Abstract
Coho salmon are a critical Pacific salmon species that undergo complex physiological transformations as they migrate towards seawater and enter adult life stages. During these periods, coho may receive exposure to waterborne pollutants that coincide with outmigration through contaminated waterways and return to natal streams. However, little is known regarding the ontogenic modulation of gene expression during these critical life stages. Accordingly, the purpose of the present study was to characterize the hepatic transcriptome of smolting coho, adult males, and adult females by carrying out microarray analysis with a commercially available 16,000 cDNA element platform. Quantitative PCR (Q-PCR) analysis of genes involved in chemical biotransformation (cytochrome P450 isoforms 1A, and 2M1, glutathione S-transferase pi, microsomal GST), defense against metal exposure (metallothionein-A), and reproductive function (vitellogenin receptor) were developed for the purpose of analyzing specific genes of interest and to validate the microarray data. Microarray analysis identified 842 genes that were differentially expressed between smolts and adult males or females (p<0.001 and more than 2-fold difference). These 842 genes were not differentially expressed between adult males and females and, therefore, can be interpreted as a smolt-specific transcriptional profile. Of these 842 genes, 275 were well annotated and formed the basis for further bioinformatics analysis. Many of the differentially-expressed genes were involved in basic cellular processes related to protein biosynthesis and degradation (24%), ion transport (12%), transcription (8%), cell structure (8%) and cellular energetics (6%). The majority of differentially expressed genes involved in signal transduction and energy metabolism were expressed at higher levels in adult coho relative to smolts. However, genes associated with cellular protection against chemical injury (i.e. biotransformation, DNA damage repair, and protection against oxidative stress) did not generally differ among the groups. Quantitative-PCR studies revealed extensive interindividual variation in mRNA expression, but were highly consistent with the microarray results (R2=0.74). Collectively, our results indicate differences in liver gene expression in young smolting coho salmon relative to adults and extensive interindividual variation in biotransformation gene expression. However, we did not find a global lack of hepatic biotransformation capacity or poor cellular detoxification response capacity in smolting cohos based on mRNA profiles.
Keywords: microarrays, coho salmon, gene expression, biotransformation, liver, developmental differences
1. Introduction
Coho salmon constitute an important ecological, cultural, and economic resource in the Pacific Northwest whose populations have undergone significant declines (Quinn, 2005) associated with the loss of coastal habitat and exposures to environmental chemicals (Wentz et al., 1998). Pacific salmon are most likely to encounter exposures to complex mixtures of pollutants while migrating through urban waterways, and sublethal injury consistent with exposure to PAHs, PCBs, pesticides and trace metals has been observed (Collier et al., 1998). There is also evidence to indicate that exposures to low levels of common waterborne pollutants, including pesticides and trace metals, may negatively impact critical behaviors such as predator detection and avoidance, prey selection, reproductive timing, imprinting and homing behaviors, which are not observed using traditional toxicology testing (Morgan and Kiceniuk, 1990; Scholz et al., 2000).
The physiological basis for chemical susceptibility in aquatic organisms can be complex, and may include life history factors such as age, migration, dietary habits, and physiological status. Relative to other salmonids, coho exhibit a relatively simple three-year life cycle in which adult fish typically spend two growing seasons in the ocean before returning to their natal stream to spawn (Quinn, 2005). The juveniles rear in freshwater for up to 15 months and migrate to the ocean as smolts. The smoltification of juvenile salmon involves morphological, physiological and behavioural changes of the fish from a freshwater-adapted to a salt water-adapted form to allow for downstream migration and seawater entry. Accordingly, the smoltification process allows coho and other salmon to pre-adapt for survival and growth in the marine environment. At the biochemical level, this transformation involves a complex modulation of immune and endocrine factors, as well as changes in gill Na+ K+P ATPase, the latter of which allows for sea water tolerance (Quinn, 2005). These changes involve tremendous changes in the transcriptome of smolt tissues, although generally little is known regarding global hepatic gene expression profiles in smolting coho relative to other life stages.
Juvenile salmon may be at particular risk to chemical injury due to exposures associated with anthropogenic chemicals in urban waterways prior to outmigration to the ocean (Collier et al., 1998). Juvenile salmon may be particularly susceptible to the toxic effects of metals (Hedtke et al., 1982) and organic chemicals such as 4-nonylphenol (Luo et al., 2005). Although there is not an extensive database regarding life stage susceptibility to chemicals in salmonids, the ontogenic expression of chemical biotransformation and detoxification enzymes has been demonstrated to markedly effect susceptibility to toxicity in other species (Chauhan et al., 1991; Anand et al., 2006), including salmonids (Morgan and Kiceniuk, 1990; Schlenk et al., 1995).
The production of a 16,000-gene salmonid microarray platform through the Genomic Resources on Atlantic Salmon Project (GRASP) at the University of Victoria has provided a powerful tool for studying the ontogeny of gene expression in salmonids (Rise et al., 2004). The 16,000 elements present on the platform include approximately 8000 different expressed sequence tags (ESTs) isolated from Atlantic salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss), chinook salmon (Oncorhynchus tshawytscha), sockeye salmon (Oncorhynchus nerka), and lake whitefish (Coregonus clupeaformis) cDNA libraries (Rise et al., 2004). Analyses of cross-species hybridizations to the microarray indicate that this platform is applicable for studies involving all salmonids, including coho (Rise et al., 2004). Several researchers have utilized this platform in rainbow trout for studies directed towards a better understanding of the immunological and reproductive systems (von Schalburg et al., 2005b), and the modes of action of structurally diverse environmental toxicants (Hook et al., 2006a; Hook et al., 2006b).
The purpose of the present study was to use the GRASP microarray to analyze differences in global hepatic gene expression in smolting and adult coho salmon. To this end, it is important to understand if genes involved in biotransformation of environmental chemicals (i.e. phase I and phase II biotransformation enzymes) as well as genes protecting against cell injury (i.e. induction of DNA repair, protection against oxidative stress etc.) are differentially expressed in smolting salmon relative to adults. Our hypothesis was that smolting coho salmon would display quantitatively lower expression of genes which mediate the effects of toxic chemicals relative to adults. A secondary goal of the project was to develop a battery of real-time quantitative PCR assays to analyze the expression of a subset of coho genes involved in chemical biotransformation and detoxification. Our approach was to use pooled samples from a relatively large number of individual fish in microarray experiments to minimize the relatively high costs associated with microarray experiments, and to then use mRNA samples from individual fish to validate the microarray results and observe the extent of interindividual variation in gene expression.
2. Materials and Methods
2.1. Chemicals and Biochemicals
Trizol reagent, superscript first strand synthesis kit, formamide, SSC, Denhardt's solution, CotI DNA, HPLC grade water, TaqMan polymerase, Taq antibody, sequence-specific PCR primers and probes and other molecular biology reagents were purchased from Invitrogen Inc, (Carlsbad, CA). The RNAse inhibitor and MessageAmp Kit were purchased from Ambion, Inc. (Austin, TX). Superscript enzyme, dithiothreitol, dATP, dGTP, dTTP, dCTP were purchased from Millipore Corp. (Bedford, MA). All other chemicals were obtained from Sigma Chemical Co. (St. Louis, MO) or Fisher Scientific (Orlando, FL). The 16K v.1.0 microarray slides were purchased from the Genomic Resources in Atlantic Salmon Project (GRASP) from the University of Victoria (Victoria BC, Canada),
2.2. Animals
Adult male and female coho salmon were 2.5 years of age and were raised in cylindrical tanks at the Wallace Creek fish hatchery near Seattle, WA at 11-12°C under simulated natural photoperiods. The adult females did not appear to be reproductively active based upon visual inspection of the gonads. Juvenile smolting salmon (6 months of age) were raised under similar conditions at the Northwest Fisheries Science Center in Seattle, WA. Fish were fed BioOregon diet, and the water quality conditions for the dechlorinated municipal water were typically 120 mg/L total hardness as measured by CaCO3, pH 6.6, and dissolved oxygen 8.1 mg/L. Twelve-to-fifteen fish from each group were euthanized using MS-222, and the livers were removed washed in PBS and snap frozen in liquid nitrogen.
2.3. RNA isolation, processing and array hybridization
Total RNA was extracted from snap frozen liver from each individual animal using a standard TRIzol procedure (Invitrogen Inc) with the inclusion of 1 μl RNAse inhibitor/sample. Following determination of RNA concentrations by UV absorbance, the integrity of each RNA sample was verified using an Agilent 2100 Bioanalyzer. Samples devoid of significant contamination and RNA degradation (as measured by the ratio of 28S to 18S peaks) were used for microarray analysis. RNA samples from each group were pooled for amplification via the MessageAmp kit according to the manufacturer's protocol. Briefly, total first strand cDNA synthesis was followed by a second strand cDNA synthesis and purification of the double-stranded cDNA products. In vitro transcription was used to generate multiple copies of antisense RNA (aRNA) from the double-stranded cDNA template, and aRNA was purified to improve the stability of the aRNA. The aRNAs were bioanalyzed for integrity before continuing on with microarray analysis.
