Abstract
To identify transcriptional profiles predictive of the clinical benefit of cisplatin and fluorouracil (CF) chemotherapy to gastric cancer patients, endoscopic biopsy samples from 96 CF-treated metastatic gastric cancer patients were prospectively collected before therapy and analyzed using high-throughput transcriptional profiling and array comparative genomic hybridization. Transcriptional profiling identified 917 genes that are correlated with poor patient survival after CF at P<0.05 (poor prognosis signature), in which protein synthesis and DNA replication/recombination/repair functional categories are enriched. A survival risk predictor was then constructed using genes, which are included in the poor prognosis signature and are contained within identified genomic amplicons. The combined expression of three genes—MYC, EGFR and FGFR2—was an independent predictor for overall survival of 27 CF-treated patients in the validation set (adjusted P=0.017), and also for survival of 40 chemotherapy-treated gastric cancer patients in a published data set (adjusted P=0.026). Thus, combined expression of MYC, EGFR and FGFR2 is predictive of poor survival in CF-treated metastatic gastric cancer patients.
Keywords: gastric, cancer, chemotherapy, gene, expression
Introduction
Although the emerging area of targeted anticancer agents holds great promise, cytotoxic chemotherapy remains the primary treatment option for many cancer patients. Identifying patients who likely will or will not benefit from cytotoxic chemotherapy through the use of biomarkers could greatly improve clinical management by better defining appropriate treatment options for patients. None of the molecules experimentally identified to cause chemotherapy resistance in vitro was sufficiently validated in primary tumors and thus clinically applicable,1 underscoring the importance of well-designed, clinical study to identify clinically relevant mechanisms for chemotherapy resistance. In fact, however, such predictors derived to date from high-throughput transcriptional profiling of primary tumors, especially gastrointestinal tract cancers, have not shown satisfactory performance.2, 3, 4, 5 It may be primarily owing to the high rate of false-positive discovery in high-throughput data, in addition to the high degree of genetic variation of individual tumor compared with limited number of samples available for the study.
To provide insight into clinically relevant mechanisms for chemotherapy resistance in gastric cancer, we prospectively collected and analyzed 123 endoscopic biopsy samples before cisplatin and fluorouracil (CF) chemotherapy from patients with extended follow-up, using high-throughput transcriptional profiling and comparative genomic hybridization (CGH) analyses. We could identify functional categories enriched in genes correlated with patient outcome, and develop a genomic predictor that was validated in two independent data sets.
Materials and methods
Patients
Sample collection, treatment and follow-up were performed according a protocol approved by the Institutional Review Board of the National Cancer Center Hospital in Goyang, Korea (NCCNHS01-003). All patients signed an Institutional Review Board-approved informed consent form. Eligibility for enrollment into the study included the following parameters: (1) age⩾18 years; (2) histologically confirmed gastric adenocarcinoma; (3) clinically documented distant metastasis; (4) no previous or concomitant malignancies other than the gastric cancer; (5) no previous history of chemotherapy, either adjuvant or palliative; and (6) adequate function of all major organs. Patients who were lost to follow-up before completing six cycles of chemotherapy, except for documented progressive disease, were excluded from this study.
Sample size calculation
Overall survival was the primary clinical end point of this study. As a minimum of 91 events were estimated to be required for the number of training set samples6 at α=0.001, β=0.05, τ (standard deviation of log intensity)=0.75 and δ (hazard ratio (HR) associated with one-unit change of log intensity)=2, we used the 96 samples collected until January 2005 as the training set for development of the predictor.
Ninety-six eligible patients who were treated with CF by one medical oncologist (HK) from August 2001 to January 2005 were used for the expression profiling training set. A second group of 27 eligible patients was used as the array validation cohort. Twenty-two patients in the validation cohort were treated with CF, and five patients were treated with cisplatin plus oral capecitabine (a fluorouracil pro-drug considered equivalent to fluorouracil; CX),7 by another group of medical oncologists in the same institution between February 2005 and April 2006. Tissue procurement and processing were the same for the training and validation samples.
Treatment
Patients continued therapy indefinitely until they experienced unacceptable toxicities or progressive disease was documented. CF-treated patients received cisplatin 60 mg m−2 intravenously on day 1 and fluorouracil 1000 mg m−2 intravenously on days 1–5 of a 3-week schedule. The treatment schedule for fluorouracil could be shortened at the discretion of the oncologist to 3 instead of 5 days for elderly patients (⩾70 years) or patients with poor performance status (Eastern Cooperative Oncology Group performance status ⩾2). Chemotherapy doses were reduced according to toxicities and the patient's performance status. Specific dose modification schemes for the subsequent cycle were left to the discretion of treating oncologist. Five patients (18.5%) in the validation group received oral capecitabine (Xeloda; Roche, Basel, Switzerland; 1250 mg m−2 twice a day for 2 weeks) instead of intravenous infusion of fluorouracil. Time to progression was measured from the initiation of chemotherapy to the progressive disease. In patients without any measurable lesions, time to progression was measured to the time when a change in therapy was required because unmeasurable lesions (such as ascites) unequivocally progressed.
