and all animals appeared healthy upon termination of the experiment.
3.3 Effects of cadmium on olfactory rosette GST expression
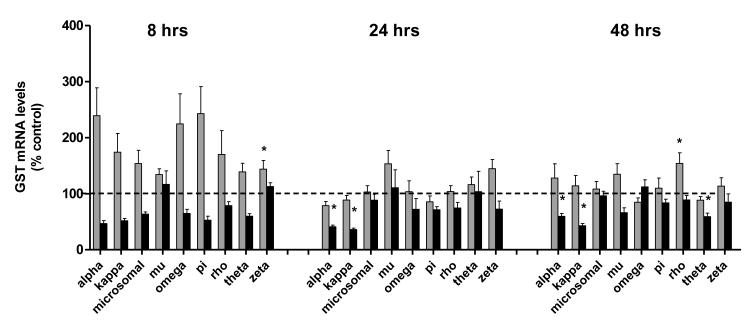
The results of cadmium on olfactory rosette GSTs are presented graphically in Figure 2. Exposure to 3.7 ppb caused transient increases (<2.5-fold) in the expression of all 9 olfactory rosette GST genes at 8 hrs, including >2-fold increases in the expression in GST alpha, omega, pi and kappa isoforms (Figure 2). However, due to interindividual variation in responses, these responses were not statistically significant. An increase in GST zeta expression was the only statistically significant effect of cadmium on olfactory GSTs observed at low environmental cadmium exposures at 8 hr. As opposed to the low 3.7 ppb cadmium concentration, exposure to 347 ppb cadmium elicited a consistent loss in several olfactory GSTs that was observed over the time course of the exposures (Figure 2). In particular, the expression of olfactory alpha, theta, and kappa GSTs were significantly reduced by 48 hrs (40%, 41 %, and 57% losses respectively, Figure 2). As observed in Figure 2, four of the 9 olfactory GST isoforms were significantly dysregulated by cadmium exposure at 48 hr, suggesting a cumulative response from exposures.
Figure 2. Effects of cadmium on olfactory GST expression.
Gene expression of 9 GST isoforms, including; pi, rho, alpha, omega, microsomal GST, zeta, kappa, mu, and theta, in the olfactory rosettes of coho salmon exposed to 0, 3.7, and 347 ppb cadmium for 8, 24 and 48 hr periods. The 3.7 ppb and 347 ppb datasets are represented by light and dark bars, respectively. Data represent the mean ± SEM of GST isoform expression normalized to β-actin from 8 juvenile coho in each treatment group and are presented as percent-change relative to the corresponding controls. Asterisks indicate statistically significant differences compared to control samples (p <0.05).
3.4 Effects of cadmium on gill and liver GST expression
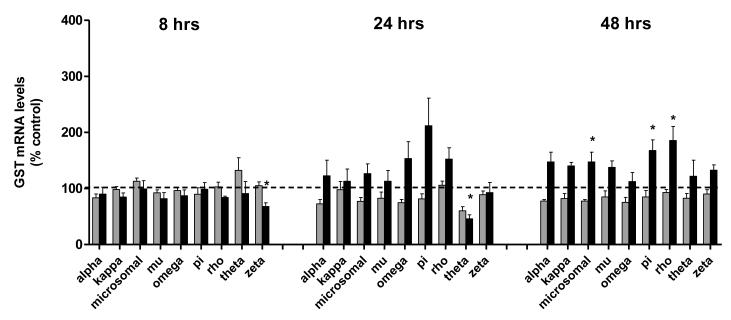
Gill GSTs were somewhat less responsive than those in the olfactory rosettes with regards to modulation by cadmium (Figure 3). For example, none of the 9 gill GSTs were significantly dysregulated by the low dose of cadmium at 8 hr, and GST zeta, an isoform that was significantly induced in the olfactory rosettes, was repressed in the gills at the early time point in the group receiving 347 ppb cadmium. As in the olfactory system, more pronounced effects were observed on gill GSTs on continued exposure, and by 48 hr, a trend toward gill GST induction was observed. For example, by 48 hrs, gill rho, pi, and microsomal GSTs were significantly increased in fish exposed to 347 ppb cadmium (Figure 3).
Figure 3. Effects of cadmium exposure on gill GSTs.
