Abstract
The composition of the cellular proteome is under the control of multiple processes, one of the most important being translation initiation. The majority of eukaryotic cellular mRNAs initiates translation by the cap-dependent or scanning mode of translation initiation, a mechanism that depends on the recognition of the m7G(5′)ppp(5′)N, known as the cap. However, mRNAs encoding proteins required for cell survival under stress bypass conditions inhibitory to cap-dependent translation; these mRNAs often harbor internal ribosome entry site (IRES) elements in their 5′UTRs that mediate internal initiation of translation. This mechanism is also exploited by mRNAs expressed from the genome of viruses infecting eukaryotic cells. In this paper we discuss recent advances in understanding alternative ways to initiate translation across eukaryotic organisms.
1. Alternative Translation Initiation Mechanisms: An Important Layer of Gene Expression Control
The coding capacity of eukaryotic genomes is much larger than anticipated. Many layers of gene expression control operate at the posttranscriptional level, as illustrated by the RNA splicing process, the noncoding RNAs regulatory elements, and the large repertoire of factors that contribute to control mRNA transport, localization, stability, and translation. Translation control is one of the posttranscriptional cellular processes that exert a profound impact on the composition of the cellular proteome. This is particularly relevant to maintain homeostasis in response to stress induced by a large variety of environmental factors, as well as during development or disease [1]. In addition, these layers of gene expression control contribute to increase the coding capacity of the genome by generating different polypeptides from the same transcriptional unit.
The majority of cellular mRNAs initiate translation by a mechanism that depends on the recognition of the m7G(5′)ppp(5′)N structure (termed cap) located at the 5′end of most mRNAs (Figure 1(a)). This manner of initiating translation involves a large number of auxiliary proteins termed eukaryotic initiation factors (eIFs) [1]. The 5′cap structure is recognized by eIF4E that, in turn, is bound to the scaffold protein eIF4G and the RNA helicase eIF4A (within a trimeric complex termed eIF4F). Additionally, eIF4G further interacts with eIF3 and the poly(A)-binding protein (PABP) that is bound to the poly(A) tail of the mRNA. Separately, the 40S ribosomal subunit associates with the ternary complex (TC) consisting of the initiator methionyl-tRNAi and eIF2-GTP, leading to the formation of the 43S complex that is stabilized by eIF1A and eIF3. Following assembly of the 43S complex into the eIF4F-bound mRNA, scanning of the 5′UTR region commences until the first AUG codon is encountered, leading to the formation of the 48S initiation complex. At this step, eIF1 is displaced and eIF5 mediates the hydrolysis of eIF2-bound GTP; joining of the 60S subunit is then mediated by eIF5B yielding the 80S ribosome that gives rise to the start of polypeptide synthesis. For a review on the translation initiation process, see [1] and references therein.
Figure 1.
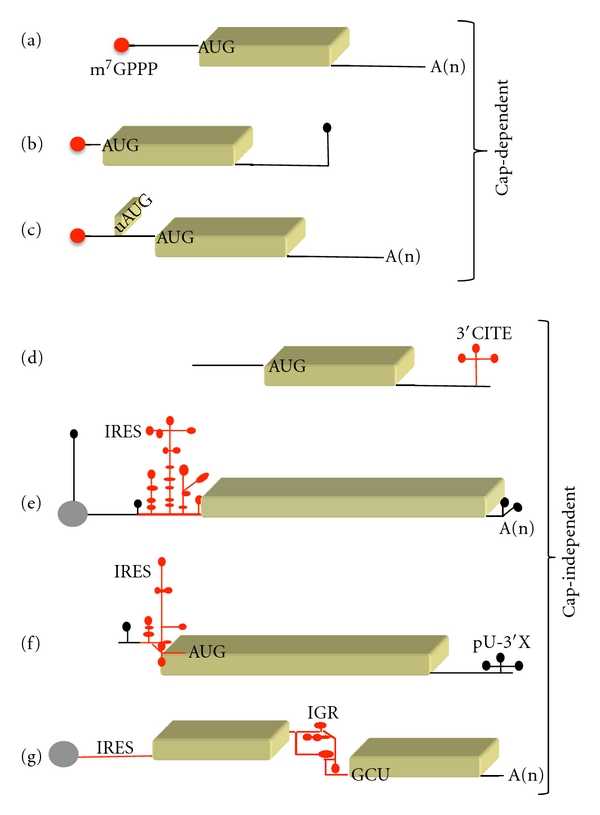
Schematic representation of eukaryotic mRNAs. (a) Features of a conventional mRNA. The red circle at the 5′end depicts the cap (m7Gppp); A(n) depicts the poly(A) tail at the 3′end. (b) (c) Schematic of atypical RNA structures, stem loops (black hairpin), or uAUGs, respectively, located in mRNAs translated via cap-dependent initiation. (d) RNA structural elements located in the 3′untranslated region of viral RNAs mediating cap-independent translation (3′CITE). Different types of IRES elements found in the viral RNA of picornaviruses (e), hepatitis C (f), and dicistroviruses (g) are schematically depicted in red.
Various alternative mechanisms to initiate translation are, however, operative during cellular stress [1, 2]. Furthermore, atypical mRNAs that lack the cap structure at the 5′end or the poly(A) tail at the 3′end employ specific mechanisms to initiate translation. Histones are among the most abundant proteins in eukaryotic cells, despite having mRNAs with an organization that is incompatible with the conventional scanning initiation model. Peculiar features of metazoan histone mRNAs are that they harbor a short 5′UTR and lack a poly(A) tail. Instead, these mRNAs harbor a conserved stem loop near the 3′end (Figure 1(b)) that is recognized by the stem-loop-binding protein (SLBP). In addition, the open reading frame of mouse histone mRNA contains two structural elements critical for translation initiation. One of them binds to eIF4E without the need of the cap, such that the 43S complex is recruited to this site and loaded near the AUG start codon [3]. This process is assisted by a second structural element located downstream of the initiator triplet that sequesters the cap, facilitating the direct loading of the ribosome on the cognate codon.
A different example of unconventional RNA organization is presented by a plant viral RNA, the pea enation mosaic virus, that initiates translation using a cap-independent mechanism. This mRNA does not possess a cap at the 5′end even though it requires eIF4E for translation initiation. In this particular case, the RNA structure adopts a pseudoknot structure that projects a guanosine residue into the cap-binding pocket of eIF4E [4].
In addition to the 5′-cap and 3′-poly(A) tail, eukaryotic mRNAs can contain upstream open reading frames (uORFs) (Figure 1(c)), 3′cap-independent translation elements (3′CITEs) (Figure 1(d)), or internal ribosome entry site (IRES) elements (Figures 1(e), 1(f), and 1(g)). These types of structural elements can act as sensors of environmental factors, mediate efficient translation of some viral mRNAs, or control translation of mRNAs that encode proteins performing critical roles during cell death, DNA-damage response, or in the differentiation process of higher eukaryotes and algae [5–10]. In general, these structural elements act as strong barriers to scanning ribosomes in the 5′UTR of mRNAs. Hence, their presence is incompatible with the scanning model of translation initiation, and the corresponding mRNAs have evolved different manners to initiate translation using nonconventional mechanisms.