Gene expression and CGH microarray analyses
Tissue samples were collected and processed for RNA and DNA extraction as described previously,8 only if samples contained at least 50% tumor cells. Affymetrix (Santa Clara, CA, USA) HG-U133A gene expression microarray data were analyzed with survival analysis algorithms of BRB-ArrayTools (version 3.6, National Cancer Institute, http://linus.nci.nih.gov/BRB-ArrayTools.html).9 The survival risk groups were constructed using a predictive index based on the supervised principal component method of Bair and Tibshirani.10 A three-gene predictive index percentile was generated based on the weighted average of the log intensities of the three genes (FGFR2 (211401_s_at), EGFR (210984_x_at) and c-MYC (202431_s_at)), using a proportional hazards regression on the first two principal components of the log intensities of those three genes, in which a high value of the predictive index corresponds to a high risk of death. If the predictive index of a sample in the validation set corresponded to the median predictive index of the training set, the sample was assigned a 50% predictive index. We specified the number of risk groups as 2 (high and low) and the predictive index percentile for defining the two risk groups as 67%, using a 67.1% rate of clinical benefit (partial response and stable disease) and 32.9% rate of progressive disease in the training set. We also performed Cox regression analyses using this three-gene predictive index percentile as a continuous variable, in which HRs for survival were calculated according to each percentile increase in three-gene predictive index percentile (from 0 to 100%). Array CGH data were generated using Agilent (Santa Clara, CA, USA) 4 × 44k HD-CGH Microarrays and analyzed using CGH Analytics software (version 3.5.14). Aberrations with average tumor/normal log2 ratio >2.0 were defined as amplifications. Experimental details are provided in Supplementary Materials and Methods.
Analyses of published DNA microarray data
The entire set of published Affymetrix U133 Plus 2.0 DNA microarray data4 (n=40) was combined with our training set data (n=96), using common probe set IDs. MAS5 data of the combined data set were log2 transformed, normalized using the median over the entire arrays and analyzed for survival risk prediction using BRB-ArrayTools 3.6, as described above.
Publicly accessible microarray data for surgically treated gastric cancer patients generated by the Stanford Functional Genomics Facility were obtained from the NCBI GEO database (GSE4007) and included about 30 300 genes common to these data sets. The microarray data were generated and normalized as described in Leung et al.11 Batch effects in gene expression were removed with probe-wise mean centering and missing data were imputed with the nearest-neighbor averaging method.12 The array cDNA clones were annotated using SOURCE (Stanford Microarray Database) and the Entrez GeneID was used as the mapping identifier for the Affymetrix HG-U133A array. A combined data set of our training set data (n=96) and GSE4007 data (n=88) was analyzed for survival risk prediction using BRB-ArrayTools 3.6 as described above.
Results
Genes correlated with poor survival after CF therapy
As primary gastric cancer lesions cannot be reliably measured by diagnostic imaging, patient survival, not radiographic response, was used as the primary clinical covariate to which gene expression was correlated to identify a predictor of response to CF therapy. To define a gene expression signature that correlates with overall survival, we used expression array data of 96 pretreatment biopsy samples as the training set to develop a predictor (Supplementary Table 1). Ninety-five out of 96 patients (99%) in the training set cohort died with follow-up for one survivor at 39.4 months. None of the clinicopathological or treatment factors listed in Table 1, including second-line chemotherapy, were significantly correlated with survival time of the patients in the training set.
Table 1. Clinicopathological characteristics of patients.
| Training set (n=96) | Validation set (n=27) | |
|---|---|---|
| Baseline clinicopathological characteristic | ||
| Age, no. (%) | ||
| <70 years | 90 (93.8%) | 25 (92.6%) |
| ⩾70 years | 6 (6.2%) | 2 (7.4%) |
| Sex, no. (%) | ||
| Male | 73 (76.0%) | 23 (85.2%) |
| Female | 23 (24.0%) | 4 (14.8%) |
| PS, no. (%) | ||
| ECOG PS 0 or 1 | 91 (94.8%) | 25 (92.6%) |
| ECOG PS 2 or 3 | 5 (5.2%) | 2 (7.4%) |
| Histological type, no. (%) | ||
| Lauren's intestinal | 40 (41.7%) | 9 (33.3%) |
| Lauren's diffuse | 56 (58.3%) | 18 (66.6%) |
| Location of primary lesion, no. (%) | ||
| Upper 1/3 | 14 (14.6%) | 2 (7.4%) |
| Middle 1/3 | 28 (29.2%) | 10 (37.0%) |
| Lower 1/3 | 49 (51.0%) | 15 (55.6%) |
| Entire stomach | 5 (5.2%) | 0 |
| Distant metastasis, no. (%) | 96 (100%) | 27 (100%) |
| Tumor cell percentage in sample (%) | ||
| Median | 60 | 70 |
| Interquartile range | 50–70 | 55–80 |
| Treatment and outcome | ||
| Chemotherapy regimen, no. (%) | ||
| Cisplatin/fluorouracil | 96 (100%) | 22 (81.5%) |
| Cisplatin/capecitabine | 0 (0%) | 5 (18.5%) |
| Relative dose intensity (%) | ||
| Median | 79 | 81 |
| Interquartile range | 73–88 | 72–87 |
| Number of chemotherapy cycles | ||
| Median | 4 | 7 |
| Interquartile range | 3–9 | 5–13 |
| Response (WHO criteria), no. (%) | ||
| PR | 38 (44.7%) | 12 (48.0%) |
| SD | 19 (22.4%) | 9 (36.0%) |
| PD | 28 (32.9%) | 4 (16.0%) |
| Non-measurable disease | 11 | 2 |
| Second-line chemotherapy, no. (%) | 69 (71.9%) | 19 (70.4%) |
| Median follow-up for survivors (months) | 39.4 | 30.4 |
| Overall survival (months) | ||
| Median | 8.1 | 12.6 |
| Interquartile range | 5.6–15.9 | 7.4–30.4 |
| Time to progression (months) | ||
| Median | 3.9 | 6.3 |
| Interquartile range | 2.2–8.3 | 3.9–14.6 |
Abbreviations: ECOG, Eastern Cooperative Oncology Group; PD, progressive disease; PR, partial response; PS, performance status; SD, stable disease; WHO, World Health Organization.