Gene expression of 9 GST isoforms, including; pi, rho, omega, alpha, microsomal, kappa, zeta, theta, and mu, in the gills of coho salmon exposed to 0, 3.7, and 347 ppb cadmium for 8, 24 and 48 hr periods. The 3.7 ppb and 347 ppb datasets are represented by light and dark bars, respectively. Data represent the mean ± SEM of GST isoform expression normalized to β-actin mRNA from 8 juvenile coho in each treatment group and are presented as percent-change relative to the corresponding controls. Asterisks indicate statistically significant differences compared to control samples (p <0.05).
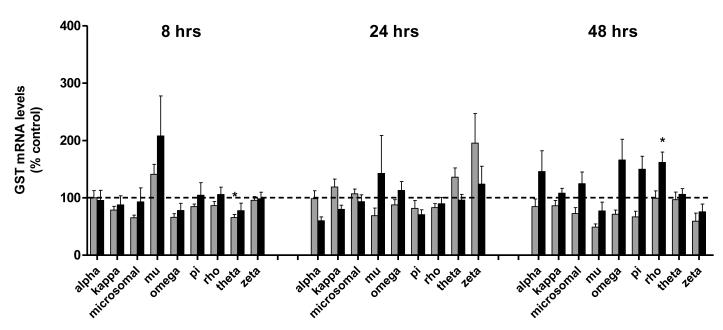
Although some transient effects on the expression of hepatic GST isoforms were noted throughout the exposures, no clear pattern of effects on liver GSTs were observed. For example, liver omega, microsomal, and theta class GSTs were generally decreased on exposure to the lower dose of cadmium (19%, 34% and 35% losses, respectively, Figure 4). However, due to individual variability in responses, only the theta GST significantly differed from that in control coho. A longer exposure stimulated a minor, albeit significant, 1.6- fold increase in liver GST rho expression by 48 hrs (Figure 4).
Figure 4. Effects of cadmium exposure on liver GSTs.
Gene expression of 9 GST isoforms, including; alpha, theta, pi, microsomal GST, rho, zeta, omega, kappa, and mu, in the livers of coho salmon exposed to 0, 3.7, and 347 ppb cadmium for 8, 24 and 48 hr periods. The 3.7 ppb and 347 ppb datasets are represented by light and dark bars, respectively. Data represent the mean ± SEM of GST isoform expression normalized to β-actin mRNA from 8 juvenile coho in each treatment group and are presented as percent-change relative to the corresponding controls. Asterisks indicate statistically significant differences compared to control samples (p <0.05).
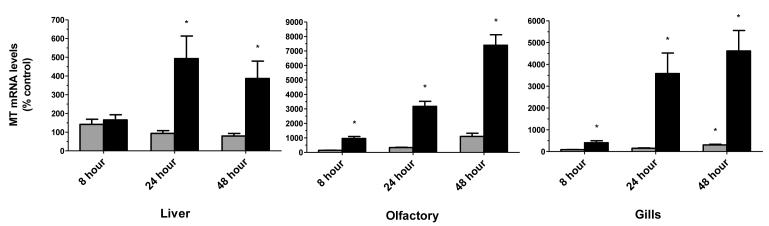
3.5. Metallothionein mRNA as a biomarker of cadmium exposure
Juvenile coho receiving exposures to the low environmental dose of cadmium resulted in 1.4-, 8.9- and 11-fold inductions of olfactory MT mRNA expression at 8, 24 and 48 hours respectively (Figure 5). Exposure to the higher concentration of cadmium more strongly stimulated the olfactory MT response, with 11-, 22-, and 74-fold inductions of olfactory MT mRNA expression occurring at 8, 24, and 48 hours respectively. Similarly, exposures to the lower environmental dose resulted in 1.0-, 1.5-, and 3.1-fold inductions of gill MT mRNA expression at 8, 24, and 48 hours respectively (Figure 5). Moreover, exposure to the higher concentration also resulted in a particularly strong MT induction response in the gills with 4-, 36-, and 46-fold inductions of gill MT mRNA expression occurring at 8, 24, and 48 hours respectively.
Figure 5. Effect of cadmium on metallothionein expression in liver, gills and olfactory rosettes.
The 3.7 ppb and 347 ppb datasets are represented by light and dark bars, respectively. Data represents the mean ± SEM of MT expression normalized to β-actin mRNA from 8 juvenile coho in each treatment group and are presented as percent-change relative to the corresponding controls. Asterisks indicate statistically significant differences compared to control samples (p <0.05).