The array hybridizations were performed in a loop design (Kerr and Churchill, 2001) consisting of six microarrays each comprising equivalent amounts of RNA pooled from the livers of 12-15 animals of each group (i.e. smolting coho, adult females, and adult males) to minimize variation. A loop design (Kerr and Churchill, 2001) was used to array the three pools of RNA samples because it provided the best balance between statistical power and cost effectiveness. Each of the three samples was labeled with both the Cy3 and Cy5 fluorescent dyes and any possible pairwise combination of samples was hybridized to an array requiring a total of six microarrays (smolt-Cy3+female-Cy5; smolt-Cy5+female-Cy3; smolt-Cy3+male-Cy5; smolt-Cy5+male-Cy3; male-Cy3+female-Cy5; male-Cy5+female-Cy3). First-strand cDNA probes were prepared by direct incorporation of CyDye labeled dCTP through reverse transcription of high quality aRNA. Two micrograms of aRNA were reverse transcribed in a 20 μl reaction volume consisting of 200 U Superscript enzyme, 0.01M dithiothreitol, 0.25mM dATP, dGTP, dTTP, and 0.05 mM dCTP, 2.0μg random 9-mers and 2 μg anchored oligo(25)dT, 20 units RNase inhibitor and 1 nmol Cy3 or Cy5 labeled dCTP. The reaction mixture was denatured at 70°C for 10 min, incubated at 42°C for 3 hrs, and the RNA templates degraded by alkaline treatment prior to purification of the single-stranded cDNA probes. The cDNA probes were purified by initially vacuum filtration of the reactions followed by removal of unincorporated nucleotides through a Sephadex G-50 column. To assess purity and labeling efficiency, a full absorption spectrum ranging from 210-700 nm of each fluorescent probe was conducted. Absorption readings at 550 and 650 nm were used to quantify Cy3 and Cy5 incorporation in cDNA probes, respectively. The Cy3 and Cy5-labeled cDNA probes were combined, dried and resuspended in a hybridization solution consisting of 50% formamide, 5X sodium chloride/sodium citrate (SSC), 5X Denhardt's solution, 0.1% SDS, 100μg/ml CotI DNA, and 20μg/ml polyA(72) primer. Probes were denatured by heating to 95°C for 3 min and then placed on ice for 30 sec. The hybridization solution was applied to the microarray slide, covered and incubated in a humid chamber at 42°C for 16 hrs. Following hybridizations, the slides were washed once in 1X SSC/0.2% SDS at 54°C for 10 min., twice in 0.1x SSC/0.2% SDS at 54°C for 10 min., and then twice in 0.1X SSC at room temperature. Slides were then dried and scanned using the ScanArray 5000XL (Packard BioMicroarray Technologies, Billerica, MA).
2.4 Microarray analysis
Statistical analysis and data normalization for the microarray experiments were carried out with R statistical software package that is specific for microarray analysis and also the microarray software analysis program Bioconductor (Gentleman et al., 2004). Within- and between group comparisons were calculated using the limma package in Bioconductor which uses a modified t-test to calculate p-values using an empirical Bayesian method to moderate the standard errors of the estimated log-fold changes (Smyth, 2004). Limma uses variance information from all the genes on the array to arrive at an estimate of per gene variance used in the t-tests. P-values were adjusted for multiplicity with the program q value (Storey and Tibshirani, 2003), which allows for selecting statistically significant genes while controlling the estimated “false discovery rate.” Initially, a p-value of <0.001 was used to identify differentially-expressed genes across the groups. Subsequently, the genes were filtered based upon a 2-fold change cutoff and p value of <0.001 to select gene lists, followed by elimination of non-annotated genes. Genes with altered expression were grouped according to gene ontology (GO) terms provided by the GRASP website http://web.uvic.ca/cbr/grasp/ using the updated version 2 annotation of the 16K microarray. In addition, we identified a number of genes encoding proteins involved in chemical biotransformation (i.e. phase I and phase II biotransformation enzymes) as well as protecting against cell injury (i.e. induction of DNA repair, protection against oxidative stress etc.) which we hypothesized would be expressed at lower levels in smolting salmon relative to adults. The list of GO terms for these genes is provided in table 1, whereas a list of their specific identities, differential expression, GRASP annotation, GenBank accession numbers, and sequences, are available as supplementary material (Appendix I).
Table 1.
Important genes by ontology involved in the detoxification of environmental chemicals present on the GRASP array1.
| Function | GO term or key word |
|---|---|
| Biotransformation | Cytochrome or cytochrome P450 |
| Glutathione transferase | |
| UDP glucuroyltransferase sulfotransferase | |
| Esterase | |
| Epoxide hydrolase | |
| Alcohol dehydrogenase | |
| Aldehyde dehydrogenase | |
| Flavin monooxygenase | |
| Defense against Oxidative stress | Glutathione |
| Superoxide dismutase | |
| Catalase | |
| Thioredoxin | |
| Peroxidase | |
| Heat shock protein | |
| Alcohol | |
| Antioxidant | |
| Metallothionein | |
| DNA repair | DNA repair endonuclease |
| DNA repair exonuclease |
see appendix I for details on the specific genes within these categories and their GRASP accession number
2.5. Real-time quantitative PCR analysis
Snap frozen liver samples (50 mg) from 6 animals of each age and sex group were used to isolate total RNA using TRIzol (Hughes and Gallagher, 2004). Two μg of RNA was used to generate first strand cDNA, which was stored at -20°C until the Q-PCR analysis. Gene-specific primers and probes specific for Coho salmon GST pi, metallothionein-A, CYP2M1, CYP1A, metallothionein-A, and the vitellogenin receptor (VTG-receptor) and microsomal GST were designed against phylogenetically similar species such as rainbow trout, Atlantic salmon, and sockeye salmon using Primer Express™ (Applied Biosystems, Foster City, CA). The resulting PCR products were electrophoretically separated, purified and sequenced. TaqMan® real-time quantitative PCR was performed using 4 μl of 1 μg/μl cDNA, Taq antibody, TaqMan polymerase, and gene-specific primers and probes (table 2.). The sequences were verified for specificity using BLAST software. Because of the extensive homology between salmonid CYP1A1 and CYP1A3 cDNAs, we could not discriminate the two sequences and will subsequently refer to this gene as CYP1A. Standard curves of β-actin were run on each plate to account for inter-plate variability and quantification of each gene of interest was determined by interpolation from the β-actin standard curves. Thermocycling was performed for 40 cycles and the increase in fluorescence during each replication cycle was plotted by the instrument against cycle number. Ct values for a series of standards (0.1 ng-1.0 pg) that were simultaneously obtained using coho β-actin cDNA as PCR template. The resulting standard curve values were generated by plotting Ct versus the log of the amount of cDNA added to the reaction. Products from RT-PCR reactions without reverse transcriptase were included as a control for undesired DNA amplification. Triplicate samples were run for each gene and sample, and the results averaged. The measured relative expression levels for the target genes were divided by the sample's β-actin mRNA level to obtain the normalized mRNA expression values presented in the figures (mean±SD). Differences in gene expression among age groups and sexes for the Q-PCR data were assessed using one-way analysis of variance followed by a Duncan's multiple range test. Differences among groups were considered significant at p<0.05.
Table 2.
Primer pairs and fluorescently labeled probes used in TaqMan analyses
| Gene | Type of oligo | Sequence (5’-3’) | (Species) Genebank Accession Number Position |
|---|---|---|---|
| β-actin | (O mykiss) AF157514 | ||
| primer (forward) | gacccacacagtgcccatct | 528-547 | |
| primer (reverse) | gtgcccatctcctgctcaaa | 767-718 | |
| probe | acggagcgaggctacagcttcacca | 631-655 | |
| CYP1A | (O mykiss) AF059711 | ||
| primer (forward) | agtgctgatggcacagaactcaa | 1441-1463 | |
| primer (reverse) | agctgacagcgcttgtgctt | 1658-1639 | |
| probe | cctcttcttggctatcctgctccaaaggc | 1548-1576 | |
| CYP2M1 | (O mykiss) OMU16657 | ||
| primer (forward) | gctgtatatcacactcacctgctttg | 1811-1836 | |
| primer (reverse) | cccctaagtgctttgcatgtatagat | 2005-1980 | |
| probe | acacctgaaacttttggtcctt | 1918-1897 | |
| GST-pi | (O nerka) AB026119 | ||
| primer (forward) | ctctgctccagttgcctggat | 490-510 | |
| primer (reverse) | gttgccattaatgggcagtttct | 615-593 | |
| probe | agatgtcagcccgtcccaaaatcaagg | 542-568 | |
| mGST | (O mykiss) CF752713 | ||
| primer (forward) | gggtgaggcctgggatga | 522-539 | |
| primer (reverse) | cacaagtacggatgcccacaa | 674-654 | |
| probe | ctttccagctgccattcctgctaccattc | 564-592 | |
| (O mykiss) M81800 | |||
| MT-A | primer (forward) | tggatccttgtgaatgctcca | 2-22 |
| primer (reverse) | ggacagcagtcgcagcaact | 113-94 | |
| probe | ctccaactgcgcatgcaccagttgtaa | 60-86 | |
| VTG- Receptor | (O mykiss) X92804 | ||
| primer (forward) | cagagaggggaggccctgat | 1264-1283 | |
| primer (reverse) | catttgggcagctcctgacat | 1407-1387 | |
| probe | tggcctccaagatcagagcaccattgt | 1348-1322 | |
2.6 Microarray validation by Q-PCR
In addition to providing quantitative data on the levels of gene expression in individual animals, the relative –fold change in gene expression by Q-PCR among different life stages were compared to results of the microarray analysis. In order to make a direct comparison to microarray results, individual Q-PCR results from samples pooled in the micoarray were averaged to obtain a group average. As these results were normalized to β-actin, Q-PCR data were then compared to the microarray results for the corresponding gene(s) on the microarray normalized to the β-actin fold change measured on the microarray. Due to the inclusion of several spots on the microarray representing the same gene product, an average of the expression of the array spots corresponding to the same gene products based upon nucleotide sequence homology was calculated from microarray data. Spots were chosen based on their homology to the gene of interest as measured by alignment to the complete nucleotide sequence of the gene (if known), as well as homology to the QPCR probe. At least 94% homology was attained in most cases. A β-actin sequence on the array was included because it exhibited 91% homology to the complete coho β-actin sequence obtained by PCR. In another case, ESTs on the array corresponding to human microsomal GST shared 81% similarity and were included.