To identify a transcriptional profile related to clinical benefit from CF therapy, the survival times of patients in the array training set were correlated with the mRNA expression levels measured by microarray. One thousand five hundred and sixty-five genes were significantly correlated with the overall survival of the 96 patients (P-value <0.05). Among them, 917 genes had an HR higher than 1 (poor prognosis signature) and 648 genes had an HR lower than 1 (good prognosis signature). We performed gene ontology analyses on this ‘poor prognosis signature' using Ingenuity Pathway Analysis (www.ingenuity.com). The role of BRCA1 in DNA damage response (BRCA2, E2F5, FANCE, MSH2, NBN, PLK1, RFC, SMARCA4, SLC19A1), nucleotide excision repair (ERCC2, POLR2C, POLOR2J, RAD23A, RAD23B) and estrogen receptor signaling were highly represented canonical pathways. Many of these poor prognosis signature genes belonging to these three pathways are previously linked to in vitro cisplatin resistance.13, 14, 15 Overexpression of ERCC2 (P=0.007 in our data) is associated with cisplatin resistance in lung cancer cell lines.13 Silencing of hHR23A (P=0.022 in our data) decreases the nuclear DRP1 level and cisplatin resistance in lung adenocarcinoma cells.14 Disruption of the Fanconi anemia–BRCA pathway is reported in cisplatin-sensitive ovarian tumors.15 Thus, this gene ontology analysis supports the clinical relevance of these DNA repair canonical pathways, which were shown to be associated with in vitro cisplatin resistance.
Ingenuity Pathway Analysis functional categories enriched in poor prognosis signature were: protein synthesis, DNA replication/recombination/repair and cancer (Supplementary Table 2). The protein synthesis category includes ribosomal subunit mRNAs (RPL13, RPL18, RPL24, RPL30, RPL38, RPL5, RPL7, RPL7A, RPL8, RPS2, RPS5) and eukaryotic translation initiation factors (EIF1, EIF2B2, EIF2B4, EIF2S1, EIF3B, EIF3C, EIF3D, EIF3E, EIF3F, EIF3H, EIF3I, EIF4A1, EIF4A3, EIF4B, EIF4EBP1, EIF5, EIF5B). This result suggests that the most prominent feature of poor prognosis signature is increased protein synthesis, presumably resulting from activation of oncogenes, such as EGFR, FGFR2 and MYC (Supplementary Table 2). MYC-induced transcriptional activation of protein synthesis-related genes is previously shown by a microarray report that the majority of genes responsive to MYC overexpression are involved in macromolecular synthesis, protein turnover and metabolism, including 30 ribosomal protein genes.16
Infinitesimal perturbation analysis canonical pathways enriched in 648 genes in good prognosis signature were antigen presentation pathway, B-cell development and interleukin-15 production. Enriched functional categories were gastrointestinal disease, inflammatory disease and genetic disorder.
Development of the three-gene predictor
Although such a gene ontology analysis of the whole signature provides some insight into clinically relevant mechanisms for chemotherapy resistance, this large number of genes is not readily amenable to clinical application. Therefore, we wished to narrow down 917 genes in the whole poor prognosis signature to the smaller number of genes, which may have driven the expression of majority of genes in the signature. Focusing on such ‘driver gene' candidates would also minimize the chance of including false-positive discovery in a genomic predictor. For this purpose, a second tier of genomic analysis was performed to identify genes that could be functionally important in gastric cancer cells.
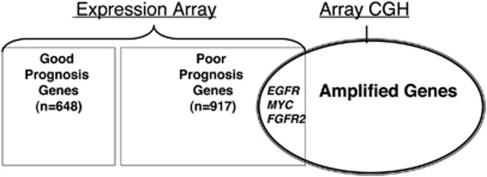
Genomic DNA from samples available from the training set patients was analyzed by array CGH to identify gene amplifications. Age, sex and overall survival were similar between the 30 patients (31.3%) whose samples were analyzed by array CGH and the other patients in the training set. Using very conservative criteria (average tumor/normal log2 ratio >2.0 for ⩾5 consecutive CGH probes), nine amplicons were identified in 11 patients (Table 2). We identified genes found in both the 1565 gene expression signature whose transcriptional levels correlated with poor survival of 96 training set patients (P-value <0.05) and that are also located within the nine amplicons identified by the array CGH. Three genes—MYC (8q24.13–24.21), EGFR (7p11.2) and FGFR2 (10q26)—were identified in the amplicons (Table 2) whose expression array signal values significantly correlated with the survival time of the 96 patients in the training set (Figure 1). Patients with EGFR and FGFR2 amplifications had higher expression levels of each gene (8.4 and 10.2±0.8 (mean±s.d.), for EGFR and FGFR2, respectively) than tested patients without the amplification of these genes (5.9±1.0 and 5.2±1.1, for EGFR and FGFR2, respectively). One of the two patients with MYC amplification had higher expression than patients without amplification (10.9 vs 9.5±0.9).
Table 2. Amplicons identified using array CGHa.
| Cytoband | Start | End | Target gene | No. of patients |
|---|---|---|---|---|
| 3q27.1 | 185 763 900 | 185 763 959 | EPHB3 | 1 |
| 5q33.1 | 149 481 646 | 149 514 673 | PDGFRB | 1 |
| 7p11.2 | 54 746 103 | 55 363 004 | EGFR | 1 |
| 8q24.13–24.21 | 126 357 675 | 128 822 455 | MYC | 2 |
| 9p13.3 | 33 745 689 | 33 961 753 | PRSS3, UBE2R2, UBAP2 | 1 |
| 10q26 | 123 264 724 | 13 123 458 467 | FGFR2 | 2 |
| 17q12 | 35 046 052 | 35 282 145 | ERBB2 | 2 |
| 17q21.2 | 36 110 139 | 36 230 022 | KRT24, KRT25A, KRT25C, KRT25D, KRT10 | 2 |
| 17q21.2 | 36 569 493 | 36 888 515 | KRTAP4-4, KRTAP4-10, KRTAP9-9, KRTAP9-4, KRTAP17-1, KRTHA3A, KRTHA3B, KRTHA4, KRTHA1, KRTHA7, KRTHA8, KRTHA2, KRTHA5 | 1 |
Abbreviation: CGH, comparative genomic hybridization.