One extensively analyzed uORF-containing mRNA is that encoding the yeast transcription factor GCN4. Translation of this particular mRNA is strongly enhanced during nutrient deprivation, an event that induces eIF2α-phosphorylation leading to low levels of TC and, thus, inhibiting cap-dependent translation. However, GCN4 mRNA overcomes the translation inhibitory effects of four uORFs under low TC levels by allowing a fraction of posttermination 40S subunits to reinitiate at the authentic GCN4 start codon [1]. In mRNAs subjected to polyamine-responsive translation regulation, uORFs act as sensors of polyamine levels [10]. Finally, in many other mRNAs, uORFs are obstacles that block or delay scanning ribosomes causing a strong reduction of protein synthesis, as exemplified in the 5′ terminal region of p53 mRNA or the embryonic form of the chicken proinsulin mRNA [5, 6].
Consistent with the fact that mRNA translation operates on functionally circularized molecules, regulatory elements also are located at the 3′UTR; this is the case of 3′CITE elements which are particularly abundant in plant RNA viruses [7, 11, 12] and promote cap-independent translation by means of complex RNA structures that generate functional bridges between the 5′ and 3′UTRs of the mRNA.
In summary, translation initiation mechanisms affecting the efficiency of protein synthesis of a given mRNA are diverse and, importantly, more frequent than anticipated, sometimes giving rise to the expression of different polypeptides from a single transcriptional unit. Therefore, presence of any of these regulatory elements can seriously complicate efforts to accurately define the sites of translation initiation at the genomewide scale.
2. IRES Elements in Eukaryotic Organisms and Their Viruses
IRES elements are specialized RNA regulatory sequences governing cap-independent translation initiation in eukaryotic mRNAs that are translated during cellular stress, that is, when cap-dependent translation is compromised [2, 13, 14]. IRES elements, initially reported in the genomic RNA of two picornaviruses (namely, poliovirus (PV) and encephalomyocarditis virus (EMCV)), drive internal initiation of translation in the mRNA of all members of the Picornaviridae family [15–20] (Table 1). Soon after their discovery, IRES elements were also found in other RNA viruses infecting mammals, such as hepatitis C (HCV), pestiviruses [21, 22], or retroviruses [23–27], as well as in RNA viruses infecting invertebrates [28–33], plants [34–39], and protozoa [40, 41]. Recently, IRES-dependent translation in mRNAs transcribed from DNA viruses belonging to the Herpesviridae family has been reported [42–45] (Table 1).
Table 1.
Distribution of IRES elements in viral mRNAs.
| Host | Virus family/genus | Virus/IRES name | Reference |
|---|---|---|---|
| Mammals | Picornaviridae/Enterovirus | Poliovirus (PV) | [15] |
| Picornaviridae/Cardiovirus | Encephalomyelitis virus (EMCV) | [16] | |
| Picornaviridae/Aphthovirus | Foot-and-mouth disease virus (FMDV) | [17] | |
| Picornaviridae/Teschovirus | Porcine teschovirus-1 (PTV-1) | [18] | |
| Picornaviridae/Kobuvirus | Aichivirus (AiV) | [19] | |
| Picornaviridae/Senecavirus | Seneca Valley virus (SVV) | [20] | |
| Flaviviridae/Hepacivirus | Hepatitis C virus (HCV) | [21] | |
| Flaviviridae/Pestivirus | Classical swine fever virus (CSFV) | [22] | |
| Retroviridae/Lentivirus | Human immunodeficiency virus-2 (HIV-2) | [23] | |
| Retroviridae/Lentivirus | Human immunodeficiency virus-1 (HIV-1) | [24] | |
| Retroviridae/Retrovirus | Moloney murine leukemia virus (MoMLV) | [25] | |
| Retroviridae/Lentivirus | Feline immunodeficiency virus (FIV) | [26] | |
| Retroviridae/Retrovirus | Mouse mammary tumor virus (MMTV) | [27] | |
| Herpesviridae/Cytomegalovirus | Human cytomegalovirus latency (pUL138) | [42] | |
| Herpesv./Lymphocryptovirus | Epstein-Barr virus (EBNA-1) | [44] | |
| Herpesv./Mardivirus | Herpes virus Marek's disease (MDV RLORF9) | [45] | |
| Papovaviridae/Polyomavirus | SV40 polycistronic 19S (SV40 19S) | [43] | |
|
| |||
| Insects | Dicistroviridae/Cripavirus | Rhopalosiphum padi virus (RhPV) | [28] |
| Dicistroviridae/Cripavirus | Cricket paralysis virus (CrPV) | [29] | |
| Dicistroviridae/Cripavirus | Ectropis obliqua picorna-like virus (EoPV) | [31] | |
| Dicistroviridae/Cripavirus | Plautia stali intestine virus (PSIV) | [32] | |
| Dicistroviridae/Cripavirus | Triatoma virus (TrV) | [33] | |
| Dicistroviridae/Aparavirus | Bee paralysis dicistrovirus (IAPV, KBV) | [30] | |
|
| |||
| Plants | Comoviridae/Nepovirus | Black currant reversion virus (BRV) | [34] |
| Tombusviridae/Carmovirus | Pelargonium flower break virus (PFBV) | [35] | |
| Tombusviridae/Carmovirus | Hibiscus chlorotic ringspot virus (HCRSV) | [38] | |
| Tobamovirus | Crucifer-infecting tobamovirus (CrTMV) | [36] | |
| Luteoviridae/Polerovirus | Potato leaf roll polerovirus (PLRV) | [37] | |
| Potyviridae/Potyvirus | Tobacco etch virus (TEV) | [39] | |
|
| |||
| Protozoa | Totiviridae/Giardiavirus | Giardiavirus (GLV) | [40] |
| Totiviridae/Leishmaniavirus | Leishmania RNA virus-1 (LRV-1) | [41] | |
As expected from the fact that viruses require components of the host machinery to translate their genome, IRES elements have been found in mRNAs encoded in the genome of the host (Table 2). Thus, IRES-dependent translation initiation has been described in mRNAs expressed in animal cells, both vertebrate and invertebrate [46–69], plants, and yeasts [70–73]. Not surprisingly, various examples of IRES elements reported in animal cells, plants, or yeasts drive internal initiation of translation in mRNAs that encode proteins performing similar functions or belonging to the same regulatory pathway, for example, nutrient deprivation, apoptosis, or heat-shock (see Table 2). Published IRES elements are available at (http://iresite.org/) (http://140.135.61.9/ires/) [74, 75].
Table 2.