The following spots on the microarray were used for Q-PCR validation of the microarray data: cytochrome P4502M1 (five spots correlating to Q92088 and aligning to U16657), β-actin (two spots correlating to O42161, and two spots correlating to AJ438158, all aligning to AF157514), cytochrome P4501A (one spot each correlating to AAD45967, AF364076, both correlating to AF015660), vitellogenin receptor (one spot correlating to AAL29923), microsomal GST (two spots correlating to NP_002404 and aligning to NM_002413), and GST-pi (two spots correlating to BAA76974 and aligning to AB026119). A Pearson's correlation analysis as well as ANOVA analysis was conducted to obtain a correlation coefficient and p-value for the relationship among microarray and Q-PCR gene expression data.
3. Results
3.1 Microarray analysis
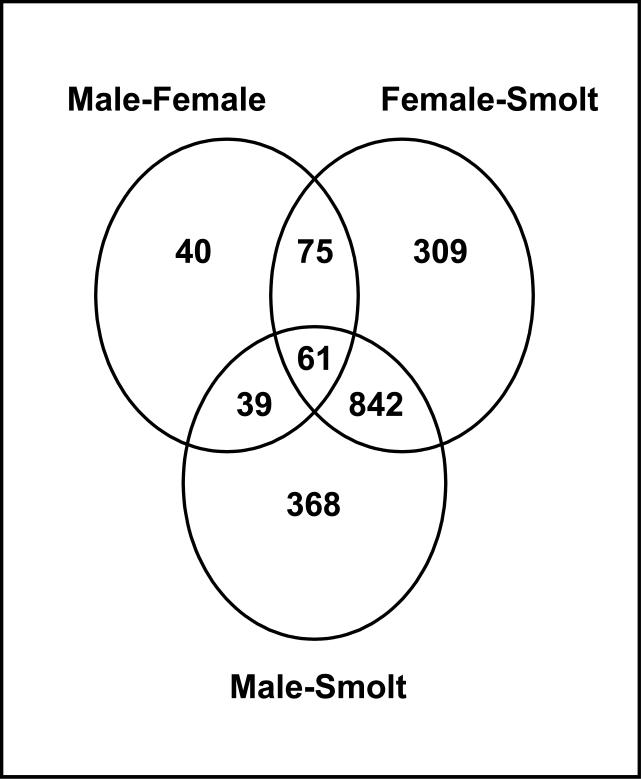
In order to determine a smolt specific transcriptional profile, we identified differential gene expression (p<0.001) between a) smolts and adult males (1310 genes), b) smolts and adult females (1287 genes), and c) adult males and adult females (210). The Venn diagram depicted in figure 1 shows the total number of genes that were differentially expressed when comparing the different groups. It also indicates the number of genes that were specific for each comparison as well as those that were shared between any of the three comparisons. This approach identified 309 and 368 genes that were uniquely differentially expressed between smolts and females and smolts and males respectively. It also identified 842 genes that were differentially expressed between both smolts and adult males and smolts and adult females, but not between adult males and females. An additional 75 genes were differentially expressed among adult males and females only (Figure 1). Since the focus of this study was identification of a smolt specific transcriptional profile, these 75 genes are not further discussed. A complete list of the identities of the differentially expressed genes and their fold change values with associated p values are available as supplemental material (appendix II).
Fig 1.
Venn diagram of the number of differentially expressed genes among adult males, adult females and smolting salmon using a cutoff of p<0.001 only. As observed, the diagram depicts 842 genes that were differentially expressed among males and females relative to smolts, 309 genes that were differentially expressed among females and smolts, 368 genes differentially expressed among males and smolts, and 75 genes that were differentially expressed among males and females.
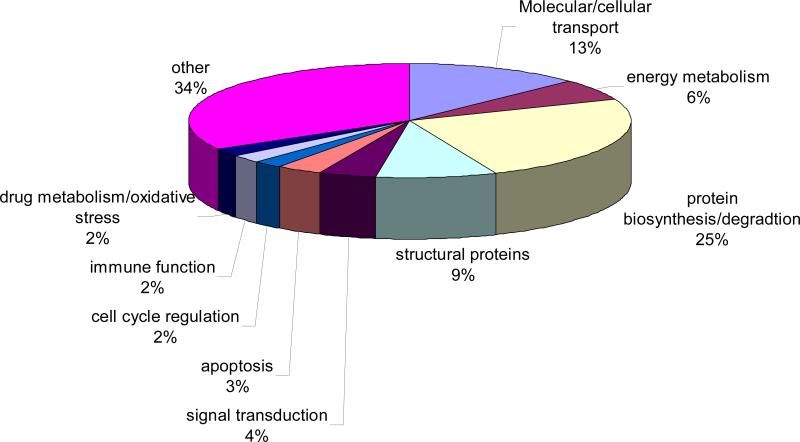
The 842 smolt-specific genes (p<0.001) were further filtered by applying a >2 fold differential expression cutoff and also eliminating poorly annotated genes. This filtering strategy resulted in 275 annotated smolt-specific genes, and figure 2 shows a pie chart summary of the major biological functions of these genes. The biological functions of most of these genes were associated with protein biosynthesis or protein degradation (68 genes, 25%), molecular/cellular transport (35 genes, 13%), energy metabolism (17 genes, 6%), structural proteins (24 genes, 9%), and signal transduction (12 genes, 4%). Other pathways that differed among the age groups to a lesser degree included genes involved in regulating apoptosis (9 genes, 3%), cell cycle regulation (6 genes, 2%), immune function (6 genes, 2%), and a combined category of drug metabolism/oxidative stress (6 genes, 2%). In addition, the 92 annotated genes (34%) that did not clearly belong to any of the aforementioned categories were referred to as “others” (figure 2).
Fig 2.
Pie chart with biological function of annotated genes that were differentially expressed among smolting salmon in adults at p<0.001 and using a 2-fold cutoff value.
In total, 112 genes (41% of the 275 differentially-expressed annotated genes) were expressed at higher levels in smolts relative to adult males and females. Table 3 presents a subset of these genes, including their identities, biological functions and fold changes. Several genes involved in regulation of cell cycle, including calmodulin, chromatin assembly factor 1 subunit C, and 26S proteasome nonATPase regulatory subunit were higher in smolts relative to adult fish (table 3). In addition, 2 genes involved in immune function, including H-2 class histocompatibility antigen L-D alpha chain precursor and CA037346 plasma protease C1 inhibitor precursor were expressed at markedly higher levels in smolts relative to adults (table 3). Several structural proteins including troponin T, dynein light chain 2, and myosin light polypeptide 3 were expressed at least 4 fold higher levels in smolts relative to adult males or adult females. NADH cytochrome b5 reductase, which functions in electron transport, was expressed at markedly higher levels in smolts relative to adults males (>11 fold change, table 3).
TABLE 3.