Defined by aberrations with average tumor/normal log2 ratio >2.0 for ⩾5 consecutive probes.
Figure 1.
Three genes—EGFR, FGFR2 and MYC—overlap between genes whose array expression levels correlated with survival times (96 training set patients, P<0.05) and gene copy number changes determined by array comparative genomic hybridization (CGH) (tumor/normal log2 ratio >2 for ⩾5 consecutive probes).
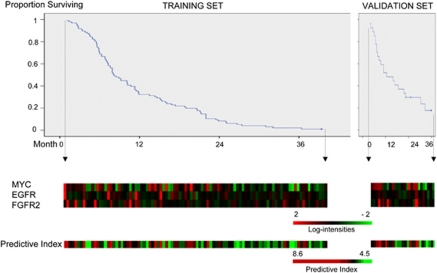
The mRNA expression array signal values of these three genes were correlated with the short survival time with P-values of 0.0154, 0.0096 and 0.0057, for MYC, EGFR and FGFR2, respectively. The expression patterns of these three genes along with the cumulative survival data for all patients are depicted in the heatmap in Figure 2. None of the three genes had significantly different expression levels between those patients who received second-line chemotherapy and those who did not. Quantitative real-time RT-PCR and immunohistochemical staining for the three genes validated the array expression data (Supplementary Figures 1 and 2).
Figure 2.
Affymetrix array expression levels of MYC, EGFR and FGFR2 in 96 training set samples (left) and 27 validation set samples (right), shown with Kaplan–Meier plots for overall survival. Samples are ordered by the increasing survival period of patient from left to right, for the training and validation sets, respectively. A three-gene predictive index for each patient based on the three-gene predictor is indicated below.
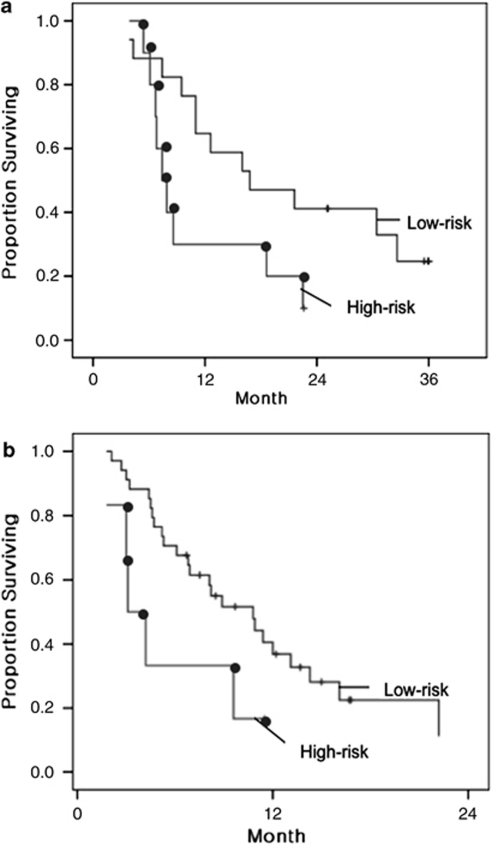
A three-gene predictive index percentile was then calculated for each of the 27 patients in the validation cohort, based on the weighted average of the log intensities of these three genes for each sample (designated as the three-gene predictor). Patterns of MYC, EGFR and FGFR2 expression in these 27 patients, together with the predictive index, are graphically displayed in Figure 2. As a continuous variable, the three-gene predictive index percentile is an independent predictor for poor survival in the validation set by Cox regression analyses, after considering age, performance status, histological type and second-line chemotherapy (adjusted P=0.017) (Table 3). Patients predicted to have poor survival after CF using a predictive index percentile ⩾67% had a significantly shorter median survival than patients with a predictive index percentile <67% (7.4 months for the high-risk group vs 16.8 months for the low-risk cohort; P=0.047) (Figure 3a). As a class, the high-risk group predicted by the three-gene predictor (patient group with a predictive index percentile ⩾67%) was associated with an adjusted HR of 3.1 (95% CI, 1.2–8.4; P=0.022). In addition, the three-gene predictive index percentile is also an independent predictor for the time to progression, which is a more specific indicator of the clinical responsiveness to systemic therapy than overall survival17 (adjusted P=0.014) (Table 3). We therefore show that, independent of old age (⩾70 years), poor performance status (Eastern Cooperative Oncology Group performance status ⩾2) and second-line chemotherapy, the three-gene predictive index is predictive of the benefit from CF to metastatic gastric cancer patients. An adjusted HR for time to progression according to each percentile increase in three-gene predictive index percentile was 1.023 (95% CI, 1.005–1.043) (that is, 100, 75 and 50% predictive indices are associated with an HR of 9.7 (=1.023100), 5.5 (=1.02375) and 3.1 (=1.02350), respectively, compared with a 0% predictive index).