Conversely, exposures to the low environmental dose of cadmium only resulted in a 1.4-fold induction of liver MT mRNA expression at 8 hours of exposure. Furthermore, at 24 and 48 hours of exposure, the liver MT response was inversely affected with a 6.5% and 21% loss in liver MT mRNA expression respectively (Figure 5). Exposure to the higher concentration of cadmium continued to elicit a MT response, albeit less substantial, with 1.6-, 4.9-, and 3.9-fold inductions of liver MT mRNA expression occurring at 8, 24, and 48 hours respectively. In general, the induction response of MT expression, and in particular in the olfactory rosettes, showed the most dynamic range of all genes examined on response to cadmium exposures (Figure 5).
Discussion
A study from our laboratory previously identified 5 GST protein isoforms from GSH affinity-purified coho liver cytosol that included members of the alpha, mu, pi, theta, and rho class GSTs (Trute et al., 2007). Since that report, advances in salmon genomics have provided a considerable enhancement of genome coverage for which we were able to mine databases for the presence of additional GST isoforms. Utilizing this approach, we identified 4 additional GST isoforms not reported in our original study and which collectively encompassed eight cytosolic classes and one microsomal GST. The fact that we did not detect three cytosolic GST isoforms (omega, theta, and kappa) in our previous report may have been due to a poor ability of these isoforms to bind to a GSH affinity column or to a low constitutive expression, which has been noted for theta class GST in other species (Hayes et al., 2005). Additionally, in our previous study, we reported relatively low recovery of an alpha class GST subunit in coho liver, whereas in the current study we observed that alpha GST mRNA was highly expressed relative to other liver GST mRNAs. This discrepancy could have been due to several factors, including that there are multiple isoforms of the alpha class GST subfamily that can have high structural homology and are difficult to discriminate at the protein level. Accordingly, we cannot ensure that the alpha class GST mRNA measured in the present study encoded the alpha GST protein in our previous report. Alternatively, such differences could be due to the different binding affinities of the GST protein isoforms for the GSH affinity column. The fact that our present study demonstrated the presence of GST mRNAs encoding eight cytosolic subunits as well as an additional microsomal GST suggests a considerable complexity of coho salmon GST expression.
It is noteworthy that the peripheral olfactory system of salmon contained a full complement of GSTs, including isoforms that maintain redox status and protect against cellular oxidative damage. Specifically, pi class GSTs are active in the detoxification of byproducts of oxidative damage, including DNA and RNA hydroperoxides, base propenals, and lipid hydroperoxides (Hayes and Pulford, 1995). GST pi is among the most predominant of the salmon GST isoforms, and is expressed in high amounts in the olfactory epithelium of sockeye salmon, where it has been suggested to serve as a biomarker of olfactory function (Kudo et al., 1999). Similarly, a rho class GST was highly expressed in the coho olfactory rosettes. Recombinant GST rho enzymes from several aquatic species can rapidly detoxify cytotoxic α,β-unsaturated aldehydes (e.g. 4-hydroxynonenal, 4-HNE) generated during lipid peroxidation (Carletti et al., 2008; Doi et al., 2004). Such substrate specificity is important with regards to cell injury, as 4-HNE levels rapidly increase during cellular oxidative stress and lead to the formation of covalent adducts with sulfhydryl groups of glutathione; cysteine, lysine and histidine residues of proteins; and nucleophilic sites of nucleic acids (Chen et al., 2000; Keller et al., 1997; Raza and John, 2006). In addition to the present study, an earlier microarray analysis of zebrafish olfactory tissues containing olfactory rosettes, olfactory bulb and the telencephalon revealed the presence of a variety of antioxidant enzymes, suggesting that the olfactory system is protected against oxidative damage by antioxidant pathways (Tilton et al., 2008). Accordingly, olfactory GSTs may participate with other protective antioxidant enzymes to maintain olfactory signal transduction under conditions of oxidative stress.