Sample genes expressed at higher levels in smolting coho relative to adults*
| Accession and Functional Category | Gene name | Smolts vs Females-fold change | Smolt vs males-fold change |
|---|---|---|---|
| Apoptosis | |||
| CA057721 | Caspase-activated deoxyribonuclease (CAD). | 2.12 | 2.51 |
| CA044877 | Cell death activator CIDEB | 2.33 | 2.54 |
| Cell cycle | |||
| CB492422 | Calmodulin (CaM). | 3.16 | 2.23 |
| CB486360 | Chromatin assembly factor 1 subunit C (CAF1 subunit C) | 2.67 | 2.84 |
| CA042758 | EB1 [Ictalurus punctatus] | 2.41 | 2.83 |
| CB498253 | 26S proteasome nonATPase regulatory subunit 8 | 4.95 | 5.25 |
| Immune system defense | |||
| NAC | H2 class I histocompatibility antigen, LD alpha chain precursor. | 25.46 | 21.78 |
| CA037346 | Plasma protease C1 inhibitor precursor (C1 Inh) | 5.86 | 5.37 |
| Drug metabolism and oxidative stress | |||
| CB491960 | Cytochrome P450 2M1 (CYPIIM1) | 8.13 | 8.08 |
| CA036995 | RE56416p (RE65881p). | 4.33 | 6.06 |
| Transcription | |||
| CA037026 | Transcription factor 15 (bHLHEC2 protein) | 2.00 | 2.19 |
| CB488346 | Zinc finger protein 593 (Zinc finger protein T86). | 2.36 | 2.24 |
| CA052340 | Zinc finger protein 239 (Zfp239) | 2.13 | 2.28 |
| CB496981 | DNAdirected RNA polymerases I, II, and III 14.4 kDa polypeptide). | 2.78 | 2.30 |
| CB514260 | Homeobox protein Nkx2.5 | 3.28 | 3.66 |
| CB492800 | Transcription initiation factor IIF, alpha subunit | 4.75 | 4.76 |
| CB510616 | Nuclease sensitive element binding protein 1 (Ybox binding protein1) | 2.06 | 3.16 |
| Structural protein | |||
| CB511888 | Tubulin beta2 chain (Beta2 tubulin). | 3.32 | 3.64 |
| CK990263 | Collagen alpha 2(I) chain precursor. | 2.24 | 2.81 |
| CA049982 | Spectrin alpha chain, brain | 3.74 | 3.20 |
| CB498116 | Troponin T, fast skeletal muscle isoforms. | 14.77 | 11.41 |
| CB492803 | Gamma crystallin B | 2.21 | 2.70 |
| CB510411 | Beta crystallin B1 | 2.58 | 2.39 |
| CA770307 | 40S ribosomal protein S8 | 2.04 | 2.04 |
| CB507561 | Dynein light chain 2 | 5.94 | 4.16 |
| CB497762 | Myosin light polypeptide 3 | 4.29 | 3.94 |
| Transport | |||
| CA053755 | Vacuolar ATP synthase subunit H (VATPase H subunit) | 3.02 | 2.31 |
| CA052539 | Ferritin heavy chain (Ferritin H subunit). | 12.19 | 15.61 |
| CA044952 | AcylCoAbinding protein homolog (ACBP) | 2.19 | 2.03 |
| CN442492 | Cytochrome c oxidase subunit 2 (Cytochrome c oxidase polypeptide II) | 3.25 | 3.53 |
| CB488683 | Probable NADH cytochrome B5 reductase | 12.82 | 10.90 |
| CN442514 | Cytochrome c oxidase subunit 1 (Cytochrome c oxidase polypeptide I). | 2.32 | 2.37 |
| CA054504 | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 4 | 4.27 | 3.98 |
| CK990360 | ATP synthase gamma chain, mitochondrial precursor | 5.44 | 6.68 |
| CB497160 | ATP synthase oligomycin sensitivity conferral protein | 5.72 | 5.88 |
| CB497468 | ProstaglandinH2 D isomerase precursor | 3.75 | 3.24 |
| CA043696 | ADPribosylation factor 1. | 2.22 | 2.15 |
| CB498010 | Mitochondrial import inner membrane translocase subunit Tim17 A | 4.86 | 3.87 |
| CA044887 | Lysosomal associated transmembrane protein 4A | 2.03 | 2.30 |
| CA054321 | Potassium channel tetramerisation domain containing protein 2. | 7.15 | 7.08 |
| Protein biosynthesis | |||
| CB505988 | Probable phenylalanyltRNA synthetase alpha chain | 3.46 | 2.91 |
| CB507602 | Nuclear protein Hcc1. | 3.66 | 4.06 |
| CA061718 | 40S ribosomal protein S26. | 2.12 | 2.41 |
| CB492789 | 60S ribosomal protein L31. | 9.41 | 13.76 |
| CB507058 | large subunit ribosomal protein L36a | 2.42 | 2.99 |
| CB514542 | 60S ribosomal protein L7a. | 4.09 | 4.74 |
| CK990280 | 60S acidic ribosomal protein P1. | 2.21 | 2.19 |
| CA038334 | 60S ribosomal protein L37a. | 2.57 | 2.91 |
| CA046196 | 60S ribosomal protein L36. | 2.75 | 2.94 |
| CA769603 | Ubiquitin. | 5.37 | 5.71 |
| CA044959 | 60S ribosomal protein L22 (Heparin binding protein HBp15). | 3.39 | 2.66 |
| CK991326 | 40S ribosomal protein S7. | 3.06 | 2.26 |
| CA046895 | 60S ribosomal protein L37. | 2.27 | 2.62 |
| CB497256 | 60S ribosomal protein L19. | 2.54 | 3.63 |
| CB492750 | 60S ribosomal protein L7. | 4.26 | 3.42 |
| CB494045 | Eukaryotic translation initiation factor 2 subunit 2 (eIF2beta). | 3.68 | 2.97 |
| CA059038 | Eukaryotic translation initiation factor 3 subunit 5 (eIF3 epsilon) | 4.33 | 3.44 |
| CK990945 | 39S ribosomal protein L46, mitochondrial precursor (L46mt) | 2.21 | 2.15 |
| CA038035 | 60S acidic ribosomal protein P2. | 4.37 | 4.48 |
| CB491302 | Metalloproteinase inhibitor 2 precursor | 2.84 | 3.70 |
| CA051033 | S phase kinase associated protein 1A | 4.32 | 4.16 |
| CB498369 | ribosome associated membrane protein 4 | 3.87 | 2.07 |
table reflects a subset of annotated genes that were expressed at higher levels in smolts relative to adults, p≤0.001
Of the 275 differentially-expressed annotated genes among the two age classes, 59% (163 genes) were expressed at lower levels in smolts relative to adult coho, including males and females. Selected genes from this list are presented in table 4. Some discernible trends were evident. Among the more highly expressed genes in adults relative to smolts were genes associated with cellular energy metabolism. These genes included NADH-ubiquinone reductase 19 kDa subunit (7.7- and 12.7- fold higher levels in adult males and females, respectively), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 9.6- and 11.5- fold higher levels in adult males and females, respectively, table 4). Several genes involved in protein biosynthesis were expressed at markedly higher levels in adults relative to smolts. These genes included ribosomal protein L31 (10.3- and 15.3-fold higher levels in adult males and females, respectively, table 4), ribosomal protein S27 (8.3- and 7.3- fold higher in adult males and females, respectively), and 40S ribosomal protein S24 7.6- and 7.9- fold higher levels in adult females and males, respectively, table 4). Other under-expressed genes in smolts included an actin-like protein 3 (6.3- and 7.9-fold higher levels in adult females and males, respectively), and myosin heavy chain (8.0- and 4.3-fold higher levels in females and males, respectively, table 4.). Overall β-actin mRNA levels did not differ among groups.
TABLE 4.
Genes expressed at lower levels in smolting coho relative to adults*
| Accession | Gene name | Females vs Smolts-fold change | Males vs Smolts fold change |
|---|---|---|---|
| Apoptosis | |||
| CB511941 | Translationally controlled tumor protein (TCTP) | 2.95 | 2.90 |
| CA046385 | Phosphatidylinositol 3-kinase regulatory alpha subunit (PI3-kinase p85-alpha subunit) | 2.54 | 2.47 |
| CB490176 | Egl nine homolog 3 (EC 1.14.11) | 2.11 | 2.19 |
| CA047477 | RING-box protein 2 (Rbx2) | 2.26 | 2.12 |
| Immune system defense | |||
| CA041338 | Beta-2-microglobulin precursor. | 3.46 | 2.09 |
| CA058303 | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) | 4.22 | 3.01 |
| CA061305 | Complement C1r subcomponent precursor (EC 3.4.21.41) | 3.85 | 2.93 |
| Drug metabolism and oxidative stress | |||
| CB506298 | Superoxide dismutase [Cu-Zn] (EC 1.15.1.1). | 2.08 | 2.12 |
| CA057296 | Thioredoxin (ATL-derived factor) | 7.11 | 9.89 |
| CB512686 | Glyoxalase II) (Glx II). | 4.16 | 3.01 |
| Energy metabolism | |||
| CB498267 | Phosphofructo-1-kinase isozyme A) (PFK-A) | 2.72 | 2.40 |
| BU965756 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH). | 9.64 | 10.53 |
| CB511022 | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (GAPDH-2). | 2.36 | 2.92 |
| CA054447 | Malate dehydrogenase, mitochondrial precursor | 4.18 | 8.43 |
| CN442494 | NADH-ubiquinone oxidoreductase chain 4 | 2.07 | 2.90 |
| CB492590 | Ubiquinol-cytochrome c reductase complex 9.5 kDa protein (Complex III sub unit VII). | 3.47 | 2.59 |
