Abstract
Purpose
Programmed cell death ligand 1 (PD-L1) is a molecule expressed on antigen-presenting cells that engages the PD-1 receptor on T cells and inhibits T-cell receptor signaling. The PD-1 axis can be exploited by tumor cells to dampen host anti-tumor immune responses and foster tumor cell survival. PD-1 blockade has shown promise in multiple malignancies but should be directed towards patients in whom it will be most effective. In recent studies, we found that the chromosome 9p24.1 amplification increased the gene dosage of PD-L1 and its induction by JAK2 in a subset of patients with classical Hodgkin lymphoma (cHL). However, cHLs with normal 9p24.1 copy numbers also expressed detectable PD-L1, prompting analyses of additional PD-L1 regulatory mechanisms.
Experimental Design
Herein, we utilized immunohistochemical, genomic and functional analyses to define alternative mechanisms of PD-L1 activation in cHL and additional EBV+ lymphoproliferative disorders.
Results
We identified an AP-1-responsive enhancer in the PD-L1 gene. In cHL Reed Sternberg cells, which exhibit constitutive AP-1 activation, the PD-L1 enhancer binds AP-1 components and increases PD-L1 promoter activity. In addition, we defined EBV infection as an alternative mechanism for PD-L1 induction in cHLs with diploid 9p24.1. PD-L1 was also expressed by EBV-transformed lymphoblastoid cell lines as a result of latent membrane protein 1-mediated, JAK/STAT-dependent promoter and AP-1-associated enhancer activity. In addition, over 70% of EBV+ post-transplant lymphoproliferative disorders expressed detectable PD-L1.
Conclusions
AP-1 signaling and EBV infection represent alternative mechanisms of PD-L1 induction and extend the spectrum of tumors in which to consider PD-1 blockade.
Introduction
Classical Hodgkin’s lymphoma (cHL) is a tumor of crippled germinal center B (GCB)-cells that occurs frequently in Western countries and often affects young adults (1). The two most common subtypes of cHL, Nodular Sclerosis (NSHL) and Mixed Cellularity (MCHL), make up 90% of cases. Primary cHLs include a small number of malignant Reed-Sternberg (RS) cells within a profuse, but ultimately ineffective, inflammatory infiltrate. This suggests that the RS cells possess one or more mechanisms to escape or suppress anti-tumor immunity. Understanding the cellular mechanisms that promote tumor immune escape is important for identifying patients who may benefit from immunomodulatory therapy.
Epstein Barr virus (EBV) has been implicated in ≈40% of cHLs(1) and in additional B-cell disorders such as post-transplant lymphoproliferative disorders (PTLD) that occur in the setting of host immunosuppression (2). Expression of the EBV latent membrane proteins, LMP1 or LMP2a, in normal germinal center (GC) B-cells is sufficient to mimic a HL RS cell-like phenotype (3, 4), highlighting the shared features of certain EBV-driven malignancies. Recent data indicate that EBV-associated malignancies, such as PTLD and cHL, also share certain mechanisms of immune evasion (5). For example, both cHL RS cells and EBV-driven PTLDs express the immunoregulatory glycan-binding protein, galectin-1, which promotes Th2/Tregulatory cell skewing in the tumor microenvironment (5–7).
In addition to galectin-1, we and others have identified the immunomodulatory programmed cell death ligand 1 (PD-L1/B7-HI/CD274) as a contributor to the immunosuppressive microenvironment of cHL (8, 9). The natural function of PD-1 signaling is to limit certain T-cell mediated immune responses(10). Normal antigen-presenting cells express PD-1 ligands that trigger PD-1 receptors on activated T cells (10). PD-1 engagement triggers Src homology 2 domain-containing protein tyrosine phosphatase-2 (SHP2)-mediated dephosphorylation of proximal T-cell receptor (TCR) signaling molecules and attenuates the TCR signal (10). PD-L1 also blocks CD28 costimulation by competitively binding to the CD28 ligand, CD80 (B7-1) (11) and regulates the function of PD-1+ regulatory T cells (12). As a consequence, PD-1 signaling results in “T-cell exhaustion”, a functional phenotype that can be reversed by PD-1 blockade (10).
Recent studies indicate that viruses and certain tumors also utilize the PD-1 signaling pathway to avoid immune detection (13–18). In addition, PD-1 ligand expression has adverse prognostic significance in multiple human tumors including melanoma, and pancreatic, hepatocellular and ovarian carcinoma (19–24).