While the majority of fish GST biomarker studies have focused on liver GST expression, we found that coho liver GSTs were the least responsive of the three tissues to cadmium. Moreover, induction of MT expression in the liver proved to be a less reliable indicator of short term cadmium exposure. By contrast, olfactory GSTs were a more sensitive indicator of cadmium exposure and were sensitive to a loss of steady-state mRNA expression. In contrast to other aquatic studies that employed much higher levels of cadmium (i.e. ppm), the present study investigated lower doses that are more representative of environmental exposures (Adiele et al., 2010; Kim et al., 2010; Scott et al., 2003; Sloman et al., 2003). However, other reports involving relatively high toxicological cadmium doses (>1 ppm) not employed here suggest that GSTs in aquatic species may not be responsive to cadmium. For example, 24 hr exposure to high cadmium concentrations (up to 11.2 ppm) in Atlantic cod primary hepatocyte cultures elicit only minor (<1.5-fold) increases in GST expression (Softeland et al., 2010). Similar to our study, MT expression responded in a more dose dependent manner to Cd exposure relative to GST (Softeland et al., 2010). In another study, Cao et al. demonstrated a loss of GST activity in the liver and gills of Japanese flounder exposed to ≥ 4 ppm Cd over a 28 day period (Cao et al., 2011). Although the doses employed by Cao et al exceeded those employed by us, the resulting loss of GST is consistent and suggests that cadmium induced oxidative stress may compromise GST expression, as opposed to inducing GSTs, in aquatic organisms.
As opposed to the relatively minor quantitative cadmium-mediated effects on olfactory rosette GSTs, we observed that olfactory MT mRNA was a highly sensitive and reliable dosimeter marker of cadmium, particularly at the higher cadmium dose. Similarly, MT expression in the gills dramatically increased over time at the higher cadmium dose but was less responsive at the lower, more environmentally relevant dose. The observed induction of olfactory MT, concomitant with significant repression of several GST isoforms, suggests that GST detoxification may be compromised during exposure to cadmium induced oxidative stress. These observations suggest that monitoring the expression of GSTs, MT, and potentially other antioxidant enzymes in the olfactory system of fish may have utility in assessing effects of chemical exposures.
Functionally, cellular injury to the peripheral olfactory system of salmon can have consequences with regards to fish survival. This is because the olfactory rosettes are covered with a sensory epithelium containing receptor neurons that detect odorant molecules in the aquatic environment such as reproductive hormones, bile acids, and amino acids (Hara, 1994; Laberge and Hara, 2001). Chemical cues in salmon habitats convey important information about the surrounding environment, including detection of mates, prey capture and predator detection, and migration (Quinn, 2005). The rosettes are in direct contact with the surrounding environment and are thus vulnerable to dissolved contaminants; hence, chemicals that interfere with odorant detection and signal processing can impair olfactory-mediated behaviors crucial for survival. Metals such as cadmium are relevant in this regard as extremely low waterborne concentrations can inhibit olfactory-driven behaviors (Beauvais et al., 2000; Beauvais et al., 2001; Cripe et al., 1984; Little et al., 1990; Morgan and Kiceniuk, 1990; Scholz et al., 2000; Scott et al., 2003).
As discussed, a criterion for using cadmium as a model compound in the present study was its relevance as a gene inducer via activation of the Nrf2 pathway. Whereas this phenomenon has not been extensively explored in aquatic animals, GST pi in zebrafish is regulated by Nrf2 through an ARE-like regulatory element, suggesting conservation of this pathway among vertebrates (Suzuki et al., 2005). It remains to be shown if Nrf2 activators are potent GST inducers in other fish, or if there are species differences in responsiveness. Although we observed transient induction of several GST isoforms, including pi, omega and zeta GST, we did not observe a marked increase in GST induction in any coho tissue reflective of a potent Nrf2 response observed in rodents or in mammalian cells (Hayes et al., 2000; Hayes et al., 2005; Schlenk et al., 2008). It is possible that Nrf2 induction of fish cytosolic GSTs is not active relative to that in mammalian cells, or is less responsive in cold water species such as salmonids. The microsomal GSTs differ from their cytosolic counterparts in that they function as integral membrane proteins and are not typically associated with the Nrf2-responsive GSTs (Hayes and Pulford, 1995). The mGSTs are relatively understudied in aquatic animals, although an induction of liver mGST has been reported in pufferfish (Takifugu obscurus) exposed to high levels of cadmium (5 ppm) (Kim et al., 2010). Common carp (Cyprinus carpio) express at least three mGSTs, including an isoform (GST3) that is inducible by microcystin LR (Fu and Xie, 2006). However, other carp mGSTs were decreased by microcystin LR exposure, similar to our results with cadmium and to a previous field study from our laboratory showing decreased hepatic mGST in juvenile Chinook salmon (Onchorhynchus tshawytscha) exposed to sediment chemicals (Browne et al., 2010). Although we did not analyze for the presence of additional mGSTs in our study, the results of this study suggest that, similar to their cytosolic counterparts, mGST regulation may differ among aquatic species and by chemical classes.