Distribution of IRES elements in cellular mRNAs.
| Organism | Protein function | IRES name | Reference |
|---|---|---|---|
| Mammals | Apoptotic proteins | Apaf-1 | [60] |
| XIAP | [56] | ||
| HIAP2/c-IAP1 | [57] | ||
| DAP5 | [58] | ||
| Bcl-2 | [61] | ||
| Oncogene | c-myc | [77] | |
| Amino acid starvation | CAT-1 | [46] | |
| Nutrient signaling | INR | [59] | |
| Differentiation | LEF-1 | [65] | |
| PDGF2 | [66] | ||
| Hypoxia | HIF-1a | [50] | |
| VEGF | [55] | ||
| FGF2 | [53] | ||
| Heat shock | BiP | [51] | |
| BAG-1 | [49] | ||
| Cold shock | CIRP | [48] | |
| DNA damage response | p53 | [47] | |
| SHMT1 | [54] | ||
| Mitosis | PITSLREp58 | [50] | |
| CDK1 | [61] | ||
|
| |||
| Insects | Apoptotic proteins, | Rpr, hid | [62] |
| Heat shock | hsp70 | [63] | |
| grim, skl | |||
| Homeotic protein | Antennapedia | [64] | |
| Insulin signaling | dFoxO | [67] | |
| dInR | [68] | ||
| Alcohol dehydrogenase | Adh-Adhr | [69] | |
|
| |||
| Plants | Heat shock | HSP101 | [70] |
| Alcohol dehydrogenase | ADH | [71] | |
|
| |||
| Yeast | Nitrogen assimilation | URE-2 | [72] |
| glucose starvation | GPR1, NCE102 | [73] | |
| YMR181a, MSN1 | |||
| BOI1, FLO8, GIC1 | |||
The ability of being translated under conditions inhibitory to cap-dependent initiation, though with different efficiency, is a general feature of all IRES-containing mRNAs. With notable exceptions [23, 37], IRES elements are located in the 5′UTR of mRNAs upstream of the initiator codon. Other features such as long length of the 5′UTR (200 to 500 nucleotides), heavy RNA structure, high GC content, initiation at non-AUG codons, ignored AUGs upstream of the functional start codon are often but not universally found [46, 53, 76, 77]. For instance, some IRES elements found in plant RNA viruses, D. melanogaster and S. cerevisiae, have a high AU content [37, 62, 63, 73]. In this regard, the 5′UTR sequences of mRNAs are highly divergent [78]. This is consistent with the great sequence diversity of the currently known IRES elements (http://rfam.sanger.ac.uk/) found in mRNAs of different organisms, including viruses, protozoa, yeast, plants, and animals. Furthermore, this lack of sequence conservation creates serious problems to predict the presence of IRES elements in eukaryotic mRNAs using computational methods. Therefore, defining critical hints (short primary sequences, tertiary structure elements, unique RNA-binding protein motifs, etc.) of model IRES elements is crucial to accurately predict putative IRES elements at the genomic level.
3. Types of IRES Elements
Two picornavirus RNAs, PV and EMCV, contain the first reported IRES elements [15, 16]. This property was later extended to all picornavirus RNAs as well as to several positive-strand RNA viruses, such as HCV, pestiviruses, and dicistroviruses [79]. Nevertheless, it is remarkable that despite performing the same function, viral IRES elements differ in nucleotide sequence, RNA secondary structure, and trans-acting factors requirement. A distinctive feature of the picornavirus IRES is their long length that varies between 350 to 450 nucleotides, depending on the virus genera. Furthermore, picornavirus IRES elements are classified in four types (termed I, II, III, and HCV-like) according to their RNA structure organization. The genome of picornaviruses consists of a single-stranded RNA of positive polarity that harbors a short poly(A) tail at the 3′end (Figure 1(e)). However, picornavirus RNAs differ from cellular mRNAs in having a long, heavily structured 5′UTR and a viral-encoded protein (VPg) covalently linked to the 5′end instead of cap, hence, incompatible with the cap-dependent mechanism of translation initiation. Not surprisingly, translation of the viral genome is governed by the IRES element using a cap-independent mechanism that is resistant to the action of viral proteases. Picornavirus-encoded proteases execute the processing of the viral polyprotein but also recognize as substrates several host factors. Among the host factors proteolyzed during infection are eIF4G and PABP [80, 81], which are key components of the cap-dependent translation initiation machinery. Thereby, cleavage of host factors induces the shut-off of cap-dependent translation in infected cells.
Picornavirus elements IRES belonging to types I and II require the C-terminal end of eIF4G, eIF4A, and eIF3 to assemble 48S initiation complexes [82–84]. Type III IRES require intact eIF4G, and, in contrast, the HCV-like IRES does not need eIF4G to assemble 48S complexes [18]. In addition to eIFs, auxiliary factors termed IRES transacting factors (ITAFs) contribute to modulate (either stimulate or repress) picornavirus IRES activity. In support of the relevance of factors different than eIFs for internal initiation, transcripts encompassing the region interacting with eIFs do not possess IRES activity [85], indicating that interaction with eIFs is necessary but not sufficient for IRES function.
The HCV viral RNA does not possess poly(A) tail; instead, a poly(U) tract and a complex RNA structure are located near the 3′end (Figure 1(f)). The HCV IRES element is located close to the 5′end of the viral genome and differs profoundly in RNA structure organization from picornavirus IRES belonging to types I, II, and III. Specifically, the HCV IRES (340 nucleotides) is arranged in structural domains II, III, and IV, including a pseudoknot upstream of the AUG start codon [86] which is conserved with pestivirus IRES elements [22]. Domain III participates in the interaction with eIF3 and the 40S subunit, while domain II helps to accommodate the mRNA in the tRNA-exit site of the ribosome and mediates eIF2 release during 80S assembly [87, 88]. Interestingly, HCV-like IRES elements with similar eIF requirement have been found in some picornavirus genera [18, 89, 90], which presumably arose by recombination events.
A unique type of IRES element is located in the intergenic region (IGR) of the genome of dicistroviruses (Figure 1(g)). This RNA region spans about 200 nucleotides and adopts a tertiary structure including three pseudoknots that functionally substitute the initiator tRNA during internal initiation [29, 91]. The IGR mimics a tRNA anticodon loop base-paired to mRNA and a translation elongation factor, facilitating initiation without the help of eIFs. A unique feature of these IRES elements is to initiate protein synthesis at non-AUG codons (CUU, GCU, CCU, CUC, depending on the dicistrovirus genus) [92, 93], with preference for alanine-coding triplets.
IRES elements were reported in various cellular mRNAs that remained attached to polysomes under conditions inhibitory to cap-dependent translation [51, 94]. These mRNAs contain a cap at the 5′end although they are translated at very low levels and have the capacity to switch to an IRES-dependent mechanism when cap-dependent initiation is impaired. This process is assisted by ITAFs, a group of RNA-binding proteins that are thought to help in the proper folding of the IRES region facilitating the mRNA recruitment to the translation machinery.
Thus, attending to the essential requirements for internal initiation, IRES elements can be grouped in two main categories: (a) those that do not need proteins to assemble the initiation complex (e.g., the IGR of dicistroviruses that adopts a docking structure capable of fitting in the ribosomal subunit [95]) and (b) those that do need factors to recruit the ribosome (typically, picornaviruses, HCV, and cellular IRES elements [60, 96, 97]). Within the second category, distinct groups can be made depending on the RNA structural motifs and proteins required for activity.
4. RNA Structural Motifs Found in IRES Elements
RNA structure plays a fundamental role in viral IRES-dependent translation initiation [13]. In support of this, mutations leading to the disruption of specific RNA structure motifs impaired IRES activity while the corresponding compensatory mutations restored IRES function [21, 98]. Furthermore, RNA structure of viral IRES elements is organized in modules which are phylogenetically conserved [99–102], providing evidence in favour of a distribution of functions among the different RNA domains [103, 104].