Table 3. Cox regression analyses of the three-gene predictive index percentile, as a continuous variable, for 27 patients in the validation set.
|
Overall survival |
Time to progression |
|||
|---|---|---|---|---|
| P-value | HR (95% CI) | P-value | HR (95% CI) | |
| Univariate | ||||
| Three-gene predictive index percentilea | 0.050 | 1.015b (1.000–1.030) | 0.026 | 1.017 (1.002–1.031) |
| Multivariate | ||||
| Three-gene predictive index percentile | 0.017 | 1.023 (1.004–1.042) | 0.014 | 1.023 (1.005–1.043) |
| Age ⩾70 yearsc | 0.027 | 7.614 (1.257–46.130) | 0.144 | 3.605 (0.646–20.112) |
| Poor performance status (ECOG PS 2 or 3) | 0.346 | 2.130 (0.442–10.258) | 0.074 | 4.829 (0.861–27.086) |
| Second-line chemotherapy | 0.041 | 4.231 (1.064–16.831) | 0.011 | 5.992 (1.502–23.902) |
| Diffuse histological type | 0.773 | 1.164 (0.415–3.263) | 0.280 | 1.774 (0.626–5.025) |
Abbreviations: CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; HR, hazard ratio.
Computed based on weighted average of log intensities of the three genes (EGFR, FGFR2 and MYC) obtained using a proportional hazards regression on the first two principal components of the log signal intensities of those three genes.
HR for each percentile increase in three-gene predictive index percentile. For example, a predictive index percentile of 100 (the highest predictive index) is associated with an HR of 4.4 (=1.015100), compared with a predictive index percentile of 0 (the lowest predictive index). The median predictive index (50%) is associated with HRs of 2.1 (=1.01550), compared with the lowest predictive index.
For patients aged ⩾70 years, the treatment schedule for fluorouracil could be shortened at the discretion of the oncologist to 3 instead of 5 days.
Figure 3.
(a) Kaplan–Meier survival curves for the two risk groups of the validation cohort predicted by three-gene predictor. Patients at a high risk (predictive index percentile ⩾67% n=10) had significantly shorter median survival than patients at a low risk (n=17) (7.4 vs 16.8 months; log rank P=0.047). Green and blue lines represent overall survival curves for the predicted high- and low-risk groups, respectively. (b) Kaplan–Meier survival curves for the two risk groups of the published microarray data set from 40 metastatic gastric cancer patients treated with either fluorouracil-based regimens or cisplatin/irinotecan combination chemotherapy regimen. Patients at a high risk (predictive index percentile ⩾67% n=6) had shorter median survival than patients at a low risk (n=34), at a borderline significance (3.1 vs 10.8 months; log rank P=0.056). Green and blue lines represent overall survival curves for the predicted high- and low-risk groups, respectively. The color reproduction of the figure is available on the html full text version of the manuscript.
Three-gene predictor predicts survival of patients in the second validation set
To extend these results, we wished to test the predictive power of the three-gene predictor in other independent data sets. After the three-gene predictor was validated in 27 patient samples in our validation set, another microarray study with a comparable study design to our study was published in the literature.4 These data were only one published microarray data set that could be used to determine whether the three-gene predictor could predict the outcome of metastatic gastric cancer patients treated with either cisplatin or fluorouracil. This data set contains pretreatment expression array data for 40 patients who subsequently received either fluorouracil-based chemotherapy (n=24) or cisplatin/irinotecan combination chemotherapy (n=16) and patient survival data. We applied the same three-gene predictor to this published microarray data set, just as we did to our 27 patient data in the first validation set. The three-gene predictive index percentile, as a continuous variable, was found to be significantly associated with poor survival of these 40 patients (P=0.047; HR according to each percentile increase in three-gene predictive index percentile=1.014 (95% confidence interval, 1.000–1.027)). Cox multivariate analysis showed that the three-gene predictive index percentile is an independent predictor for poor survival, after considering performance status, age, sex and the chemotherapy regimen (adjusted P=0.026; adjusted HR=1.017 (1.002–1.032)) (Table 4, Figure 3b). Thus, the predictive power of the three-gene predictor is consistent across two validation sets, that is, one from our study patients and the other from published data.
Table 4. Cox regression analyses of the three-gene predictive index percentile, as a continuous variable, for published DNA microarray data from 40 metastatic gastric cancer patients treated with either FU-based chemotherapy or cisplatin/irinotecan combination chemotherapy.
|
Overall survival |
||
|---|---|---|
| P-value | HR (95% CI) | |
| Univariate | ||
| Three-gene predictive index percentile | 0.047 | 1.014 (1.000–1.027) |
| Multivariate | ||
| Three-gene predictive index percentile | 0.026 | 1.017a (1.002–1.032) |
| Performance status ⩾1 | 0.028 | 3.008 (1.129–8.016) |
| Ageb | 0.766 | 0.995 (0.961–1.030) |
| Male | 0.538 | 1.359 (0.512–3.605) |
| FU-based chemotherapy regimenc | 0.744 | 0.854 (0.332–2.199) |
Abbreviations: CI, confidence interval; FU, fluorouracil; HR, hazard ratio.
Adjusted HR for each percentile increase in three-gene predictive index percentile. For example, a predictive index percentile of 100 (the highest predictive index) is associated with an HR of 5.4 (=1.017100), compared with a predictive index percentile of 0 (the lowest predictive index).
As a continuous variable.
As compared with the irinotecan/cisplatin combination chemotherapy regimen.
Interestingly, the three-gene predictor was found to be an independent predictor for poor survival, when the same Cox regression analysis was performed only on a subset of these patients (n=16) treated with cisplatin in combination with irinotecan, a topoisomerase I inhibitor (adjusted P=0.011; adjusted HR=1.038 (1.008–1.068)). Patients treated with irinotecan were not included in the original training set patients. Hence, the predictive power of three-gene predictor may not be specifically associated with only CF therapy, although further large-scale studies need to be performed to address the predictive value of the three-gene predictor for other therapeutic regimens.
Three-gene predictive index and radiographic response
Although the radiographic tumor response was not the main end point of this study, we also evaluated the association between the three-gene predictive index and radiographic response of patients with measurable disease. When published data4 were also included, 104 patients had either partial response or stable disease (clinical benefit) as the best response, whereas 46 patients had progressive disease. The three-gene predictive index was significantly associated with radiographic response at a univariate P-value of 0.039, which is higher than the Cox regression P-value for the overall survival of all study patients (Table 5). This statistical association was at borderline significance in a multivariate regression analysis.