Summary and conclusions
In summary, we have developed qPCR assays to discriminate and analyze GST expression in coho salmon tissues and have observed transient modulation of several GST isoforms by cadmium. Although our results revealed overall perturbations in GST pathway homeostasis by cadmium, we did not clearly identify a particular GST class or isoform that could reliably serve as a biomarker of cadmium exposure. However, it must be pointed out that there were temporal limitations to our study in that we did not investigate chronic cadmium exposures for more than 48 hrs, and for several isoforms, a trend toward an increased GST expression was observed by the end of this experiment. In this regard, it would be of interest to determine if a more dynamic response in GST, or of particular GST isoforms, might occur under chronic exposures representative of field studies. Under the acute exposure conditions of our study, the metal exposure biomarker MT strongly outperformed all of the GST biomarkers examined, although the reduced expression of several GSTs that were concomitant with MT induction provided a more complete analysis of the effects of cadmium on coho antioxidant responses. The fact that the majority of the effects of cadmium on GST and MT occurred in the peripheral olfactory system indicates that this sensory organ should be further investigated in fish biomarker studies. This conclusion is supported by the responsiveness of olfactory MT, which among all genes and tissues analyzed, was the most sensitive marker of cadmium exposure. From a comparative biochemical perspective, although cadmium is a potent Nrf2 activator of GST in mammals, the effects observed in coho suggest that salmon GSTs are not as Nrf2-responsive as mammals. Conversely, a reduction or loss of fish GST, a common occurrence in field and laboratory exposures, is of consideration as this phenomenon can reflect an overwhelming of antioxidant responses that can lead to cell and tissue injury.
Supplementary Material
Appendix 1. Intended and measured cadmium concentrations in water samples.
Highlights of the current research.
Developed qPCR assays to distinguish closely related GST isoforms in salmon.
Examined the effect of cadmium on GST and metallothionein genes in 3 tissues.
Modulation of GST varied among isoforms, tissues, and included a loss of expression.
Metallothionein outperformed, but generally complemented, GSTs as biomarkers.
Salmon olfactory genes were among the most responsive to cadmium
Acknowledgements
This work was supported in part by the University of Washington NIEHS Superfund Basic Sciences Grant NIEHS P42004696. The authors appreciate the technical assistance of Valerie McClain, and also Dr. Brian Beckman and Abby Tillotson at NOAA fisheries who provided the juvenile coho salmon.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adiele RC, Stevens D, Kamunde C. Features of cadmium and calcium uptake and toxicity in rainbow trout (Oncorhynchus mykiss) mitochondria. Toxicol In Vitro. 2010 doi: 10.1016/j.tiv.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Beauvais SL, Jones SB, Brewer SK, Little EE. Physiological measures of neurotoxicity of diazinon and malathion to larval rainbow trout (Oncorhynchus mykiss) and their correlation with behavioral measures. Environmental Toxicology and Chemistry. 2000;19:1875–1880. [Google Scholar]
- Beauvais SL, Jones SB, Parris JT, Brewer SK, Little EE. Cholinergic and behavioral neurotoxicity of carbaryl and cadmium to larval rainbow trout (Oncorhynchus mykiss) Ecotoxicol Environ Saf. 2001;49:84–90. doi: 10.1006/eesa.2000.2032. [DOI] [PubMed] [Google Scholar]
- Blahova J, Havelkova M, Kruzikova K, Hilscherova K, Halouzka R, Modra H, Grabic R, Halirova J, Jurcikova J, Ocelka T, Harustiakova D, Svobodova Z. Assessment of contamination of the Svitava and Svratka rivers in the Czech Republic using selected biochemical markers. Environ Toxicol Chem. 2010;29:541–549. doi: 10.1002/etc.89. [DOI] [PubMed] [Google Scholar]