Examples of structural motifs found in IRES elements are the pseudoknots (Pks). These are tertiary motifs that play important role in the IRES of HCV, bovine viral diarrhea virus (BVDV), and classical swine fever virus (CSFV) [22]. Three different Pks conform the IGR of cricket paralysis virus (CrPV) and plautia stali intestine virus (PSIV), as well as other dicistroviruses [91, 93]. Giardiavirus (GLV) and tobacco etch virus (TEV) IRES elements also contain Pk structures [39, 40]. While these IRES elements are located in genetically distant RNA viruses, the Pk structure is conserved, indicating that the RNA organization is biologically relevant for internal initiation. Some cellular IRES elements were reported to contain Pk structures, as illustrated by c-myc and L-myc [77, 105]. In further support of the role of RNA structure for IRES-dependent translation, the zipper model proposed for the cationic amino acid transporter CAT-1 mRNA suggested that RNA structure modification via translation of an upstream uORF induces the formation of the active IRES [46].
Evidence for tight links between RNA structure and biological function are provided by the conservation of structural motifs within IRES elements of highly variable genomes. Specifically, the purine-rich GNRA and RAAA motifs (N stands for any nucleotide, and R, purine), as well as the G : C-rich stems that hold these motifs are conserved between aphthovirus and cardiovirus [106, 107]. In the case of foot-and-mouth disease virus (FMDV), the prototype of the aphthovirus genus, the central IRES domain is a self-folding region that has been proposed to instruct the functional conformation of the whole IRES element [85, 108, 109]. Along this idea, RNA structural analysis provided evidence for stem loops whose structural conformation depends on distant interactions within this domain, involving residues of the GNRA motif [110]. Thus, it is likely that RNA structural motifs located in the apical region of the central domain could constitute a signature of picornavirus type II IRES elements.
The GNRA motif of picornavirus IRES adopts a tetraloop conformation at the tip of a stem loop [111–113]. This motif is essential for IRES activity in FMDV and EMCV [114, 115], showing a strong preference for GUAA in the case of the FMDV IRES [108]. This observation, together with the lack of genetic variability within the apical stem and the covariation observed in the adjacent stems, pointed towards their joint contribution to IRES activity. Importantly, mutational analysis of the invariant apical stem revealed a better performance of G : C than C : G base-pairs, demonstrating the relevance of the three-dimensional RNA conformation for IRES activity [106].
Although related RNA viruses share the overall organization of their genomic RNA, viral IRES is organized in high-order structures that differ between distant families. Cryo-electron microscopy studies of the HCV IRES and the IGR provided information on the capacity of these RNAs to be accommodated in the interface of the ribosomal subunits [88, 95]. Even though the IGR of CrPV and the HCV IRES exhibit different structural organization, they interact with the ribosomal protein RpS25 [116] and induce similar conformational changes in the 40S ribosomal subunit. This finding opens the possibility that IRES elements could possess a universal structural motif mediating its direct interaction with the 40S subunit. This putative universal RNA motif still remains elusive, but it could be a promising tool to search for unidentified IRES elements at the genomic level.
Concerning the identification of structural motifs conserved between genetically distant RNAs, the IRES region of FMDV, EMCV, CrPV, HCV, CSFV, and BVDV contains a structural element recognized as substrate of the RNase P ribozyme [117–119]. RNase P is a nuclear structure-dependent endonuclease involved in the processing of the tRNA precursor, that also recognizes as substrate viral RNAs containing tRNA-like structures at the 3′end. The RNase P cleavage site in the FMDV IRES maps within an internal region that is involved in tertiary interactions; in addition, defective IRES mutants bearing modified RNA structures exhibited a differential response to ribozyme cleavage both in vitro and in transfected cells [119, 120]. The significance of the RNase P recognition motif in IRES elements is unknown since there is no proof for its direct involvement in the translation process. However, it does not constitute an RNA processing motif in transfected cells [121], consistent with the fact that the picornavirus infection cycle, as well as that of HCV and pestivirus, occurs in the cytoplasm of infected cells; therefore, the viral RNA has no access to RNase P.
The possibility that this structural motif constitutes a remnant of an ancient tRNA-like structure, similar to that found in the IGR IRES, is open to further investigations. Indeed, the evolutionary origin of IRES elements is unknown, but it has been proposed that this mode of initiating protein synthesis could be operating earlier than the cap-dependent [122]. In keeping with this hypothesis, the IRES property of self-interacting with the ribosome is a very attractive idea, also consistent with the finding that IRES activity is sensitive to changes in ribosome composition [123–126].
Another possibility to explain the presence of tRNA-like motifs within viral IRES elements is that they were inherited from RNA replication signals accommodated to assist in the translation process. In plant RNA viruses, tRNA-like structures located at the 3′end of the viral genome control cap-independent translation initiation and viral RNA replication [11, 127]. RNA-RNA interactions between the 3′and the 5′UTR of the viral genome assist in these processes. In this regard, long-range RNA-RNA interactions between the 5′ and the 3′end of some viral genomes have been observed [128, 129]. Consistent with a functional link between the ends of the viral RNA, IRES activity is stimulated by the 3′UTR [130, 131]. In picornavirus RNAs, the 3′UTR is composed of two stem loops and a short poly(A) tail that are required for replication and infectivity [132]. Furthermore, the insulin-like growth factor II mRNA-binding protein 1 (IGF2BP1) was identified among the proteins identified in complexes assembled with RNAs that contained the HCV IRES and the 3′UTR. This protein coimmunoprecipitates with eIF3 and the 40S subunit [133], suggesting that it enhances HCV IRES activity by recruiting the ribosomal subunits to a pseudo circularized RNA. Thus, bridging of 5′ and 3′ends involves direct RNA-RNA contacts and RNA-protein interactions. These results provide a mechanistic basis for translation stimulation and replication of the viral RNA resembling the synergistic stimulation of cap-dependent translation.
5. RNA-Protein Interactions Controlling IRES Activity
The lack of conserved features among distantly related IRES elements has led to the view that different IRES elements could recruit the ribosomal subunits assisted by unique sets of ITAFs. Along this idea, riboproteomic approaches have facilitated the identification of various proteins interacting with different IRES elements [134]. Most ITAFs are RNA-binding proteins previously identified as transcription regulators, splicing factors, RNA transport, RNA stability, or translation control proteins [133, 135, 136]. Typical examples of multifunctional proteins that act as ITAFs are the polypyrimidine tract-binding protein (PTB), the poly-r(C) binding protein (PCBP2), the SR splicing factor (SRp20), the far upstream element binding protein 2 (FBP2), the lupus La antigen (La), or Gemin5, among others [136–140].
The IRES elements of HCV and HIV-1 differ from those of picornaviruses not only in RNA structure but also in some factor requirement. Assembly of the HCV IRES-48S initiation complex requires eIF3, but not eIF4G [96]. Consistent with this, eIF3 has been identified by mass spectrometry of IRES-bound protein complexes [135, 141]. Other proteins bound to HCV and picornavirus IRES are PTB, PCBP 2, nucleolin, Gemin5, upstream of n-ras (unr), heterogeneous nuclear RNA-binding protein (hnRNP) A1/A2, La autoantigen (La), NS1-associated protein, as well as several RNA helicases DEAH-box polypeptide 9 (DHX9) [134]. Gemin5 binds directly to FMDV and HCV IRES regions and down regulates translation efficiency. Additionally, Gemin5 binds m7GTP [142], explaining its down regulation of cap-dependent translation [140]. In contrast, the HIV-1 IRES is stimulated by hnRNP A1/2, the RNA helicase DEAD/H Box 3 (DDX3), the human Rev-interacting protein (hRIP), and the nuclear RNA-binding protein Src-associated in mitosis (Sam68) [143].