We recently integrated high-resolution copy number data with transcriptional profiles and identified the PD-1 ligands, PD-L1 and PD-L2, as key targets of the recurrent 9p24.1 amplification in NSHL and the related lymphoid malignancy, primary mediastinal large cell lymphoma (MLBCL) (9). In these tumors, the extended 9p24.1 region of amplification also included the Janus kinase 2 (JAK2) locus (9). JAK2 amplification increased JAK2 expression and augmented JAK/STAT signaling and associated PD-L1 promoter activity and transcription (9). These studies defined the 9p24.1 amplicon as a structural alteration that increased both the gene dosage of PD-1 ligands and their induction by JAK2 (9). These findings also highlighted the potential therapeutic benefit of PD-1 blockade in cHL patients with 9p24.1 amplification.
However, we also noted that cHL cell lines with only low-level 9p24.1 amplification expressed detectable cell surface PD-L1 as did primary cHLs with diploid 9p24.1 (9). These observations suggested that the PD-1 pathway might be targetable in a larger group of cHL patients and prompted analyses of other mechanisms of PD-L1 induction in cHL and additional EBV-associated lymphoproliferative disorders.
Materials and Methods
Primary Tumor Specimens and Immunohistochemistry
Primary tumor samples included 22 cHLs (7 MCHL and 15 NSHL) with known 9p24.1 copy numbers (9) and 26 EBV+ PTLDs (5). PD-L1 immunostaining was performed on slides with 4μm thick formalin-fixed, paraffin-embedded tissue sections which were deparaffinized in xylene, passed through graded alcohols, and subsequently put in distilled water. All further steps were performed at room temperature in a hydrated chamber. Slides were pre-treated with Peroxidase Block (DAKO USA, Carpinteria, CA) for 5 minutes to quench endogenous peroxidase activity, and then washed in 50-mM Tris-Cl, pH 7.4. Slides were blocked using an Avidin/Biotin Blocking Kit (Vector, Burlingame, CA) as per manufacturer’s instructions. For PD-L1, monoclonal mouse anti-human PD-L1 antibody (clone 29E.2A3, cat.# 329702, BioLegend, San Diego, CA) was applied at 1:1000 (final concentration of 500ng/ml) in DAKO diluent for 1 hour. For the isotype control, monoclonal IgG2b,κ from murine myeloma (Sigma-Aldrich, Saint Louis, MO) was applied at 1:400 (final concentration of 500ng/ml) in DAKO diluent for 1 hour. Slides were washed in Tris buffer and treated with Labelled Polymer-HRP Anti-Mouse EnVision+ (DAKO) for 30 minutes. After another wash, slides were treated with a Tyramide kit (PerkinElmer, Boston, MA) at 1:250 for 10 minutes. Slides were then thoroughly washed and treated with LSAB2 Streptavidin-HRP (DAKO) as per manufacturer’s instruction. After further washing, immunoperoxidase staining was developed using a DAB chromogen (DAKO) for 5 minutes. Slides were counterstained with hematoxylin, dehydrated in a series of alcohols and xylene, and mounted and coverslipped. PD-L1 protein expression was quantified in individual RS cells using a previously described protocol (9) and Aperio ScanScope XT (Aperio Technologies Inc., Vista, CA) ImageScope Analysis software (Aperio Technologies), and an Aperio Color Deconvolution v9 optimized algorithm. The final qIHC score per RS cell consists of the average optical density (OD) of DAB staining within the annotated area after color deconvolution. Photomicrographs were taken with an Olympus BX41 microscope, 100x/1.30 Olympus UPlanFL objective lens (final magnification 1000x), Olympus QColor5 digital camera, and QCapture Pro 6.0 (QImaging) and Adobe Photoshop 6.0 software (Adobe).
In cHLs and PTLDs, EBV status was assessed by EBV-encoded RNA in situ hybridization (EBER-ISH), as previously described (25). Images were acquired with an Olympus QColor5 and analyzed with QCapture Pro 6.0 software (QImaging) and Adobe Photoshop 6.0 (Adobe).