- Browne E, Kelley M, Zhou GD, He LY, McDonald T, Wang S, Duncan B, Meador J, Donnelly K, Gallagher E. In situ biomonitoring of juvenile Chinook salmon (Onchorhynchus tshawytscha) using biomarkers of chemical exposures and effects in a partially remediated urbanized waterway of the Puget Sound, WA. Environ Res. 2010;110:675–683. doi: 10.1016/j.envres.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Huang W, Shan X, Ye Z, Dou S. Tissue-specific accumulation of cadmium and its effects on antioxidative responses in Japanese flounder juveniles. Environ Toxicol Pharmacol. 2011;33:16–25. doi: 10.1016/j.etap.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Carletti E, Sulpizio M, Bucciarelli T, Del Boccio P, Federici L, Di Ilio C. Glutathione transferases from Anguilla anguilla liver: identification, cloning and functional characterization. Aquat Toxicol. 2008;90:48–57. doi: 10.1016/j.aquatox.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Casalino E, Calzaretti G, Landriscina M, Sblano C, Fabiano A, Landriscina C. The Nrf2 transcription factor contributes to the induction of alpha-class GST isoenzymes in liver of acute cadmium or manganese intoxicated rats: comparison with the toxic effect on NAD(P)H:quinone reductase. Toxicology. 2007;237:24–34. doi: 10.1016/j.tox.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Chen J, Petersen DR, Schenker S, Henderson GI. Formation of malondialdehyde adducts in livers of rats exposed to ethanol: role in ethanol-mediated inhibition of cytochrome c oxidase. Alcohol Clin Exp Res. 2000;24:544–552. [PubMed] [Google Scholar]
- Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, Crispin DA, Goodlett DR, Aebersold R, Bronner MP. Quantitative proteomics analysis reveals that proteins differentially expressed in chronic pancreatitis are also frequently involved in pancreatic cancer. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- Coyle P, Philcox JC, Carey LC, Rofe AM. Metallothionein: the multipurpose protein. Cell Mol Life Sci. 2002;59:627–647. doi: 10.1007/s00018-002-8454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crago J, Corsi SR, Weber D, Bannerman R, Klaper R. Linking biomarkers to reproductive success of caged fathead minnows in streams with increasing urbanization. Chemosphere. 2011;82:1669–1674. doi: 10.1016/j.chemosphere.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Cripe GM, Goodman LR, Hansen DJ. Effect of chronic exposure to EPN and to guthion on the critical swimming speed and brain acetylcholinesterase activity of Cyprinodon variegatus. Aquatic Toxicology. 1984;5:255–266. [Google Scholar]
- Doi AM, Pham RT, Hughes EM, Barber DS, Gallagher EP. Molecular cloning and characterization of a glutathione S-transferase from largemouth bass (Micropterus salmoides) liver that is involved in the detoxification of 4-hydroxynonenal. Biochem Pharmacol. 2004;67:2129–2139. doi: 10.1016/j.bcp.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Durou C, Smith BD, Romeo M, Rainbow PS, Mouneyrac C, Mouloud M, Gnassia-Barelli M, Gillet P, Deutch B, Amiard-Triquet C. From biomarkers to population responses in Nereis diversicolor: assessment of stress in estuarine ecosystems. Ecotoxicol Environ Saf. 2007;66:402–411. doi: 10.1016/j.ecoenv.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Ferreira M, Caetano M, Antunes P, Costa J, Gil O, Bandarra N, Pousao-Ferreira P, Vale C, Reis-Henriques MA. Assessment of contaminants and biomarkers of exposure in wild and farmed seabass. Ecotoxicol Environ Saf. 2010;73:579–588. doi: 10.1016/j.ecoenv.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Fu J, Xie P. The acute effects of microcystin LR on the transcription of nine glutathione S-transferase genes in common carp Cyprinus carpio. L. Aquat Toxicol. 2006;80:261–266. doi: 10.1016/j.aquatox.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Hara TJ. The diversity of chemical stimulation in fish olfaction and gustation. Reviews in Fish Biology and Fisheries. 1994;4:1–35. [Google Scholar]
- Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, Wolf CR, Yamamoto M. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Judah DJ, McLellan LI, Kerr LA, Peacock SD, Neal GE. Ethoxyquin-induced resistance to aflatoxin B1 in the rat is associated with the expression of a novel alpha-class glutathione S-transferase subunit, Yc2, which possesses high catalytic activity for aflatoxin B1-8,9-epoxide. Biochem. J. 1991a;279:385–398. doi: 10.1042/bj2790385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Judah DJ, McLellan LI, Neal GE. Contribution of the glutathione S-transferases to the mechanisms of resistance to aflatoxin B1. Pharmac. Ther. 1991b;50:443–472. doi: 10.1016/0163-7258(91)90053-o. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- He X, Chen MG, Ma Q. Activation of Nrf2 in defense against cadmium-induced oxidative stress. Chem Res Toxicol. 2008;21:1375–1383. doi: 10.1021/tx800019a. [DOI] [PubMed] [Google Scholar]