Cellular IRES elements are typically present in mRNAs encoding stress response proteins, such as those needed during nutrient deprivation, temperature shock, hibernation, hypoxia, cell cycle arrest, or apoptosis (Table 2) [2, 14, 57, 144]. However, with the exception of polypyrimidine tracts, conservation of primary sequence is not readily detected between viral and cellular IRES elements [145]. The observation that cellular IRES elements do not share overall structural similarity [146] has led to the view that short motifs may control the interaction with transacting factors needed to recruit the mRNA to 40S subunits. In favor of this hypothesis, PTB stimulates the IRES of the apoptotic protease-activating factor 1 (apaf-1), BCL2-associated athanogene (BAG)-1, and the hypoxia-inducible factor (HIF1a) [50, 147], allowing the synthesis of proteins that mediate cell survival under apoptosis, hypoxia, nutrient deprivation, or cell growth dysregulation. Proteins interacting with the lymphoid enhancer factor (LEF-1) IRES recently identified using biotin-tagged RNAs combined with stable isotope labeling with amino acids in cell culture (SILAC)-based quantitative mass spectrometry [65] include the splicing-related protein proline and glutamine-rich SFPQ/PSF, the non-POU domain-containing octamer-binding nuclear RNA-binding protein (nonO/p54nrb), PCBP 2, HuR, and the oncoprotein DEK (named by the initials of a patient affected of acute myeloid leukemia). Investigating whether the proteins identified in intracellular IRES-ribonucleoprotein complexes perform the same or different functions from those found in IRES complexes assembled in vitro requires further work.
Since ITAFs usually act in large complexes with various factors within the cellular compartments, proteins interacting with different targets may lead to distinct effects depending on the target RNA and the other partners of the complex. Thus, changes in the abundance, posttranslational modifications, or subcellular location of ITAFs could be responsible for the distinct IRES response to stress conditions. For example, the kinase PITSLREp58 IRES is specifically activated during mitosis, while mPer 1 translation oscillates during circadian rhythmic period [148, 149]; the Apaf IRES is activated during apoptosis, while the X-linked inhibitor of apoptosis protein (XIAP) is inhibited [150]. Relocalization of hnRNP A1 mediates internal initiation of c-myc, unr, cyclin D1, vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF-2), Apaf-1, and XIAP mRNAs [151]. In contrast, death-associated protein (DAP 5), nuclear factor NF45, G-rich RNA sequence binding factor (GRSF-1), fragile-X mental retardation protein (FMRP), dyskeratosis congenita (DKC1), heterogeneous nuclear ribonucleoprotein D-like protein (JKTBP1), or zinc-finger protein (ZNF9) are IRES-specific [57, 61, 152–154]. Hence, individual mRNAs seem to use different mechanisms to evade the global repression of protein synthesis.
6. Perspectives towards the Identification of IRES Elements at Genomewide Scale in Eukaryotes
A functional assay testing the cap-independent capacity and the ability to resist cap-inhibitory conditions is the usual way to identify IRES elements in mRNAs. This is a cumbersome task in terms of genomewide scale identification of IRES elements in eukaryotic genomes. To facilitate this task, short conserved structural motifs identified in model IRES elements could provide a tool to search for putative IRES at a genomic level. In considering critical features of IRES elements, signals that may be suggestive of functional IRES could be the presence of polypyrimidine tracts, pseudoknots near the start codon, or hairpin-loops mimicking those present in the IRES of picornavirus, HCV, or the IGR of dicistrovirus RNAs. Although none of these features individually are sufficient to define a functional IRES element, the presence of one (or more) of these motifs may provide hints to select potential IRES in mRNAs.
The question that remains unresolved is what are the distinctive features of IRES elements that may allow their accurate prediction at the genomewide scale, even though computer programs have being designed to predict IRES elements using bioinformatics tools (http://140.135.61.9/ires/) [75]. From our point of view this question is still far from being answered; however, the detailed molecular, biochemical, and structural characterization of model IRES elements will provide critical hints to reveal the presence of similar elements within eukaryotic genomes.
Acknowledgment
This work was supported by Grants BFU2008-02159, CSD2009-00080, and by an Institutional grant from Fundación Ramón Areces.
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Molecular Cell. 2010;40(2):228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 3.Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, Eriani G. Cap-assisted internal initiation of translation of histone H4. Molecular Cell. 2011;41:197–209. doi: 10.1016/j.molcel.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Parisien M, Scheets K, Miller WA. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure. 2011;19:868–880. doi: 10.1016/j.str.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez-Sanchez C, Mansilla A, de la Rosa EJ, Pollerberg GE, Martínez-Salas E, de Pablo F. Upstream AUGs in embryonic proinsulin mRNA control its low translation level. The EMBO Journal. 2003;22(20):5582–5592. doi: 10.1093/emboj/cdg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaszczyk L, Ciesiolka J. Secondary structure and the role in translation initiation of the 5′-terminal region of p53 mRNA. Biochemistry. 2011;50:7080–7092. doi: 10.1021/bi200659b. [DOI] [PubMed] [Google Scholar]
- 7.Rakotondrafara AM, Polacek C, Harris E, Miller WA. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006;12(10):1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powley IR, Kondrashov A, Young LA, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes and Development. 2009;23(10):1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babinger K, Hallmann A, Schmitt R. Translational control of regA, a key gene controlling cell differentiation in Volvox carteri. Development. 2006;133(20):4045–4051. doi: 10.1242/dev.02582. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Research. 2009;38(2):353–359. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stupina VA, Yuan X, Meskauskas A, Dinman JD, Simon AE. Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′ untranslated region in cap-independent translation of turnip crinkle virus. Journal of Virology. 2011;85:4638–4653. doi: 10.1128/JVI.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan X, Shi K, Meskauskas A, Simon AE. The 3′ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA. 2009;15(10):1849–1864. doi: 10.1261/rna.1708709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends in Microbiology. 2008;16(5):230–237. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 16.Jang SK, Krausslich HG, Nicklin MJH, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. Journal of Virology. 1988;62(8):2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Salas E, Saiz JC, Davila M, Belsham GJ, Domingo E. A single nucleotide substitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. Journal of Virology. 1993;67(7):3748–3755. doi: 10.1128/jvi.67.7.3748-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. Journal of Virology. 2004;78(9):4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. The EMBO Journal. 2011;30:4423–4436. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willcocks MM, Locker N, Gomwalk Z, et al. Structural features of the Seneca valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES. Journal of Virology. 2011;85:4452–4461. doi: 10.1128/JVI.01107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda M, Ping LH, Rijnbrand RCA, et al. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222(1):31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 22.Rijnbrand R, van der Straaten T, van Rijn PA, Spaan WJM, Bredenbeek PJ. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. Journal of Virology. 1997;71(1):451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locker N, Chamond N, Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Research. 2011;39(6):2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallejos M, Deforges J, Plank TD, et al. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Research. 2011;39:6186–6200. doi: 10.1093/nar/gkr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vagner S, Waysbort A, Marenda M, Gensac MC, Amalric F, Prats AC. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. The Journal of Biological Chemistry. 