Cell lines
The HL lines, L428 and L540, were originally obtained from the DSMZ Cell Line Bank (Braunschweig, Germany) and characterized by short tandem repeat (STR) DNA typing to confirm identity (26). The HL lines and the EBV-transformed lymphoblastoid cell lines (LCLs), NOR-LCL, RIC-LCL, STA-LCL, FW-LCL, SC-LCL and VS-LCL, were maintained as described in (5). In certain experiments, the NOR-LCL was treated for 48 hr with 250 nM or 1250 nM JAK3 inhibitor VI (Calbiochem) or the inactive control (JAK3 inhibitor control, Calbiochem).
Regulatory element analysis and AP-1 element validation
The region within the first intron of PD-L1 was assessed for potential regulatory elements with the USCS Genome Browser (http://www.genome.usc.edu) the publicly available MatInspector module of the Genomatix suite (http://www.genomatix.de) and the open-source TOUCAN software (27) as described (9). Binding of cJUN to predicted AP-1 sites was assessed using publicly available Chromatin Immunoprecipitation (ChIP)-sequencing data for cJUN (Supplemental Information). In addition, ChIP was performed for cJUN and JUN-B in the L428 and L540 HL cell lines and the SUDHL4 DLBCL cell line, as previously described (5). Immunoprecipitation of the predicted AP-1-responsive element was performed following 25 cycles of PCR amplification (Primer sequences: TGC CTA GCA CAG AAG AGG TG and TAA CAG TGG GGA GTG GGA AG) and the band was visualized by agarose gel electrophoresis. Activity of the predicted enhancer element was assessed using pGL3 basic luciferase constructs (Promega) containing the PD-L1 promoter element (9) upstream of the luciferase gene (PDL1-P), the promoter element with the additional wild-type sequence of the predicted enhancer element (chr9:5445267-5445817) downstream of the luciferase gene (PDL1-P+E), or the promoter element with the predicted enhancer element with a deletion of the most highly conserved AP-1 binding site (chr9:5445434-5445455; PDL1-P+Emt). Luciferase assays were performed as described using L428 and L540 HL cell lines which have only low-level 9p24.1 amplification (9).
Dominant-negative inhibition of cJUN activity
AP-1 activity was inhibited in two HL cell lines, L428 and L540, by over-expression of dominant-negative cJUN which lacks the transactivation domain (6). Cells were transfected with 1μg of either pFLAGCMV2-cJUN-DN (cJUNDN) or empty pFLAGCMV2 vector (EV) using an Amaxa nucleofector apparatus (Lonza). Following 48h recovery at 37°C, cells were harvested for preparation of whole cell lysates using NP-40 lysis buffer, and total RNA using an RNeasy mini kit (Qiagen) according to the manufacturer’s protocol. cDNA was synthesized from total RNA using Superscript III First Strand Synthesis system (Invitrogen) and the abundance of PD-L1 and PD-L2 transcripts was evaluated by quantitative real-time PCR (qPCR) using commercially available TaqMan assays (Hs01125299_m1, Hs00228839_m1; Applied Biosystems) and internally normalized to HRPT (4326321E; Applied Biosystems). Relative gene transcript levels were calculated by the comparative CT method (28).
Lymphoblastoid cell lines and flow cytometry
PD-L1 expression was evaluated on EBV-transformed lymphoblastoid cell lines (5) by flow cytometry using phycoerythrin-conjugated anti-PD-L1 (Clone 29E.A3, BioLegend), as described (9). STAT activity in these cell lines was assessed by intracellular phospho-specific flow cytometry using anti-phospho(Tyr701)-STAT1, anti-phospho(Ser727)-STAT3, anti-phospho(Tyr694)-STAT5 and anti-phospho(Tyr641)-STAT6 (all from BD Biosciences), as previously described (9), with reference to fixed and permeabilized unstained cells.
PD-L1 promoter induction by LMP1
The effect of LMP1 on PD-L1 promoter activity was measured using a previously described LMP1 expression vector (5) co-transfected into 293T cells with either PDL1-P or empty pGL3 vector with Fugene 6 (Roche) according to the manufacturer’s protocol. Following 48h incubation at 37°C, cells were harvested for luciferase assays as described above.
Western blots
LMP1 expression and cJUN activity were interrogated in empty vector and LMP1-transfected 293T cells by western blot using the LMP-1 monoclonal antibody, S12 (29) and cJUN-specific antibodies described above. PhosphoJAK3, total JAK3 and PD-L1 expression were evaluated in NOR-LCLs treated with the JAK3 inhibitor or control compound by western blot using the anti-phospho(Tyr980/981)-JAK3 and anti-JAK3 antibodies (Cell Signaling) and anti PD-L1 (BioLegend).