- Hoffman RS, Capel PD, Larson SJ. Comparison of pesticides in eight US urban streams. Environmental Toxicology and Chemistry. 2000;19:2249–2258. [Google Scholar]
- Keller JN, Mark RJ, Bruce AJ, Blanc E, Rothstein JD, Uchida K, Waeg G, Mattson MP. 4-Hydroxynonenal, an aldehydic product of membrane lipid peroxidation, impairs glutamate transport and mitochondrial function in synaptosomes. Neuroscience. 1997;80:685–696. doi: 10.1016/s0306-4522(97)00065-1. [DOI] [PubMed] [Google Scholar]
- Kim JH, Dahms HU, Rhee JS, Lee YM, Lee J, Han KN, Lee JS. Expression profiles of seven glutathione S-transferase (GST) genes in cadmium-exposed river pufferfish (Takifugu obscurus) Comp Biochem Physiol C Toxicol Pharmacol. 2010;151:99–106. doi: 10.1016/j.cbpc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Konishi T, Kato K, Araki T, Shiraki K, Takagi M, Tamaru Y. A new class of glutathione S-transferase from the hepatopancreas of the red sea bream Pagrus major. Biochem J. 2005;388:299–307. doi: 10.1042/BJ20041578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo H, Ueda H, Mochida K, Adachi S, Hara A, Nagasawa H, Doi Y, Fujimoto S, Yamauchi K. Salmonid olfactory system-specific protein (N24) exhibits glutathione S-transferase class pi-like structure. J Neurochem. 1999;72:1344–1352. doi: 10.1046/j.1471-4159.1999.721344.x. [DOI] [PubMed] [Google Scholar]
- Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res Brain Res Rev. 2001;36:46–59. doi: 10.1016/s0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- Leaver MJ, Scott K, George SG. Cloning and characterization of the major hepatic glutathione S-transferase from a marine teleost flatfish, the plaice (Pleuronectes platessa), with structural similarities to plant, insect and mammalian Theta class isoenzymes. Biochem J. 1993;292(Pt 1):189–195. doi: 10.1042/bj2920189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little EE, Archeski RD, Flerov BA, Kozlovskaya VI. Behavioral indicators of sublethal toxicity in rainbow trout. Archives of Environmental Contamination and Toxicology. 1990;19:380–385. doi: 10.1007/BF01054982. [DOI] [PubMed] [Google Scholar]
- Maceda-Veiga A, Monroy M, de Sostoa A. Metal bioaccumulation in the Mediterranean barbel (Barbus meridionalis) in a Mediterranean River receiving effluents from urban and industrial wastewater treatment plants. Ecotoxicol Environ Saf. 2011;76:93–101. doi: 10.1016/j.ecoenv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Man AK, Woo NY. Upregulation of metallothionein and glucose-6-phosphate dehydrogenase expression in silver sea bream, Sparus sarba exposed to sublethal levels of cadmium. Aquat Toxicol. 2008;89:214–221. doi: 10.1016/j.aquatox.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Moreira SM, Guilhermino L. The use of Mytilus galloprovincialis acetylcholinesterase and glutathione S-transferases activities as biomarkers of environmental contamination along the northwest Portuguese coast. Environ Monit Assess. 2005;105:309–325. doi: 10.1007/s10661-005-3854-z. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Kiceniuk JW. Effect of fenitrothion on the foraging behavior of juvenile Atlantic salmon. Environmental Toxicology and Chemistry. 1990;9:489–495. [Google Scholar]
- Nahrgang J, Camus L, Gonzalez P, Goksoyr A, Christiansen JS, Hop H. PAH biomarker responses in polar cod (Boreogadus saida) exposed to benzo(a)pyrene. Aquat Toxicol. 2009;94:309–319. doi: 10.1016/j.aquatox.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Pathiratne A, Hemachandra CK. Modulation of ethoxyresorufin O-deethylase and glutathione S-transferase activities in Nile tilapia (Oreochromis niloticus) by polycyclic aromatic hydrocarbons containing two to four rings: implications in biomonitoring aquatic pollution. Ecotoxicology. 2010;19:1012–1018. doi: 10.1007/s10646-010-0482-3. [DOI] [PubMed] [Google Scholar]
- Quinn TP. The behavior and ecology of Pacific salmon and trout. University of Washington press; Seattle: 2005. [Google Scholar]