| CA060625 | Ubiquinol-cytochrome-c reductase complex core protein | 2.79 | 2.57 |
| CB498293 | Creatine kinase, B chain (B-CK). | 3.30 | 2.17 |
| CB496473 | NADH-ubiquinone oxidoreductase 19 kDa subunit | 7.73 | 12.69 |
| CB497546 | Transaldolase | 5.81 | 6.78 |
| Transcription | |||
| CA051239 | similar to Cofactor required for Sp1 transcriptional activation, subunit 6 | 8.61 | 7.06 |
| CA057271 | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | 2.47 | 3.17 |
| CA057291 | No-on-transient A protein. | 2.14 | 2.53 |
| CK990915 | DNA topoisomerase III beta-1 | 2.43 | 2.72 |
| CB493965 | THO complex subunit 3 (Tho3) | 11.51 | 9.08 |
| CB502666 | DNA-directed RNA polymerase II largest subunit (RPB1) | 3.70 | 4.43 |
| CB516494 | TGFB-inducible early growth response protein 2) (TIEG-2) | 4.72 | 5.31 |
| CA059823 | TGFB-inducible early growth response protein 3) (TIEG-3) | 2.61 | 3.32 |
| CB494556 | Nuclease sensitive element binding protein 1 (Y-box binding protein-1 | 21.22 | 15.35 |
| CA053876 | similar to CSRP2 binding protein isoform a | 3.48 | 4.06 |
| CB497076 | CCAAT/enhancer binding protein delta (C/EBP delta) | 5.48 | 9.02 |
| Structural protein | |||
| CB514461 | Actin, cytoplasmic 2 (Gamma-actin). | 4.15 | 3.54 |
| CB509968 | Troponin T, fast skeletal muscle isoforms. | 2.99 | 3.07 |
| CB492803 | Gamma crystallin B | 2.21 | 2.70 |
| CA058602 | Actin-like protein 3 | 6.27 | 7.90 |
| CA051136 | Claudin-6 (Skullin) | 2.25 | 3.04 |
| CB502342 | Keratin, type I cytoskeletal 13 (Cytokeratin 13) | 2.08 | 2.11 |
| CB494048 | Tubulin alpha-1 chain | 2.19 | 2.59 |
| CB493415 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform. | 5.25 | 4.31 |
| CB497013 | Myosin heavy chain, cardiac muscle alpha isoform | 7.95 | 4.26 |
| Signal transduction | |||
| CA052159 | Cyclooxygenase | 9.44 | 11.52 |
| CA051578 | GTP-binding nuclear protein Ran (GTPase Ran) | 6.62 | 7.34 |
| CB497163 | Guanine nucleotide-binding protein beta subunit 2-like 1 | 3.99 | 5.15 |
| CA042130 | Fatty-acid amide hydrolase | 5.28 | 3.41 |
| CB517167 | Dual specificity protein phosphatase 6 ((Mitogen-activated protein kinase phosphatase 3) | 2.52 | 2.10 |
| CB496992 | cAMP-dependent protein kinase, beta-catalytic subunit (PKA C-beta). | 4.36 | 3.67 |
| CA052159 | Prostaglandin G/H synthase 1 precursor (Cyclooxygenase- 1) | 9.44 | 11.52 |
| CA041082 | TGF-beta receptor type III precursor (TGFR-3) | 2.16 | 2.30 |
| CB511660 | Asialoglycoprotein receptor 1 (Hepatic lectin 1) | 2.40 | 3.06 |
| CA049880 | Polyposis locus protein 1 homolog (TB2 protein homolog) | 2.15 | 2.04 |
| CA046385 | Phosphatidylinositol 3-kinase regulatory alpha subunit | 2.54 | 2.47 |
| Transport | |||
| CB510731 | Ferritin heavy chain (Ferritin H subunit). | 2.73 | 3.67 |
| CB510912 | ADP,ATP carrier protein, heart/skeletal muscle isoform T1 | 3.64 | 3.75 |
| CB491550 | Fatty acid-binding protein, adipocyte (AFABP) | 3.81 | 2.40 |
| CA064198 | Clathrin light chain A (Lca). | 6.67 | 6.62 |
| CB511915 | Voltage-dependent anion-selective channel protein 2 (VDAC-2) | 2.12 | 2.14 |
| CA047666 | ATP synthase e chain, mitochondrial | 4.49 | 3.79 |
| CB516797 | Vacuolar ATP synthase subunit G 1 (V-ATPase G subunit 1) | 3.18 | 2.72 |
| CB494032 | Carbonic anhydrase XIII(Carbonate dehydratase XIII) | 4.57 | 2.82 |
| CA050893 | P2X purinoceptor 7 (ATP receptor) (P2X7) | 6.44 | 4.47 |
| CB493984 | Alpha-fetoprotein precursor (Alpha-fetoglobulin) | 6.24 | 3.53 |
| CB505164 | Adipophilin (Adipose differentiation-related protein) | 4.46 | 2.73 |
| Protein biosynthesis | |||
| CA063412 | Eukaryotic initiation factor 4A-II (eIF4A-II) | 6.20 | 5.57 |
| CB490852 | Ribonuclease P protein subunit p30 (RNaseP protein p30) | 2.43 | 2.63 |
| CB509809 | Alanyl-tRNA synthetase (Alanine--tRNA ligase) | 2.27 | 2.11 |
| CA054662 | 60S ribosomal protein L31. | 10.32 | 15.33 |
| CB505864 | 40S ribosomal protein S27-like protein. | 8.35 | 7.29 |
| CB514237 | 60S ribosomal protein L28. | 5.93 | 3.50 |
| CB498121 | Mitochondrial 28S ribosomal protein S21 (MRP-S21). | 2.14 | 2.73 |
| CK991117 | 40S ribosomal protein S16. | 2.24 | 2.42 |
| CB516607 | 60S ribosomal protein L22 (Heparin binding protein HBp15). | 2.26 | 2.18 |
| CB515229 | 40S ribosomal protein S24. | 7.63 | 7.92 |
| CA058008 | 40S ribosomal protein S6 (Phosphoprotein NP33). | 2.79 | 4.74 |
| CB494481 | 60S ribosomal protein L32. | 2.24 | 2.29 |
| CB504457 | 40S ribosomal protein S7. | 2.21 | 2.22 |
| CB509649 | 60S ribosomal protein L23a. | 3.59 | 4.34 |
| CK990739 | 40S ribosomal protein S29. | 8.67 | 9.14 |
| CB497023 | Gamma-glutamyl hydrolase precursor | 2.19 | 2.05 |
| CB512539 | Cathepsin L precursor | 5.15 | 6.77 |
| CA050484 | similar to Ubiquitin carboxyl-terminal hydrolase 5 | 4.81 | 4.57 |
| CB508017 | Placental thrombin inhibitor (Protease inhibitor 6) | 2.73 | 2.43 |
| CA037310 | Plasminogen activator inhibitor-2, macrophage (PAI-2). | 2.35 | 2.68 |
| CB498673 | Cathepsin E precursor | 4.18 | 3.71 |
| CB499697 | Matrix metalloproteinase-9 precursor (MMP-9) | 3.44 | 2.45 |
| CB493844 | Cathepsin L precursor (Cysteine proteinase 1). | 3.48 | 3.32 |
| CB502976 | Angiotensin-converting enzyme precursor | 5.91 | 4.81 |
| CB512385 | Light protein. | 5.09 | 8.48 |
| CA060241 | Ubiquitin-like 1 activating enzyme E1A (SUMO-1 activating enzyme subunit | 2.82 | 2.49 |
| CA770294 | Ubiquitin. | 2.31 | 2.69 |
| CB488006 | 26S protease regulatory subunit 8 (Proteasome subunit p45) | 2.37 | 2.56 |
| CB494281 | Proteasome subunit alpha type 1 (Proteasome component C2) | 5.90 | 4.07 |
| CA060381 | Ornithine decarboxylase antizyme (ODC-Az). | 7.36 | 7.06 |
table reflects a subset of annotated genes that were expressed at lower levels in smolts relative to adults, p≤0.001
Although a number of mRNAs encoding structural proteins were differentially expressed among the groups, there was no clear pattern with regards to the expression of these genes. For example, approximately half of the differentially expressed genes with ontology relating to cell structure were expressed at higher levels in smolts, with other 50% of differentially expressed ontology genes being at higher levels higher in adults (see appendix II for a complete list of genes in this category). Interestingly, thioredoxin, which plays an important role in protecting against oxidative stress and maintaining cellular redox status and is also a potential B cell growth factor in fish (Khayat et al., 2001), was expressed at 7.0- and 9.9- fold higher levels in adult females and males, respectively (table 4). Cyclooxygenase-1, which has numerous cellular functions mediating signal transduction and oxidation reactions (Liu et al., 2006) was expressed at 11.5- and 9.4- fold higher levels in adult males and females, respectively (table 4). In this regard, signal transduction genes were almost uniformly expressed at higher levels in adult fish relative to smolts (11/12 of the differentially expressed genes).
3.2. Quantitative-PCR analysis of targeted genes of toxicological significance
Initial RT-PCR analysis of coho salmon liver cDNA using the oligonucleotide primers in table 2 generated PCR products with expected molecular weights of the target gene products. Sequencing of the PCR products confirmed extensive identity to the target genes of other species. Specifically, the CYP1A primers amplified a 218 bp fragment with 100% percent identity to Onchorhynchus mykiss CYP1A1 (AF157514) and CYP1A3 (AF059711). The CYP2M1 primers amplified a 195 bp fragment exhibiting 100% identity to O. mykiss CYP2M1 (OMU16657). MT-A (M81800) amplified a 205 bp fragment exhibiting 100% identity to O mykiss MT-A (CB492197), and GST pi (AB026119) amplified a 678 bp fragment exhibiting 97% identity to O. nerka GST pi (CB497579).