Results
cHLs with normal 9p24.1 copy numbers express PD-L1
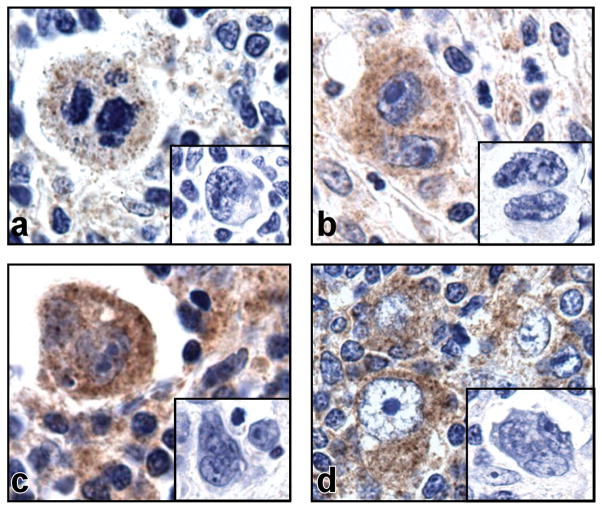
We have previously shown that expression of PD-L1 on cHL RS cells is increased by 9p24.1 amplification, but also noted detectable expression of PD-L1 on tumors with diploid 9p24.1 (9). In an extended series of primary cHLs with diploid 9p24.1, all of the tumors (9/9 NSHL and 7/7 MCHL) had detectable PD-L1 expression in RS cells (Fig. 1). In a publicly available gene expression data set, PD-L1 transcripts were more abundant in almost all cHLs in comparison with diffuse large B-cell lymphomas (DLBCLs) or normal B-cells (30) (Fig. S1). These data, which suggested that cHL RS cells with diploid 9p24.1 might have additional mechanisms of increasing PD-L1 expression, prompted us to further characterize PD-L1 regulatory elements.
Figure 1. PD-L1 expression in primary cHL tumors.
Immunohistochemical staining for PD-L1 on four representative primary cHLs with diploid 9p24.1. PD-L1 expression (brown) is detected in large multinucleated RS cells which are unstained with the isotype control (inset).
Constitutive AP-1 activity supports basal PD-L1 expression in cHL
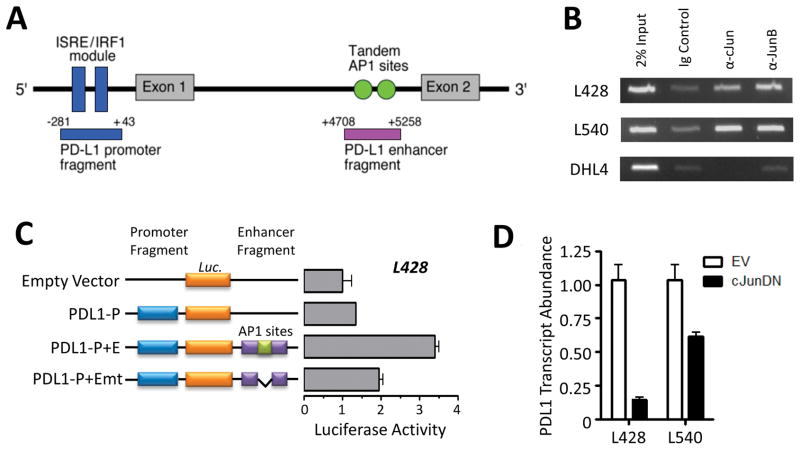
We previously described a PD-L1 promoter element containing an ISRE/IRF1 module that is responsive to JAK/STAT signaling (9). Further analyses of the PD-L1 gene revealed a highly conserved candidate enhancer element in intron 1, ~5kb downstream from the transcription start site (Fig. 2A). Of note, this sequence contained tandem candidate AP-1 binding sites (Fig. 2A). A similar AP-1-containing element was not found in the neighboring PD-1-ligand gene, PD-L2 (Fig. S2A). We confirmed binding of cJUN to the predicted PD-L1 AP-1-responsive region in non-lymphoid cells using public ChIP-seq data (Fig. S2B). Thereafter, we utilized ChiP-coupled PCR to demonstrate cJUN and JUN-B binding to the candidate PD-L1 enhancer region in two HL cell lines (L428 and L540) but not in a DLBCL cell line (SUDHL4; Fig. 2B).