1995;270(35):20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 26.Camerini V, Decimo D, Balvay L, et al. A dormant internal ribosome entry site controls translation of feline immunodeficiency virus. Journal of Virology. 2008;82(7):3574–3583. doi: 10.1128/JVI.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallejos M, Ramdohr P, Valiente-Echeverría F, et al. The 5′-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Research. 2009;38(2):618–632. doi: 10.1093/nar/gkp890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolaway KE, Lazaridis K, Belsham GJ, Carter MJ, Roberts LO. The 5′ untranslated region of Rhopalosiphum padi virus contains an internal ribosome entry site which functions efficiently in mammalian, plant, and insect translation systems. Journal of Virology. 2001;75(21):10244–10249. doi: 10.1128/JVI.75.21.10244-10249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson JE, Pestova TV, Hellen CUT, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102(4):511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 30.Firth AE, Wang QS, Jan E, Atkins JF. Bioinformatic evidence for a stem-loop structure 5′-adjacent to the IGR-IRES and for an overlapping gene in the bee paralysis dicistroviruses. Virology Journal. 2009;6, article 193 doi: 10.1186/1743-422X-6-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, Hu Y, Hu L, et al. Ectropis obliqua picorna-like virus IRES-driven internal initiation of translation in cell systems derived from different origins. Journal of General Virology. 2007;88(10):2834–2838. doi: 10.1099/vir.0.83201-0. [DOI] [PubMed] [Google Scholar]
- 32.Shibuya N, Nakashima N. Characterization of the 5′ internal ribosome entry site of Plautia stali intestine virus. Journal of General Virology. 2006;87(12):3679–3686. doi: 10.1099/vir.0.82193-0. [DOI] [PubMed] [Google Scholar]
- 33.Czibener C, Alvarez D, Scodeller E, Gamarnik AV. Characterization of internal ribosomal entry sites of Triatoma virus. Journal of General Virology. 2005;86(8):2275–2280. doi: 10.1099/vir.0.80842-0. [DOI] [PubMed] [Google Scholar]
- 34.Karetnikov A, Lehto K. The RNA2 5′ leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. Journal of General Virology. 2007;88:286–297. doi: 10.1099/vir.0.82307-0. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez-Miragall O, Hernandez C. An internal ribosome entry site directs translation of the 3′-gene from pelargonium flower break virus genomic RNA: implications for infectivity. PLoS ONE. 2011;6, article e22617 doi: 10.1371/journal.pone.0022617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorokhov YL, Ivanov PA, Komarova TV, Skulachev MV, Atabekov JG. An internal ribosome entry site located upstream of the crucifer-infecting tobamovirus coat protein (CP) gene can be used for CP synthesis in vivo. Journal of General Virology. 2006;87(9):2693–2697. doi: 10.1099/vir.0.82095-0. [DOI] [PubMed] [Google Scholar]
- 37.Jaag HM, Kawchuk L, Rohde W, Fischer R, Emans N, Prüfer D. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(15):8939–8944. doi: 10.1073/pnas.1332697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh DCY, Wong SM, Liu DX. Synergism of the 3′-untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. The Journal of Biological Chemistry. 2003;278(23):20565–20573. doi: 10.1074/jbc.M210212200. [DOI] [PubMed] [Google Scholar]
- 39.Zeenko V, Gallie DR. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. The Journal of Biological Chemistry. 2005;280(29):26813–26824. doi: 10.1074/jbc.M503576200. [DOI] [PubMed] [Google Scholar]
- 40.Garlapati S, Wang CC. Structural elements in the 5′-untranslated region of giardiavirus transcript essential for internal ribosome entry site-mediated translation initiation. Eukaryotic Cell. 2005;4(4):742–754. doi: 10.1128/EC.4.4.742-754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maga JA, Widmer G, LeBowitz JH. Leishmania RNA virus 1-mediated cap-independent translation. Molecular and Cellular Biology. 1995;15(9):4884–4889. doi: 10.1128/mcb.15.9.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grainger L, Cicchini L, Rak M, et al. Stress-inducible alternative translation initiation of human cytomegalovirus latency protein pUL138. Journal of Virology. 2010;84(18):9472–9486. doi: 10.1128/JVI.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y, Alwine JC. 19S late mRNAs of simian virus 40 have an internal ribosome entry site upstream of the virion structural protein 3 coding sequence. Journal of Virology. 2006;80(13):6553–6558. doi: 10.1128/JVI.00517-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isaksson A, Berggren M, Ekeland-Sjoberg K, Samuelsson T, Ricksten A. Cell specific internal translation efficiency of Epstein-Barr virus present in solid organ transplant patients. Journal of Medical Virology. 2007;79(5):573–581. doi: 10.1002/jmv.20854. [DOI] [PubMed] [Google Scholar]
- 45.Tahiri-Alaoui A, Smith LP, Baigent S, et al. Identification of an intercistronic internal ribosome entry site in a Marek’s disease virus immediate-early gene. Journal of Virology. 2009;83(11):5846–5853. doi: 10.1128/JVI.02602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez J, Yaman I, Huang C, et al. Ribosome stalling regulates ires-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Molecular Cell. 2005;17(3):405–416. doi: 10.1016/j.molcel.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 47.Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25(33):4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
- 48.Al-Fageeh MB, Smales CM. Cold-inducible RNA binding protein (CIRP) expression is modulated by alternative mRNAs. RNA. 2009;15(6):1165–1176. doi: 10.1261/rna.1179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Molecular and Cellular Biology. 2004;24(12):5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S. The polypyrimidine tract-binding protein stimulates HIF-1α IRES-mediated translation during hypoxia. Nucleic Acids Research. 2005;33(21):6884–6894. doi: 10.1093/nar/gki1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353(6339):90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 52.Stoneley M, Subkhankulova T, Le Quesne JPC, et al. Analysis of the c-myc IRES; A potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Research. 2000;28(3):687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonnal S, Schaeffer C, Créancier L, et al. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. The Journal of Biological Chemistry. 2003;278(41):39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fox JT, Shin WK, Caudill MA, Stover PJ. A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair. The Journal of Biological Chemistry. 2009;284(45):31097–31108. doi: 10.1074/jbc.M109.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. Journal of the American Chemical Society. 2010;132(50):17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 56.Riley A, Jordan LE, Holcik M. Distinct 5′ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Research. 2010;38(14):4665–4674. doi: 10.1093/nar/gkq241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graber TE, Baird SD, Kao PN, Mathews MB, Holcik M. NF45 functions as an IRES transacting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death and Differentiation. 2010;17(4):719–729. doi: 10.1038/cdd.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(8):5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spriggs KA, Cobbold LC, Ridley SH, et al. The human insulin receptor mRNA contains a functional internal ribosome entry segment. Nucleic Acids Research. 2009;37(17):5881–5893. doi: 10.1093/nar/gkp623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Molecular Cell. 2003;11(3):757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 61.Marash L, Liberman N, Henis-Korenblit S, et al. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Molecular Cell. 2008;30(4):447–459. doi: 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 62.Hernández G, Vázquez-Pianzola P, Sierra JM, Rivera-Pomar R. Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA. 2004;10(11):1783–1797. doi: 10.1261/rna.7154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vazquez-Pianzola P, Hernández G, Suter B, Rivera-Pomar R. Different modes of translation for hid, grim and sickle mRNAs in Drosophila. Cell Death and Differentiation. 2007;14(2):286–295. doi: 10.1038/sj.cdd.4401990. [DOI] [PubMed] [Google Scholar]