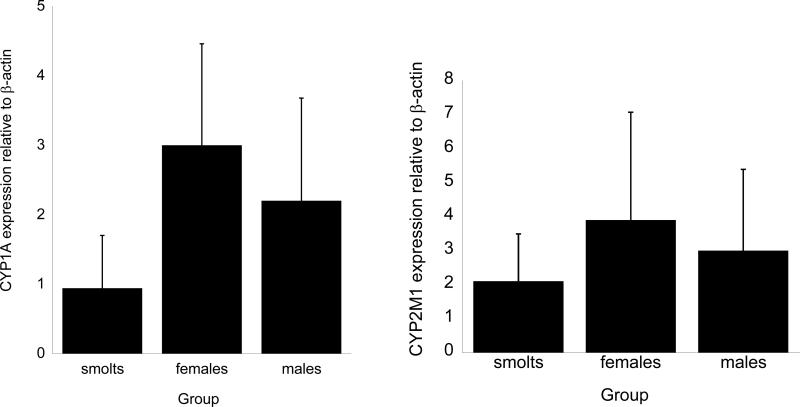
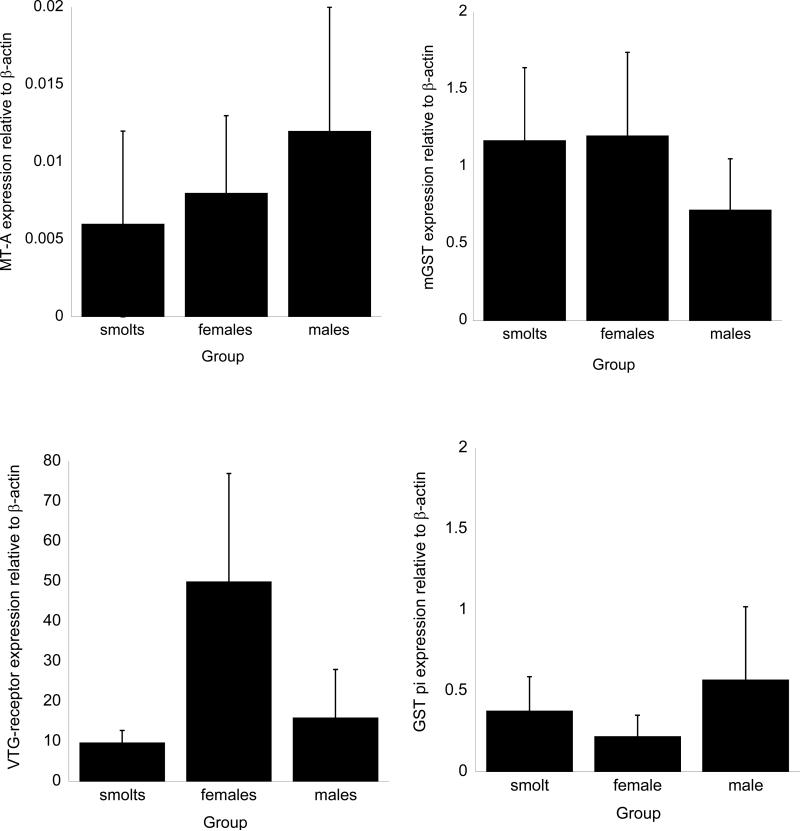
The results of the Q-PCR analysis of individual coho are presented in figures 3 and 4. Among the cytochrome P450 genes, CYP2M1 mRNA exhibited the highest constitutive expression followed by CYP1A1 (figure 3). Of the two GSTs measured, microsomal GST mRNA expression in all groups exceeded that for cytosolic GST pi (figure 4). Relatively small amounts of metallothionein-A mRNA were detected in the samples. In all three groups, the level of expression of the VTG-receptor exceeded that for other genes (figure 4). Assuming equal annealing and amplification properties of the PCR primers, the relative amounts of individual mRNAs normalized to the expression of β-actin for smolts was VTG-receptor>CYP2M1>mGST>CYP1A>GST pi>MT-A. The expression pattern somewhat differed in males and females, with the quantitative level of mRNA expression for the genes analyzed in both males and females being VTG-receptor>CYP2M1>CYP1A>mGST>GST pi>MT-A.
Figure 3.
Q-PCR analysis of CYP1A and 2M1 mRNA gene expression in coho salmon of different life stages. Values represent the mean ±SD for n=4-6 animals for each group with all Q-PCR assays conducted in triplicate incubations. Values sharing different letter superscript are significantly different than their corresponding values atp≤ 0.05. Full gene names for gene labels are found in Table 2.
Figure 4.
Q-PCR analysis of GST pi, mGST, metallothionein A, and vitellogenin receptor mRNA gene expression in coho salmon of different life stages. Values represent the mean±SD for n=4-6 animals for each group with all Q-PCR assays conducted in triplicate incubations. Values sharing different letter superscript are significantly different than their corresponding values at p≤ 0.05. Full gene names for gene labels are found in Table 1.
Despite some observable trends in mRNA expression by Q-PCR among the salmon of different age groups and sexes, the extensive individual differences in gene expression led to non-significant differences in gene expression among groups for any of the genes tested at p<0.05. For example, adult female coho tended to have higher expression of microsomal GST relative to the other groups, however these differences were not statistically significant (figure 4), whereas the expression of metallothionein A, despite being extremely low in all samples, was somewhat higher in male coho than either adult female samples or smolt samples (figure 4). Interestingly, the levels of vitellogenin receptor mRNA were generally higher in females relative to the other groups, but some individuals showed lower vitellogenin receptor mRNA expression which resulted in a lack of statistical significance among the samples.
3.4. Correlation of gene expression data obtained by Q-PCR and microarray analysis
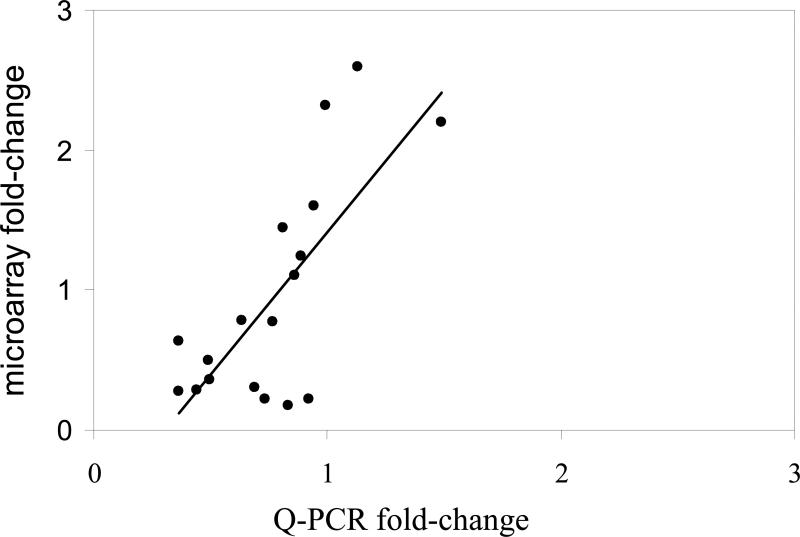
As described above, mRNA levels of CYP1A, CYP2M1, MT-A, mGST, VTG-receptor and GST pi were measured by Q-PCR in individual animals. The Q-PCR measurements for each gene were averaged for all animals belonging to one of the three groups and correlated with the corresponding microarray data. As shown in figure 5, there was strong agreement between the Q-PCR and the microarray data (R2=0.74,p<0.05).
Figure 5.
Correlation among gene expression data obtained by microarray analysis and Q-PCR. Values represent the mean mRNA expression values for CYP1A, CYP2M1, mGST, GST pi and VTG-receptor normalized to the expression of β-globin for all individuals and groups, with the Q-PCR data on the X-axis and the microarray data on the y-axis. A correlation value of R2=0.74 (p≤0.05) was obtained by linear regression analysis.
4. Discussion
A key challenge in using genomics technologies in assessing susceptibility to chemical toxicity is demonstrating that a particular profile of gene expression may lead to or underlie an adverse response of the organism. Implicit in the assumption is that there are characteristic patterns of change in gene expression that can be used to discriminate susceptible and resistant species. However, there are additional challenges in constructing accurate gene ontology, orthology and annotational relationships across species. For example, the literature can be confusing with regards to gene nomenclature and orthology across animal species, although for rats and mice these shortcomings have been substantially reduced with increased knowledge of these genomes. As one becomes further removed phylogenetically from the target species of interest, the more difficult it can be to assign proper orthology of genes/proteins from the test species to the extrapolated species in question. While this is not a problem for genes that are conserved across species (e.g. transcription factors, genes involved in intermediary metabolism), it can be problematic for multigene families such as the cytochrome P450s and glutathione S-transferases, which show genetic divergence and include isoforms that can be difficult to distinguish, but that may have markedly different chemical substrate specificities (Eaton and Gallagher, 1994; Buetler et al., 1995; Buhler and Wang-Buhler, 1998).
As discussed, our approach was to use pooled samples for microarray analysis followed by Q-PCR analysis of a subset of key genes among individuals in an attempt to analyze life stage differences in chemical biotransformation capacity. In designing our Q-PCR studies, we were especially interested in including analysis of genes from the multi-genic biotransformation families such as cytochrome P450 and GST, especially since a number of corresponding cDNA sequences were present on the array but have not been well characterized in coho. We encountered no notable hybridization issues with the arrays using coho mRNA and the results generally tracked Q-PCR analysis of gene expression in individuals (figure 5). A notable exception was that 1 of the 5 elements on the array corresponding to a salmonid CYP2M1 (GRASP accession number CB491960) showed significantly higher expression in smolts relative to adults by microarray analysis, but did not differ by Q-PCR. Interestingly, the four other cDNAs on the array corresponding to CYP2M1 (GRASP accession numbers CB488811, CA053315, CB491764, and CA056952) were not differentially expressed in smolts relative to adults. This discrepancy may have been due to the CYP2M1-like probes on the array hybridizing to different CYP isoforms. This is likely due to stretches of high sequence similarity to CYP2M1.