Figure 2. Constitutive AP-1 activity supports PD-L1 expression in cHL.
A) Diagramatic representation of the PD-L1 regulatory elements, including the previously described ISRE/IRF1 module of the JAK/STAT-responsive promoter and the predicted enhancer element containing dual AP-1 binding sites. Fragments cloned into luciferase constructs are annotated below, with positions relative to the PD-L1 transcription start site. B) Binding of cJUN and JUN-B to the candidate PD-L1 enhancer region following ChiP-coupled PCR. L428 and L540, cHL cell lines; DHL4, DLBCL cell line. Data are representative of two independent experiments. C) PD-L1 promoter and enhancer-driven luciferase activity in the L428 cHL cell line. Constructs include the empty pGL3 vector, the pGL3 vector with the promoter cloned upstream of the luciferase gene alone (PDL1-P), the promoter with the wild-type enhancer cloned downstream of the luciferase gene (PDL1-P+E), and the promoter with an enhancer lacking AP-1 binding sites (PDL1-P+Emt). Data are the average of three independent experiments. D) PD-L1 transcript abundance in cHL cell lines following overexpression of a cJUN dominant-negative (cJUNDN) construct. The effect of cJUNDN expression on PD-L1 abundance was greater in L428, which has lower level 9p24.1 amplification than L540. Data are the average of three independent experiments.
To assess the activity of the predicted AP-1 enhancer element in HL cells, we utilized pGL3 luciferase constructs containing either the PD-L1 promoter alone (PDL1-P), the promoter plus the candidate wild-type enhancer (PDL1-P+E), or the promoter plus an enhancer with deleted AP-1 sites (PDL1-P+Emt) (Fig. 2C). The luciferase assays were performed in the L428 and L540 HL cell lines because these lines have the lowest levels of 9p24.1 amplification (9). In both of these HL cell lines, the AP-1 containing enhancer element increased PD-L1 promoter-driven luciferase activity in an AP-1 dependent manner (Fig. 2C and S2C). In addition, over-expression of a dominant-negative mutant form of cJUN (cJUNDN) in the L428 and L540 HL cell lines significantly decreased PD-L1 expression (Fig. 2D). In contrast, cJUNDN had no effect on the expression of PD-L2, which lacks the AP-1-responsive enhancer element (Fig. S2D).
EBV promotes PD-L1 expression
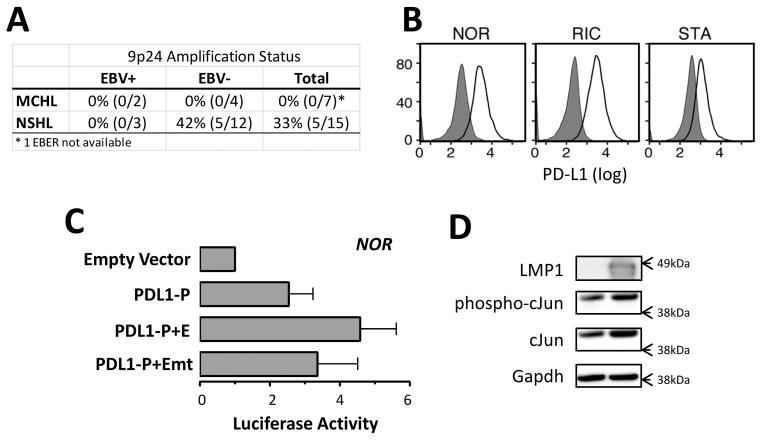
Given the detectable PD-L1 expression in primary cHLs with diploid 9p24.1 and previous evidence that viral infection can induce PD-L1 (31–33), we assessed a potential role for EBV in cHL PD-L1 expression. For the initial analysis, we used a series of primary cHL tumors with known PD-L1 copy numbers (determined with a qPCR assay of laser capture-microdissected RS cells (9)). We determined EBV status by EBER-ISH and found that 23% of the cHLs were EBV+ (2/6 MCHL, 3/16 NSHL) (Fig. 3A). 9p24.1 amplification and EBV infection were mutually exclusive in this series of primary cHLs (Fig. 3A), prompting speculation regarding EBV infection as an alternative mechanism of inducing PD-L1. Consistent with this hypothesis, EBV-transformed lymphoblastoid cell lines (LCLs) were found to express high levels of cell surface PD-L1 (Fig. 3B and S3A).