- 64.Oh SK, Scott MP, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes and Development. 1992;6(9):1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 65.Tsai BP, Wang X, Huang L, Waterman ML. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Molecular & Cellular Proteomics. 2011;10(4, article M110 007385) doi: 10.1074/mcp.M110.007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sella O, Gerlitz G, Le SY, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Molecular and Cellular Biology. 1999;19(8):5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villa-Cuesta E, Sage BT, Tatar M. A role for drosophila dFoxO and dFoxO 5′UTR internal ribosomal entry sites during fasting. PLoS ONE. 2010;5(7, article e11521) doi: 10.1371/journal.pone.0011521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marr MT, D’Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes and Development. 2007;21(2):175–183. doi: 10.1101/gad.1506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramanathan P, Guo J, Whitehead RN, Brogna S. The intergenic spacer of the Drosophila Adh-Adhr dicistronic mRNA stimulates internal translation initiation. RNA Biology. 2008;5(3):149–156. doi: 10.4161/rna.5.3.6676. [DOI] [PubMed] [Google Scholar]
- 70.Dinkova TD, Zepeda H, Martinez-Salas E, Martinez LM, Nieto-Sotelo J, de Jimenez SE. Cap-independent translation of maize Hsp101. Plant Journal. 2005;41(5):722–731. doi: 10.1111/j.1365-313X.2005.02333.x. [DOI] [PubMed] [Google Scholar]
- 71.Mardanova ES, Zamchuk LA, Skulachev MV, Ravin NV. The 5′ untranslated region of the maize alcohol dehydrogenase gene contains an internal ribosome entry site. Gene. 2008;420(1):11–16. doi: 10.1016/j.gene.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 72.Reineke LC, Merrick WC. Characterization of the functional role of nucleotides within the URE2 IRES element and the requirements for eIF2A-mediated repression. RNA. 2009;15(12):2264–2277. doi: 10.1261/rna.1722809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317(5842):1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 74.Mokrejs M, Vopalensky V, Kolenaty O, et al. IRESite: the database of experimentally verified IRES structures ( http://www.iresite.org/) Nucleic Acids Research. 2006;34:D125–D130. doi: 10.1093/nar/gkj081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu TY, Hsieh CC, Hong JJ, Chen CY, Tsai YS. IRSS: a web-based tool for automatic layout and analysis of IRES secondary structure prediction and searching system in silico. BMC Bioinformatics. 2009;10, article 160 doi: 10.1186/1471-2105-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.López de Quinto S, Martínez-Salas E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255(2):324–336. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- 77.Le Quesne JPC, Stoneley M, Fraser GA, Willis AE. Derivation of a structural model for the c-myc IRES. Journal of Molecular Biology. 2001;310(1):111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 78.Grillo G, Turi A, Licciulli F, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Research. 2009;38(1):D75–D80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martínez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. Journal of General Virology. 2008;89(3):611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 80.Rodríguez Pulido M, Serrano P, Sáiz M, Martínez-Salas E. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology. 2007;364(2):466–474. doi: 10.1016/j.virol.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 81.Martinez-Salas E, Ryan M. Translation and protein processing. In: Ehrenfeld E, Domingo E, Roos R, editors. Picornaviruses. ASM Press; 2010. pp. 141–161. [Google Scholar]
- 82.Andreev DE, Fernandez-Miragall O, Ramajo J, et al. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13(8):1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CUT. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.López de Quinto S, Lafuente E, Martínez-Salas E. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7(9):1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fernandez-Miragall O, Lopez de Quinto S, Martinez-Salas E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Research. 2009;139:172–182. doi: 10.1016/j.virusres.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 86.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. The EMBO Journal. 2007;26(3):795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spahn CMT, Kieft JS, Grassucci RA, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291(5510):1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 89.de Breyne S, Yu Y, Pestova TV, Hellen CUT. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA. 2008;14(2):367–380. doi: 10.1261/rna.696508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Research. 2009;37(16):5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nature Structural and Molecular Biology. 2008;15(1):57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. Journal of Virology. 1999;73(2):1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakashima N, Uchiumi T. Functional analysis of structural motifs in dicistroviruses. Virus Research. 2009;139(2):137–147. doi: 10.1016/j.virusres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 94.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex elF4F concentrations using a cDNA microarray. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(23):13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spahn CMT, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118(4):465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initation of hepatitis C and classical swine fever virus RNAs. Genes and Development. 1998;12(1):67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lopez de Quinto S, Martinez-Salas E. Interaction of the elF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6(10):1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martinez-Salas E, Regalado MP, Domingo E. Identification of an essential region for internal initiation of translation in the aphthovirus internal ribosome entry site and implications for viral evolution. Journal of Virology. 1996;70(2):992–998. doi: 10.1128/jvi.70.2.992-998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martinez-Salas E, Fernandez-Miragall O. Picornavirus IRES: structure function relationship. Current Pharmaceutical Design. 2004;10:3757–3767. doi: 10.2174/1381612043382657. [DOI] [PubMed] [Google Scholar]
- 100.Honda M, Rijnbrand R, Abell G, Kim D, Lemon SM. Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA- RNA interaction outside of the internal ribosomal entry site. Journal of Virology. 1999;73(6):4941–4951. doi: 10.1128/jvi.73.6.4941-4951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saiz JC, Lopez de Quinto S, Ibarrola N, et al. Internal initiation of translation efficiency in different hepatitis C genotypes isolated from interferon treated patients. Archives of Virology. 1999;144:215–229. doi: 10.1007/s007050050499. [DOI] [PubMed] [Google Scholar]
- 102.Barria MI, Gonzalez A, Vera-Otarola J, et al. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Research. 2009;37:957–971. doi: 10.1093/nar/gkn1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Serrano P, Ramajo J, Martínez-Salas E. Rescue of internal initiation of translation by RNA complementation provides evidence for a distribution of functions between individual IRES domains. Virology. 2009;388(1):221–229. doi: 10.1016/j.virol.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 104.Jang CJ, Jan E. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA. 2010;16(6):1182–1195. doi: 10.1261/rna.2044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jopling CL, Spriggs KA, Mitchell SA, Stoneley M, Willis AE. L-Myc protein synthesis is initiated by internal ribosome entry. RNA. 2004;10(2):287–298. doi: 10.1261/rna.5138804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fernandez N, Fernandez-Miragall O, Ramajo J, et al. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Research. 2011;39:8572–8585. doi: 10.1093/nar/gkr560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Research. 2011;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fernandez N, Garcia-Sacristan A, Ramajo J, Briones C, Martinez-Salas E. Structural analysis provides insights into the modular organization of picornavirus IRES. Virology. 2011;409:251–261. doi: 10.1016/j.virol.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 109.Ramos R, Martínez-Salas E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES) RNA. 1999;5(10):1374–1383. doi: 10.1017/s1355838299991240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Phelan M, Banks RJ, Conn G, Ramesh V. NMR studies of the structure and Mg2+ binding properties of a conserved RNA motif of EMCV picornavirus IRES element. Nucleic Acids Research. 2004;32(16):4715–4724. doi: 10.1093/nar/gkh805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Du Z, Ulyanov NB, Yu J, Andino R, James TL. NMR structures of loop B RNAs from the stem-loop IV domain of the enterovirus internal ribosome entry site: a single C to U substitution drastically changes the shape and flexibility of RNA. Biochemistry. 2004;43(19):5757–5771. doi: 10.1021/bi0363228. [DOI] [PubMed] [Google Scholar]