As is typical with microarray platforms, our results generally tracked better with quantitative PCR when microarray analysis indicated a relatively high signal compared to background, but were less reliable at lower-end measurements. This could indicate a sensitivity issue with the platform for low expressing genes such as MT-A due to a limitated dynamic range and/or cross-species hybridization. For example, the strong correlation among the Q-PCR microarray data for the six genes analyzed (R2=0.74) was further strengthened (to R2=0.87) if the expression of MT-A was dropped from the correlation analysis. Given this limitation, the Q-PCR and microarray data were in strong agreement. As Q-PCR is a much more sensitive method than microarray analysis, our approach for our future studies will be to use Q-PCR for those limited subset of genes of high interest. The Q-PCR assays developed in this study (table 2) can be used readily by others allowing gene expression analysis of six toxicologically relevant genes in the context of chemical detoxification in coho salmon.
The fact that the Q-PCR studies generally tracked results observed by microarray analysis is noteworthy given that our microarray design did not include individual biological replicates, and supported our notion that a limited cost-saving microarray design could provide biologically meaningful results. The extremely low p values observed for many of the differentially expressed genes is likely due to only one sample (pooled from 12-15 individuals) being arrayed per group, the latter which can lead to inflated test-statistics and artificially low p-values (Storey and Tibshirani, 2005). Therefore, all the variability observed in the microarray results must be due to technical variability, which was very low. These observations suggest that all technical aspects associated with the microarray analysis, including the arrays themselves, were very consistent and highly reproducible.
Although some distinct expression patterns were observed among the three groups analyzed, the extensive interindividual variation within groups diminished our capacity to generalize with regards to overall differences in biotransformation capacity based upon the genes analyzed. Others have used cloned rainbow trout to reduce inter-individual variability with the GRASP microarray platform (Hook et al., 2006a; Hooket al., 2006b; Skillman et al., 2006). However, despite such an approach, these investigators have sometimes observed significant technical variability. In contrast, the technical variability in our microarray experiment was relatively low, as illustrated by the low p-values. Furthermore, since we only had “one biological replicate”, albeit it was a pool of samples, all the variability observed in our experiment was technically derived. However, despite these issues and also using outbred wild coho salmon, our data indicate that the GRASP platform provided an accurate reflection of global hepatic gene expression in our fish.
Our analyses demonstrated extensive differences in hepatic gene expression among smolting and adult coho. Relative to adults, smolting coho showed significantly higher expression of a number of genes involved in basic cellular processes such as molecular transport, regulation of cell cycle, and protein biosynthesis and degradation. Notable exceptions were significantly higher expression of a number of signal transduction and energy metabolism genes on the array in the adult fish. Such observations indicate potential differences among the life stages with regard to basic biochemical processes. In this regard, younger fish typically have a higher growth rate which can potentially affect protein turnover in metabolism, which could explain some of the differences observed in gene expression. We did observe a number of genes involved in protein biosynthesis and turnover were more highly expressed in smolts. Similar observations were reported by von Schalburg and coworkers in a comprehensive analysis of genes involved in ovarian maturation in rainbow trout (von Schalburg et al., 2005a).
As discussed, our primary hypothesis was that genes involved in the biotransformation of environmental chemicals and protection against oxidative stress would show a generalized pattern of lower expression in smolting coho relative to adult males, and to adult females. However, this was not the case, and for the most part, genes in this category did not differ among the age groups. Similarly, we did not observe any clear differences in expression patterns of genes involved in immune system function, DNA repair, or protection against oxidative stress between smolts and adults. Based on this data, we must reject our hypothesis of a relatively low hepatic detoxification gene expression in smolts. It must be pointed out that we did not investigate catalytic activities of any proteins encoded by the aforementioned genes.
We had previously shown that GST pi is a major GST isoform involved in chemical conjugation in coho liver (Trute et al., 2007), which was consistent with results from the present study. Although little is known regarding developmental or sex differences of GST pi expression in fish, our data indicate that this isoform may not be subject to developmental expression in coho liver. In contrast, GST pi expression is developmentally regulated in human liver and its expression is very high during fetal development but exhibits repression soon after birth (Hayes and Pulford, 1995; Hayes et al., 2005). Other studies in our laboratory are directed towards understanding the ontogenic expression of GSTs and other biotransformation genes in coho tissues, including earlier life stages such as swim-up fry. In addition, the relatively high expression of an mRNA encoding microsomal GST is consistent with the presence of a microsomal GST isoform in rainbow trout (Machala et al., 1998) and other fish (Wiegand et al., 2001; Fu and Xie, 2006), and unpublished observations from our laboratory of microsomal GST activity in the species. The microsomal GSTs from other species appear to be involved in mediating protection against oxidative stress (Ji et al., 2002; Johansson et al., 2007), but the functional significance of this isoform in fish has not been thoroughly investigated.
The two cytochrome P450 genes analyzed in detail (e.g. CYP1A, and 2M1) encode for cytochrome P450 proteins that have been studied in other samonids, but not been studied in detail in coho. The rainbow trout CYP1A1 and CYP1A3 genes share 96% amino acid identity and have similar enzymatic activity, and both genes are inducible on exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (Cao et al., 2000). In trout liver, the CYP1A1 isoform predominates (Cao et al., 2000). CYP1A1 mRNA is also constitutively expressed in Atlantic salmon and is inducible on exposure to polycyclic aromatic hydrocarbons (Buhler and Wang-Buhler, 1998; Rees and Li, 2004)}. Although we could not discriminate the two isoforms in the present study, it is reasonable to assume CYP1A1 mRNA was detected in our assays. Because of the role of CYP1A1 in the bioactivation of polycyclic aromatic hydrocarbons, and the fact that juvenile salmon migrating through polluted waterways can show high levels of PAH-DNA damage, we would have hypothesized that CYP1A mRNA expression would be higher in smolts relative to adults, which was not the case. However, constitutive CYP1A expression is relatively low in nonexposed fish such as our hatchery coho, and we did not examine the inducibility of the biotransformation isoforms which occurs on exposure to pollutants in the field.
In addition to ontogenic influences, genes governing chemical detoxification and biotransformation often show tissue-specific differences in expression. Accordingly, It is highly likely that we might have observed different gene profile comparisons if a different tissue such as the gills, olfactory rosettes, brain, or gonads would have been chosen. We selected the liver as target organ for this study because it is a major route for detoxification of dietary and waterborne xenobiotics. We did not attempt to develop Q-PCR assays for genes exhibiting greater than 3 fold change as measured by microarray, as there was a lack of gene sequences available from phylogenetically similar species to coho and we focused our efforts on genes of toxicological relevance. Further work needs to be completed in cloning these genes in order to validate the microarray platform in the upper fold-change range of detection via microarray analysis.
In summary, our study does not support the notion of a low ability of coho smolts to detoxify environmental chemicals relative to adults based upon hepatic gene expression. However, we have observed extremely low expression of several genes important in protecting against cell injury of environmental chemicals. Our array data provide valuable transcriptional profiles of smolts as well as adult male and female cohos that forms a rationale basis to generate testable hypotheses in a variety of toxicological contexts. We have also developed and validated real-time quantitative PCR assays for the analysis of several important genes involved in chemical detoxification and protection against cellular injury in coho salmon that can be used in laboratory and field studies. Our quantitative PCR based analysis of gene expression suggests considerable interindividual variability in mRNA expression, but yet is generally consistent with microarray analysis that use samples pooled from a relatively large number of individuals. Such information should be valuable to other salmonid researchers involved in toxicological studies using the GRASP platform. Our studies also underscore the importance of determining gene expression levels of individual animals before results can be extrapolated and extended to other populations of the same fish species. Of importance will be further validation studies using coho exposures to model enzyme inducers in parallel with Q-PCR, and phenotypic anchoring studies of coho gene expression with sublethal injury in vivo from contaminants of concern in Pacific Northwest waters.
Supplementary Material
Acknowledgments
The authors would like to knowledge Dr. Nathaniel Scholz and colleagues at the NOAA/National Marine Fisheries Service in Seattle for assistance in providing the salmon for use in the study. In addition, the technical assistance and continuing support of Dr. Ben Koop and colleagues at the University of Victoria GRASP project was greatly appreciated.