Table 5. Logistic regression analysis on the three-gene predictive index for radiographic response of 150 patients with measurable disease, including patients represented by the published data set.
|
Radiographic responsea |
Time to progression |
Overall survival |
||||
|---|---|---|---|---|---|---|
| P-value | OR (95% CI) | P-valueb | HR (95% CI) | P-valuec | HR (95% CI) | |
| Univariate | ||||||
| Three-gene predictive indexd | 0.039 | 2.001 (1.036–3.864) | 0.020 | 1.304 (1.042–1.631) | 0.030 | 1.288 (1.026–1.618) |
| Multivariate | ||||||
| Three-gene predictive index | 0.059 | 1.902 (0.976–3.704) | 0.019 | 1.309 (1.045–1.641) | 0.018 | 1.316 (1.048–1.654) |
| Age ⩾70 years | 0.914 | 1.069 (0.318–3.598) | 0.791 | 1.119 (0.486–2.577) | 0.113 | 1.600 (0.895–2.862) |
| Poor performance status (ECOG PS 2 or 3) | 0.336 | 0.513 (0.132–1.999) | 0.026 | 2.192 (1.097–4.381) | 0.048 | 1.921 (1.004–3.677) |
Abbreviations: CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; HR, hazard ratio; OR, odds ratio; WHO, World Health Organization
No clinical benefit (progressive disease according to the WHO criteria; n=46) vs clinical benefit (partial response and stable disease; n=104).
Result of Cox regression analysis on the three-gene predictive index for the time to progression of 123 patients in the training and the first validation sets.
Result of Cox regression analysis on the three-gene predictive index for the overall survival of all of 163 study patients including published data set.
Computed based on weighted average of log intensities of the three genes (EGFR, FGFR2 and MYC) obtained using a proportional hazards survival regression on the first two principal components of the log signal intensities of those three genes.
Three-gene predictor is not prognostic but predictive
Although we showed that the three-gene predictor predicted time to progression and overall survival for CF-treated patients, we wished to further address whether it represents a prognostic signature, using the published data set from 88 gastric cancer patients who were treated by surgery alone and not with chemotherapy.11 The three-gene predictive index percentile was not a prognostic factor in this data set as a continuous variable (P=0.506). There was no difference in survival in the surgically treated patients between the high- and low-risk groups predicted by the three-gene predictor (P=0.972). These results strongly suggest that the three-gene predictor is not a predictor of prognosis for gastric cancer patients, but is predictive of the patient response to chemotherapy.
Discussion
Cytotoxic chemotherapy prolongs the median survival of metastatic gastric cancer patients from 3–5 to 9–11 months compared with best supportive care, with a response rate of 40–50%.18, 19, 20, 21 Combination CF constitutes the backbone for chemotherapy regimens commonly used for gastric cancers.19, 22 We also reported that CF in combination with low-dose docetaxel is active for metastatic gastric cancer with tolerable toxicity profile.18 The ability to predict the primary resistance of common solid tumors to cytotoxic chemotherapy is currently lacking, but would significantly improve patient care by identifying those who would best be treated by alternative strategies. This study has identified a three-gene predictor that distinguishes gastric cancer patients likely to receive a therapeutic benefit from CF from those who will not.
Most previous studies attempting to identify predictors of chemoresistance in gastric cancer have examined only individual genes such as TS or ERCC1.23, 24 High-throughput DNA microarray analyses to identify gene expression signatures predictive of chemotherapy or chemoradiotherapy resistance in gastrointestinal cancer patients have been limited by the small number of samples,2, 3 heterogeneous treatment4 or were not prospectively designed.5 In contrast to these previous studies, our study uses high-throughput genomic approaches, is prospective with a large, pre-defined number of training set patients, separate validation cohorts and survival data during an extended follow-up period. Although previously reported TS and ERCC1 tend to be associated with poor prognosis of our patients, the association was not significant enough for them to be considered for our predictive model (P=0.073 and 0.076, for TS and ERCC1, respectively). Notably, the outcome discrimination predicted by the classifier was statistically significant on two validation groups, including the only available published microarray data set from chemotherapy-treated gastric cancer patients.4 Although the sample size of our validation set is relatively small, it is nonetheless large enough to show that our three-gene predictor provides a statistically significant discrimination of patient outcome in multivariate survival analyses. The study design we employed is consistent with an allocation of two-thirds to one-third training-to-test set sample allocation as recommended by statisticians.25
We combined analyses of gene expression changes identified by expression profiling with the identification of DNA copy number changes using array CGH to develop a predictor composed of a much smaller number of critical genes that potentially could be of clinical utility. We identified MYC, EGFR and FGFR2 in regions of amplification, as well as in the gene expression signature related to clinical outcome after CF therapy, suggesting that these genes might be functionally involved in determining resistance. Amplification of MYC, EGFR and FGFR2 have previously been observed in gastric cancer at frequencies 4.8–15.5%,26 2.3–13.3%27 and 3–10%,26, 28 respectively, suggesting that, in some cases, tumors amplify these regions for selective advantage. Combined expression of these three genes could predict overall survival and time to progression of CF-treated gastric cancer patients. Thus, combining array CGH analysis with relevant transcriptional changes is a feasible approach for building a predictive model using functionally important genes and reducing the likelihood of false biomarker discovery. Transcriptional levels of genes other than MYC, EGFR and FGFR2 identified in the amplified genomic loci were not associated with the survival of the 96 training set patients (for example, P=0.313 for ERBB2).