- Raza H, John A. 4-Hydroxynonenal induces mitochondrial oxidative stress, apoptosis and expression of glutathione S-transferase A4-4 and cytochrome P450 2E1 in PC12 cells. Toxicol Appl Pharmacol. 2006;216:309–318. doi: 10.1016/j.taap.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Oris JT. Multiple biomarker response in rainbow trout during exposure to hexavalent chromium. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138:221–228. doi: 10.1016/j.cca.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schlenk D, Celander M, Gallagher E, George S, James M, Kullman S, Van den hurk P, Willett K. Biotransformation in Fishes. In: Di Giulio RT, Hinton DE, editors. The Toxicology of Fishes. CRC press; New York: 2008. pp. 153–234. [Google Scholar]
- Scholz NL, Truelove NK, French BL, Berejikian BA, Quinn TP, Casillas E, Collier TK. Diazinon disrupts antipredator and homing behaviors in chinook salmon (Oncorhynchus tshawytscha) Can. J. Fish. Aquat. Sci. 2000;57:1911–1918. [Google Scholar]
- Scott GR, Sloman KA, Rouleau C, Wood CM. Cadmium disrupts behavioural and physiological responses to alarm substance in juvenile rainbow trout (Oncorhynchus mykiss) J Exp Biol. 2003;206:1779–1790. doi: 10.1242/jeb.00353. [DOI] [PubMed] [Google Scholar]
- Simmons SO, Fan CY, Yeoman K, Wakefield J, Ramabhadran R. NRF2 Oxidative Stress Induced by Heavy Metals is Cell Type Dependent. Curr Chem Genomics. 2011;5:1–12. doi: 10.2174/1875397301105010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloman KA, Baker DW, Ho CG, McDonald DG, Wood CM. The effects of trace metal exposure on agonistic encounters in juvenile rainbow trout, Oncorhynchus mykiss. Aquat Toxicol. 2003;63:187–196. doi: 10.1016/s0166-445x(02)00176-5. [DOI] [PubMed] [Google Scholar]
- Softeland L, Holen E, Olsvik PA. Toxicological application of primary hepatocyte cell cultures of Atlantic cod (Gadus morhua)-effects of BNF, PCDD and Cd. Comp Biochem Physiol C Toxicol Pharmacol. 2010;151:401–411. doi: 10.1016/j.cbpc.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Srinivasa Gowd S, Govil PK. Distribution of heavy metals in surface water of Ranipet industrial area in Tamil Nadu, India. Environ Monit Assess. 2008;136:197–207. doi: 10.1007/s10661-007-9675-5. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Takagi Y, Osanai H, Li L, Takeuchi M, Katoh Y, Kobayashi M, Yamamoto M. Pi class glutathione S-transferase genes are regulated by Nrf 2 through an evolutionarily conserved regulatory element in zebrafish. Biochem J. 2005;388:65–73. doi: 10.1042/BJ20041860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilton F, Tilton SC, Bammler TK, Beyer R, Farin F, Stapleton PL, Gallagher EP. Transcriptional biomarkers and mechanisms of copper-induced olfactory injury in zebrafish. Environ Sci Technol. 2008;42:9404–9411. doi: 10.1021/es801636v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni C, de Menezes CC, Loro VL, Clasen BE, Cattaneo R, Santi A, Pretto A, Zanella R, Leitemperger J. Oxidative stress biomarkers in Cyprinus carpio exposed to commercial herbicide bispyribac-sodium. J Appl Toxicol. 2010;30:590–595. doi: 10.1002/jat.1530. [DOI] [PubMed] [Google Scholar]
- Trute M, Gallis B, Doneanu C, Shaffer S, Goodlett D, Gallagher E. Characterization of hepatic glutathione S-transferases in coho salmon (Oncorhynchus kisutch) Aquat Toxicol. 2007;81:126–136. doi: 10.1016/j.aquatox.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasak M. Advances in metallothionein structure and functions. J Trace Elem Med Biol. 2005;19:13–17. doi: 10.1016/j.jtemb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Won EJ, Kim RO, Rhee JS, Park GS, Lee J, Shin KH, Lee YM, Lee JS. Response of glutathione S-transferase (GST) genes to cadmium exposure in the marine pollution indicator worm, Perinereis nuntia. Comp Biochem Physiol C Toxicol Pharmacol. 2011;154:82–92. doi: 10.1016/j.cbpc.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Woo S, Yum S, Jung JH, Shim WJ, Lee CH, Lee TK. Heavy metal-induced differential gene expression of metallothionein in Javanese medaka, Oryzias javanicus. Mar Biotechnol (NY) 2006;8:654–662. doi: 10.1007/s10126-006-6046-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1. Intended and measured cadmium concentrations in water samples.