Funding Sources. This project was funded in part by grants from the University of Washington NIEHS Superfund Basic Sciences Program (P42-ES04696), the National Oceanic and Atmospheric Administration Coastal Ocean Program (NA05NS4781253), and the University of Washington Center for Ecogenetics and Environmental Health (NIEHS P30-ES07033). H.L. was the recipient of an NIEHS predoctoral fellowship in Environmental Pathology and Toxicology (NIH-5T32-ES07032).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All animal studies were approved by the University of Washington Institutional Animal Care and Use Committee
References
- Anand SS, Kim KB, Padilla S, Muralidhara S, Kim HJ, Fisher JW, Bruckner JV. Ontogeny of hepatic and plasma metabolism of deltamethrin in vitro: role in age-dependent acute neurotoxicity. Drug Metab Dispos. 2006;34:389–397. doi: 10.1124/dmd.105.007807. [DOI] [PubMed] [Google Scholar]
- Buetler TM, Gallagher EP, Wang C, Stahl DL, Hayes JD, Eaton DL. Induction of phase I and phase II drug-metabolizing enzyme mRNA, protein, and activity by BHA, ethoxyquin, and oltipraz. Toxicol Appl Pharmacol. 1995;135:45–57. doi: 10.1006/taap.1995.1207. [DOI] [PubMed] [Google Scholar]
- Buhler DR, Wang-Buhler JL. Rainbow trout cytochrome P450s: purification, molecular aspects, metabolic activity, induction and role in environmental monitoring. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;121:107–137. doi: 10.1016/s0742-8413(98)10033-6. [DOI] [PubMed] [Google Scholar]
- Cao Z, Hong J, Peterson RE, Aiken JM. Characterization of CYP1A1 and CYP1A3 gene expression in rainbow trout (Oncorhynchus mykiss). Aquatic Toxicol. 2000;49:101–109. doi: 10.1016/s0166-445x(99)00072-7. [DOI] [PubMed] [Google Scholar]
- Chauhan DP, Miller MS, Owens IS, Anderson LM. Gene expression, ontogeny and transplacental induction of hepatic UDP-glucuronosyl transferase activity in mice. Dev Pharmacol Ther. 1991;16:139–149. [PubMed] [Google Scholar]
- Collier T, Johnson LL, Myers MS, Stehr CM, Krahn MM, Stein JE. Fish Injury in the Hylebos Waterway of Commencement Bay. NMFS-NWFSC-36; Washington: 1998. p. 576. [Google Scholar]
- Eaton DL, Gallagher EP. Mechanisms of aflatoxin carcinogenesis. Annu Rev Pharmacol Toxicol. 1994;34:135–172. doi: 10.1146/annurev.pa.34.040194.001031. [DOI] [PubMed] [Google Scholar]
- Fu J, Xie P. The acute effects of microcystin LR on the transcription of nine glutathione S-transferase genes in common carp Cyprinus carpio L. Aquat Toxicol. 2006;80:261–266. doi: 10.1016/j.aquatox.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- Hedtke JL, Robinson-Wilson E, Weber LJ. Influence of body size and developmental stage of coho salmon (Oncorhynchus kisutch) on lethality of several toxicants. Fundam Appl Toxicol. 1982;2:67–72. doi: 10.1016/s0272-0590(82)80116-4. [DOI] [PubMed] [Google Scholar]
- Hook SE, Skillman AD, Small JA, Schultz IR. Dose-response relationships in gene expression profiles in rainbow trout, Oncorhyncus mykiss, exposed to ethynylestradiol. Mar Environ Res. 2006a;62(Suppl 1):S151–155. doi: 10.1016/j.marenvres.2006.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SE, Skillman AD, Small JA, Schultz IR. Gene expression patterns in rainbow trout, Oncorhynchus mykiss, exposed to a suite of model toxicants. Aquat Toxicol. 2006b;77:372–385. doi: 10.1016/j.aquatox.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EM, Gallagher EP. Effects of 17-beta estradiol and 4-nonylphenol on phase II electrophilic detoxification pathways in largemouth bass (Micropterus salmoides) liver. Comp Biochem Physiol C Toxicol Pharmacol. 2004;137:237–247. doi: 10.1016/j.cca.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Ji Y, Toader V, Bennett BM. Regulation of microsomal and cytosolic glutathione S-transferase activities by S-nitrosylation. Biochem Pharmacol. 2002;63:1397–1404. doi: 10.1016/s0006-2952(02)00879-1. [DOI] [PubMed] [Google Scholar]
- Johansson K, Ahlen K, Rinaldi R, Sahlander K, Siritantikorn A, Morgenstern R. Microsomal glutathione transferase 1 in anticancer drug resistance. Carcinogenesis. 2007;28:465–470. doi: 10.1093/carcin/bgl148. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Experimental design for gene expression microarrays. Biostatistics. 2001;2:183–201. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- Khayat M, Stuge TB, Wilson M, Bengten E, Miller NW, Clem LW. Thioredoxin acts as a B cell growth factor in channel catfish. J Immunol. 2001;166:2937–2943. doi: 10.4049/jimmunol.166.5.2937. [DOI] [PubMed] [Google Scholar]
- Liu W, Cao D, Oh SF, Serhan CN, Kulmacz RJ. Divergent cyclooxygenase responses to fatty acid structure and peroxide level in fish and mammalian prostaglandin H synthases. Faseb J. 2006;20:1097–1108. doi: 10.1096/fj.05-5273com. [DOI] [PubMed] [Google Scholar]
- Luo Q, Ban M, Ando H, Kitahashi T, Kumar Bhandari R, McCormick SD, Urano A. Distinct effects of 4-nonylphenol and estrogen-17 beta on expression of estrogen receptor alpha gene in smolting sockeye salmon. Comp Biochem Physiol C Toxicol Pharmacol. 2005;140:123–130. doi: 10.1016/j.cca.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Machala M, Drabek P, Neca J, Kolarova J, Svobodova Z. Biochemical markers for differentiation of exposures to nonplanar polychlorinated biphenyls, organochlorine pesticides, or 2,3,7, 8-tetrachlorodibenzo-p-dioxin in trout liver. Ecotoxicol Environ Saf. 1998;41:107–111. doi: 10.1006/eesa.1998.1675. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Kiceniuk JW. Effect of fenitrothion on the foraging behavior of juvenile Atlantic salmon. Environmental Toxicology and Chemistry. 1990;9:489–495. [Google Scholar]
- Quinn TP. The behavior and ecology of Pacific salmon and trout. University of Washington press; Seattle: 2005. [Google Scholar]
- Rees C, Li W. Development and application of a real time quantitative PCR analysis of CYP1A transcripts in three genera of salmonids. Aquat Toxicol. 2004;66:357–368. doi: 10.1016/j.aquatox.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Rise ML, von Schalburg KR, Brown GD, Mawer MA, Devlin RH, Kuipers N, Busby M, Beetz-Sargent M, Alberto R, Gibbs AR, Hunt P, Shukin R, Zeznik JA, Nelson C, Jones SR, Smailus DE, Jones SJ, Schein JE, Marra MA, Butterfield YS, Stott JM, Ng SH, Davidson WS, Koop BF. Development and application of a salmonid EST database and cDNA microarray: data mining and interspecific hybridization characteristics. Genome Res. 2004;14:478–490. doi: 10.1101/gr.1687304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenk D, Peters LD, Shehin-Johnson S, Hines RN, Livingstone DR. Differential expression and activity of flavin-containing monooxygenases in euryhaline and stenohaline flatfishes. Comparative Biochemistry and Physiology. 1995;112C:179–186. doi: 10.1016/0742-8413(95)02010-1. [DOI] [PubMed] [Google Scholar]
- Scholz NL, Truelove NK, French BL, Berejikian BA, Quinn TP, Casillas E, Collier TK. Diazinon disrupts antipredator and homing behaviors in chinook salmon (Oncorhynchus tshawytscha). Can. J. Fish. Aquat. Sci./J. Can. Sci. Halieut. Aquat. 2000;57:1911–1918. [Google Scholar]
- Skillman AD, Nagler JJ, Hook SE, Small JA, Schultz IR. Dynamics of 17alpha-ethynylestradiol exposure in rainbow trout (Oncorhynchus mykiss): absorption, tissue distribution, and hepatic gene expression pattern. Environ Toxicol Chem. 2006;25:2997–3005. doi: 10.1897/05-565r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. SAM Thresholding and False Discovery Rates for Detecting Differential Gene Expression in DNA Microarrays. In: Gentleman RC, Carey VJ, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2005. [Google Scholar]
- Trute M, Gallis B, Doneanu C, Shaffer S, Goodlett D, Gallagher E. Characterization of hepatic glutathione S-transferases in coho salmon (Oncorhynchus kisutch). Aquat Toxicol. 2007;81:126–136. doi: 10.1016/j.aquatox.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schalburg KR, Rise ML, Brown GD, Davidson WS, Koop BF. A comprehensive survey of the genes involved in maturation and development of the rainbow trout ovary. Biol Reprod. 2005a;72:687–699. doi: 10.1095/biolreprod.104.034967. [DOI] [PubMed] [Google Scholar]
- von Schalburg KR, Rise ML, Cooper GA, Brown GD, Gibbs AR, Nelson CC, Davidson WS, Koop BF. Fish and chips: various methodologies demonstrate utility of a 16,006-gene salmonid microarray. BMC Genomics. 2005b;6:126. doi: 10.1186/1471-2164-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz DA, Bonn BA, Carpenter KD, Hinkle SR, Janet ML, Rinella FA, Uhrich MA, Waite IR, Laenen A, Bencala K. Water quality in the Willamette Basin, Oregon, 1991-95. U.S. Geological Survey Circular, 1161. 1998 [Google Scholar]
- Wiegand C, Krause E, Steinberg C, Pflugmacher S. Toxicokinetics of atrazine in embryos of the zebrafish (Danio rerio). Ecotoxicol Environ Saf. 2001;49:199–205. doi: 10.1006/eesa.2001.2073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.