- 114.Lopez de Quinto S, Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. Journal of Virology. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Robertson MEM, Seamons RA, Belsham GJ. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5(9):1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes and Development. 2009;23(23):2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nadal A, Martell M, Lytle JR, et al. Specific cleavage of hepatitis C virus RNA genome by human RNase P. The Journal of Biological Chemistry. 2002;277(34):30606–30613. doi: 10.1074/jbc.M203595200. [DOI] [PubMed] [Google Scholar]
- 118.Lyons AJ, Robertson HD. Detection of tRNA-like structure through RNase P cleavage of viral internal ribosome entry site RNAs near the AUG start triplet. The Journal of Biological Chemistry. 2003;278(29):26844–26850. doi: 10.1074/jbc.M304052200. [DOI] [PubMed] [Google Scholar]
- 119.Serrano P, Gomez J, Martínez-Salas E. Characterization of a cyanobacterial RNase P ribozyme recognition motif in the IRES of foot-and-mouth disease virus reveals a unique structural element. RNA. 2007;13(6):849–859. doi: 10.1261/rna.506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fernandez N, Martinez-Salas E. Tailoring the switch from IRES-dependent to 5′-end-dependent translation with the RNase P ribozyme. RNA. 16:852–862. doi: 10.1261/rna.1973710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Piron M, Beguiristain N, Nadal A, Martinez-Salas E, Gomez J. Characterizing the function and structural organization of the 5′ tRNA-like motif within the hepatitis C virus quasispecies. Nucleic Acids Research. 2005;33:1487–1502. doi: 10.1093/nar/gki290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hernández G. Was the initiation of translation in early eukaryotes IRES-driven? Trends in Biochemical Sciences. 2008;33(2):58–64. doi: 10.1016/j.tibs.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 123.Martínez-Azorín F, Remacha M, Martínez-Salas E, Ballesta JPG. Internal translation initiation on the foot-and-mouth disease virus IRES is affected by ribosomal stalk conformation. FEBS Letters. 2008;582(20):3029–3032. doi: 10.1016/j.febslet.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 124.Yang C, Zhang C, Dittman JD, Whitham SA. Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology. 2009;390(2):163–173. doi: 10.1016/j.virol.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 125.Basu A, Das P, Chaudhuri S, et al. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular Internal Ribosome Entry Sites. Molecular and Cellular Biology. 2011;31:4482–4499. doi: 10.1128/MCB.05804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Horos R, Ijspeert H, Pospisilova D, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–272. doi: 10.1182/blood-2011-06-358200. [DOI] [PubMed] [Google Scholar]
- 127.Yuan X, Shi K, Young MYL, Simon AE. The terminal loop of a 3′ proximal hairpin plays a critical role in replication and the structure of the 3′ region of Turnip crinkle virus. Virology. 2010;402(2):271–280. doi: 10.1016/j.virol.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Serrano P, Pulido MR, Saiz M, Martinez-Salas E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. Journal of General Virology. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- 129.Romero-López C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009;15(9):1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lopez de Quinto S, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Research. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dobrikova E, Florez P, Bradrick S, Gromeier M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Saiz M, Gomez S, Martinez-Salas E, Sobrino F. Deletion or substitution of the aphthovirus 3′ NCR abrogates infectivity and virus replication. Journal of General Virology. 2001;82:93–101. doi: 10.1099/0022-1317-82-1-93. [DOI] [PubMed] [Google Scholar]
- 133.Weinlich S, Hüttelmaier S, Schierhorn A, Behrens SE, Ostareck-Lederer A, Ostareck DH. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3′UTR. RNA. 2009;15(8):1528–1542. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pacheco A, Martinez-Salas E. Insights into the biology of IRES elements through riboproteomic approaches. Journal of Biomedicine and Biotechnology. 2010;2010:12 pages. doi: 10.1155/2010/458927. Article ID 458927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pacheco A, Reigadas S, Martínez-Salas E. Riboproteomic analysis of polypeptides interacting with the internal ribosome-entry site element of foot-and-mouth disease viral RNA. Proteomics. 2008;8(22):4782–4790. doi: 10.1002/pmic.200800338. [DOI] [PubMed] [Google Scholar]
- 136.Vazquez-Pianzola P, Urlaub H, Rivera-Pomar R. Proteomic analysis of reaper 5′ untranslated region-interacting factors isolated by tobramycin affinity-selection reveals a role for La antigen in reaper mRNA translation. Proteomics. 2005;5(6):1645–1655. doi: 10.1002/pmic.200401045. [DOI] [PubMed] [Google Scholar]
- 137.Jang SK, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes and Development. 1990;4(9):1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 138.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. The EMBO Journal. 2007;26(2):459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin JY, Li ML, Shih SR. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Research. 2009;37(1):47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pacheco A, Lopez de Quinto S, Ramajo J, Fernandez N, Martinez-Salas E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Research. 2009;37:582–590. doi: 10.1093/nar/gkn979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yu Y, Ji H, Doudna JA, Leary JA. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Science. 2005;14(6):1438–1446. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bradrick SS, Gromeier M. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS ONE. 2009;4(9, article e7030) doi: 10.1371/journal.pone.0007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu J, Henao-Mejia J, Liu H, Zhao Y, He JJ. Translational regulation of HIV-1 replication by HIV-1 Rev cellular cofactors Sam68, eIF5A, hRIP, and DDX3. Journal of NeuroImmune Pharmacology. 2011;6:308–321. doi: 10.1007/s11481-011-9265-8. [DOI] [PubMed] [Google Scholar]
- 144.Pan P, van Breukelen F. Preference of IRES-mediated initiation of translation during hibernation in golden-mantled ground squirrels, Spermophilus lateralis. American Journal of Physiology. 2011;301:R370–R377. doi: 10.1152/ajpregu.00748.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bushell M, Stoneley M, Kong YW, et al. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Molecular Cell. 2006;23(3):401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 146.Xia X, Holcik M. Strong eukaryotic IRESs have weak secondary structure. PLoS ONE. 2009;4(1, article e4136) doi: 10.1371/journal.pone.0004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dobbyn HC, Hill K, Hamilton TL, et al. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. 2008;27(8):1167–1174. doi: 10.1038/sj.onc.1210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schepens B, Tinton SA, Bruynooghe Y, et al. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. The EMBO Journal. 2007;26(1):158–169. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee KH, Woo KC, Kim DY, et al. Rhythmic interaction between period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Molecular and Cellular Biology. 2012;32:717–728. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ungureanu NH, Cloutier M, Lewis SM, et al. Internal ribosome entry site-mediated translation of Apaf-1, but not XIAP, is regulated during UV-induced cell death. The Journal of Biological Chemistry. 2006;281(22):15155–15163. doi: 10.1074/jbc.M511319200. [DOI] [PubMed] [Google Scholar]
- 151.Cammas A, Pileur F, Bonnal S, et al. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Molecular Biology of the Cell. 2007;18(12):5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lewis SM, Cerquozzi S, Graber TE, Ungureanu NH, Andrews M, Holcik M. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Research. 2008;36(1):168–178. doi: 10.1093/nar/gkm1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sammons MA, Antons AK, Bendjennat M, Udd B, Krahe R, Link AJ. ZNF9 activation of IRES-mediated translation of the human ODC mRNA is decreased in myotonic dystrophy type 2. PLoS ONE. 2010;5(2, article e9301) doi: 10.1371/journal.pone.0009301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. The EMBO Journal. 2010;29(11):1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]