Primary gastric tumors are not easily measurable by current radiographic techniques, and often there are no metastatic lesions that are readily quantifiable in metastatic gastric cancer patients. To develop a predictor from the general population of gastric cancer patients in an unbiased way, this study was designed to correlate gene expression profiling of the tumors with overall survival and time to progression, not radiographic response. Overall survival is the ultimate measure of the treatment benefit afforded to a patient and is a particularly appropriate gauge for patients with metastatic gastric cancer, as radiographic assessment is problematic in such patients. The fact that both the time to progression as well as overall survival are predicted by our three-gene predictor in CF-treated patients, but not surgically treated patients, suggests that the three-gene predictor is a predictive indicator for the clinical benefit from CF.
Although EGFR and FGFR2 expression have been reported to have prognostic value for gastric cancer patients treated surgically,29, 30 we did not find the three-gene predictive index to be prognostic for surgically treated patients with gastric cancer. Our findings are consistent with previously reported experimental data on chemoresistance. Inhibitors of EGFR act synergistically with cisplatin31 and fluorouracil,32 whereas an FGFR2 inhibitor is synergistic with fluorouracil.33 MYC has been linked to cisplatin resistance in several in vitro models.34, 35, 36, 37
Taken together, combined expression of MYC, EGFR and FGFR2 is predictive of poor survival in CF-treated metastatic gastric cancer patients. More focused prospective trials that are designed to test the clinical utility of this three-gene predictor are warranted.
Acknowledgments
The work was supported in part by National Institute of Health Intramural Program, Center for Cancer Research, National Cancer Institute, Korean National Cancer Center Grant 0910570 and Converging Research Center Program through the Ministry of Education, Science and Technology of Korea (2010K001121). We thank Dr Richard Simon for valuable discussions and critical reading of the manuscript, Dr Lyuba Varticovski for critical review of the manuscript and Dr Chang-Hee Kim, Ms Susie Korolevich, Ms Eunbyul Lee, Ms Eugene Kim and Ms Ilji Jeon for technical help.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Transcript Profiling
All expression microarray data is available at Gene Expression Omnibus (accession number GSE14210; http://www.ncbi.nlm.nih.gov/geo).
Supplementary Material
References
- Uchida K, Danenberg PV, Danenberg KD, Grem JL. Thymidylate synthase, dihydropyrimidine dehydrogenase, ERCC1, and thymidine phosphorylase gene expression in primary and metastatic gastrointestinal adenocarcinoma tissue in patients treated on a phase I trial of oxaliplatin and capecitabine. BMC Cancer. 2008;8:386. doi: 10.1186/1471-2407-8-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadimi BM, Grade M, Difilippantonio MJ, Varma S, Simon R, Montagna C, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol. 2005;23:1826–1838. doi: 10.1200/JCO.2005.00.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimkus C, Friederichs J, Boulesteix AL, Theisen J, Mages J, Becker K, et al. Microarray-based prediction of tumor response to neoadjuvant radiochemotherapy of patients with locally advanced rectal cancer. Clin Gastroenterol Hepatol. 2008;6:53–61. doi: 10.1016/j.cgh.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Arao T, Gotoda T, Taniguchi H, Oda I, Shirao K, et al. Identification of prognostic biomarkers in gastric cancer using endoscopic biopsy samples. Cancer Sci. 2008;99:2193–2199. doi: 10.1111/j.1349-7006.2008.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WL, Coyle VM, Jithesh PV, Proutski I, Stevenson L, Fenning C, et al. Clinical determinants of response to irinotecan-based therapy derived from cell line models. Clin Cancer Res. 2008;14:6647–6655. doi: 10.1158/1078-0432.CCR-08-0452. [DOI] [PubMed] [Google Scholar]
- Simon R, Radmacher MD, Dobbin K. Design of studies using DNA microarrays. Genet Epidemiol. 2002;23:21–36. doi: 10.1002/gepi.202. [DOI] [PubMed] [Google Scholar]
- Kang Y, Kang WK, Shin DB, Chen J, Xiong J, Wang J, et al. Randomized phase III trial of capecitabine/cisplatin vs continuous infusion of 5-FU/cisplatin as first-line therapy in patients with advanced gastric cancer: efficacy and safety results J Clin Oncol 200624(Suppl): 18s(abstract 4018). [Google Scholar]
- Kim HK, Choi IJ, Kim HS, Kim JH, Kim E, Park IS, et al. DNA microarray analysis of the correlation between gene expression patterns and acquired resistance to 5-FU/cisplatin in gastric cancer. Biochem Biophys Res Commun. 2004;316:781–789. doi: 10.1016/j.bbrc.2004.02.109. [DOI] [PubMed] [Google Scholar]
- Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-Array Tools. Cancer Informatics. 2007;2:11–17. [PMC free article] [PubMed] [Google Scholar]
- Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung SY, Chen X, Chu KM, Yuen ST, Mathy J, Ji J, et al. Phospholipase A2 group IIA expression in gastric adenocarcinoma is associated with prolonged survival and less frequent metastasis. Proc Natl Acad Sci USA. 2002;99:16203–16208. doi: 10.1073/pnas.212646299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer. 2005;4:18–25. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YY, Chen SL, Hsiao YT, Huang CH, Lin TY, Chiang IP, et al. Nuclear expression of dynamin-related protein 1 in lung adenocarcinomas. Mod Pathol. 2009;22:1139–1150. doi: 10.1038/modpathol.2009.83. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia–BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, et al. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Balko JM, Black EP. A gene expression predictor of response to EGFR-targeted therapy stratifies progression-free survival to cetuximab in KRAS wild-type metastatic colorectal cancer. BMC Cancer. 2009;9:145–154. doi: 10.1186/1471-2407-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SR, Chun JH, Yu MS, Lee JH, Ryu KW, Choi IJ, et al. Phase II study of docetaxel and irinotecan combination chemotherapy in metastatic gastric carcinoma. Br J Cancer. 2006;94:1402–1406. doi: 10.1038/sj.bjc.6603133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SR, Chun JH, Yu MS, Lee JH, Ryu KW, Choi IJ, et al. Phase II study of low-dose docetaxel/fluorouracil/cisplatin in metastatic gastric carcinoma. Am J Clin Oncol. 2005;28:433–438. doi: 10.1097/01.coc.0000162424.69631.79. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Rao S, Starling N, Iveson T, Nicolson M, Coxon F, et al. Randomised multicentre phase III study comparing capecitabine with fluorouracil and oxaliplatin with cisplatin in patients with advanced oesophagogastric (OG) cancer: The REAL 2 trial J Clin Oncol 20062418s(abstr. 4017). [Google Scholar]
- Pyrhönen S, Kuitunen T, Nyandoto P, Kouri M. Randomised comparison of fluorouracil, epidoxorubicin and methotrexate (FEMTX) plus supportive care with supportive care alone in patients with non-resectable gastric cancer. Br J Cancer. 1995;71:587–591. doi: 10.1038/bjc.1995.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff ED, Cen P, Ajani JA. Emerging drugs in the treatment of advanced gastric cancer. Expert Opin Emerg Drugs. 2008;13:135–144. doi: 10.1517/14728214.13.1.135. [DOI] [PubMed] [Google Scholar]
- Lenz HJ, Leichman CG, Danenberg KD, Danenberg PV, Groshen S, Cohen H, et al. Thymidylate synthase mRNA level in adenocarcinoma of the stomach: a predictor for primary tumor response and overall survival. J Clin Oncol. 1996;14:176–182. doi: 10.1200/JCO.1996.14.1.176. [DOI] [PubMed] [Google Scholar]
- Metzger R, Leichman CG, Danenberg KD, Danenberg PV, Lenz HJ, Hayashi K, et al. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol. 1998;16:309–316. doi: 10.1200/JCO.1998.16.1.309. [DOI] [PubMed] [Google Scholar]
- Dobbin KK, Simon RM.Optimally splitting cases for training and testing high dimensional classifiers (submitted for publication) http://linus.nci.nih.gov/~brb/reprints.htm . [DOI] [PMC free article] [PubMed]
- Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest. 1998;78:1143–1153. [PubMed] [Google Scholar]
- Moutinho C, Mateus AR, Milanezi F, Carneiro F, Seruca R, Suriano G. Epidermal growth factor receptor structural alterations in gastric cancer. BMC Cancer. 2008;8:10–15. doi: 10.1186/1471-2407-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Sakamoto H, Terada M. Amplified genes in cancer in upper digestive tract. Semin Cancer Biol. 1993;4:33–40. [PubMed] [Google Scholar]
- Lieto E, Ferraraccio F, Orditura M, Castellano P, Mura AL, Pinto M, et al. Expression of vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) is an independent prognostic indicator of worse outcome in gastric cancer patients. Ann Surg Oncol. 2008;15:69–79. doi: 10.1245/s10434-007-9596-0. [DOI] [PubMed] [Google Scholar]
- Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular–pathological prognostic factors of gastric cancer: a review. Gastric Cancer. 2005;8:86–94. doi: 10.1007/s10120-005-0320-0. [DOI] [PubMed] [Google Scholar]
- Fan Z, Baselga J, Masui H, Mendelsohn J. Antitumor effect of anti-epidermal growth factor receptor monoclonal antibodies plus cis-diamminedichloroplatinum on well established A431 cell xenografts. Cancer Res. 1993;53:4637–4642. [PubMed] [Google Scholar]
- Hiraishi Y, Wada T, Nakatani K, Uchida J, Iwasa T, Yoshida T, et al. Synergistic antitumor effect of S-1 and the epidermal growth factor receptor inhibitor gefitinib in non-small cell lung cancer cell lines: role of gefitinib-induced down-regulation of thymidylate synthase. Mol Cancer Ther. 2008;7:599–606. doi: 10.1158/1535-7163.MCT-07-0567. [DOI] [PubMed] [Google Scholar]
- Yashiro M, Shinto O, Nakamura K, Tendo M, Matsuoka T, Matsuzaki T, et al. Synergistic anti-tumor effects of FGFR2 inhibitor with 5-fluorouracil on scirrhous gastric carcinoma. Int J Cancer. 2010;126:1004–1016. doi: 10.1002/ijc.24763. [DOI] [PubMed] [Google Scholar]
- Leonetti C, Biroccio A, Candiloro A, Citro G, Fornari C, Mottolese M, et al. Increase of cisplatin sensitivity by c-myc antisense oligodeoxynucleotides in a human metastatic melanoma inherently resistant to cisplatin. Clin Cancer Res. 1999;5:2588–2595. [PubMed] [Google Scholar]
- Xie XK, Yang DS, Ye ZM, Tao HM. Recombinant antisense C-myc adenovirus increase in vitro sensitivity of osteosarcoma MG-63 cells to cisplatin. Cancer Invest. 2006;24:1–8. doi: 10.1080/07357900500449520. [DOI] [PubMed] [Google Scholar]
- Knapp DC, Mata JE, Reddy MT, Devi GR, Iversen PL. Resistance to chemotherapeutic drugs overcome by c-Myc inhibition in a Lewis lung carcinoma murine model. Anticancer Drugs. 2003;14:39–47. doi: 10.1097/00001813-200301000-00006. [DOI] [PubMed] [Google Scholar]
- Biroccio A, Benassi B, Fiorentino F, Zupi G. Glutathione depletion induced by c-Myc downregulation triggers apoptosis on treatment with alkylating agents. Neoplasia. 2004;6:195–206. doi: 10.1593/neo.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.