Figure 3. EBV increases PD-L1 expression.
A) The frequency of EBV infection and 9p24.1 amplification in a cohort of primary cHL tumors of nodular sclerosis (NSHL) and mixed cellularity (MCHL) subtypes. For each category of primary cHL (NSHL or MCHL, EBV+, EBV− or total), the numbers and percentage of 9p24+/all tumors (%) are indicated. EBV infection is mutually exclusive of 9p24.1 amplification. B) Flow cytometric analysis of cell-surface PD-L1 expression in EBV-transformed lymphoblastiod cell lines. Data are representative of three independent experiments. C) PD-L1 promoter- and enhancer-driven luciferase activity in the EBV-transformed NOR LCL. D) Expression of cJUN and phospho-cJUN following LMP1 transfection of 3T3 cells. GAPDH, loading control. Data are representative of three independent experiments.
To characterize potential mechanisms of EBV induction of PD-L1, we performed luciferase assays with the above-mentioned PD-L1 constructs in the NOR-LCL line. Both the PD-L1 promoter and enhancer were active in this EBV-positive LCL, and the enhancer was AP-1 dependent (Fig. 3C). These observations suggested that AP-1 activity may also enhance PD-L1 expression in the context of EBV infection. In this regard, we and others found that expression of the EBV-encoded antigen, LMP1, was sufficient to increase activity of the AP-1 component, cJUN (Fig. 3D) (34). Notably, LMP-1 transduced normal human GCB cells also had more abundant PD-L1 transcripts than control GCB cells in a publicly available gene expression data set (Fig. S3B) (4).
EBV latent membrane protein 1 (LMP1) increases PD-L1 promoter activity
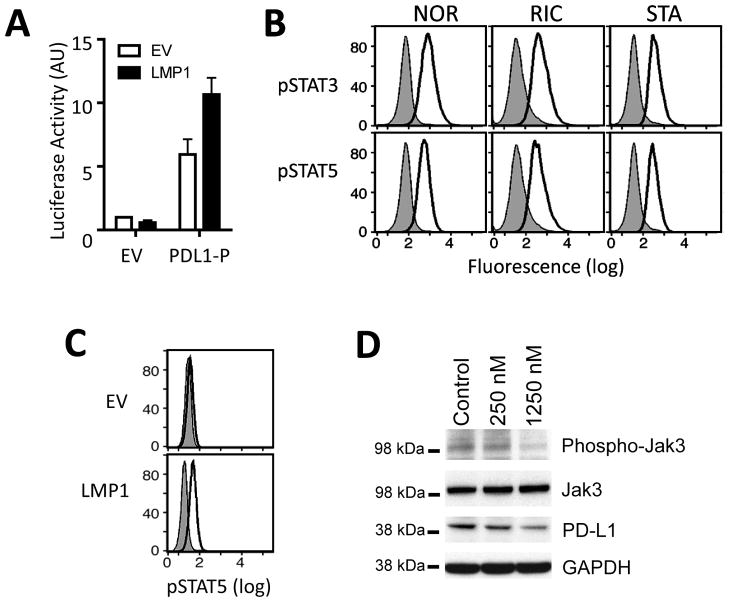
Both the PD-L1 AP-1-dependent enhancer and the PD-L1 promoter element augmented luciferase activity in the NOR-LCL (Fig. 3C). After confirming that the PD-L1 enhancer was bound by cJUN and JUN-B in the NOR-LCL (Fig. S3C) and that LMP1 increased the phosphorylation of AP-1 heterodimer component, c-Jun (Fig. 3D), we asked whether LMP1 also modulated PD-L1 promoter activity. When LMP1 and PD-L1 luciferase constructs were cotransfected into 293T cells, LMP1 increased PD-L1 promoter activity ≈ 2 fold (Fig. 4A).
Figure 4. LMP1 increases PD-L1 promoter activity.
A) LMP1-enhanced PD-L1 promoter-driven luciferase activity in 293T cells. 293T cells were cotransfected with empty pGL3 vector (EV) or PDL1-P and the LMP-FLAG expression vector (LMP1) and evaluated for luciferase activity. Data are the average of three independent experiments. B) Intracellular phosphoflow cytometric analysis of phosphorylated STAT3 and STAT5 in EBV-transformed LCLs. Data are representative of three independent experiments. C) Intracellular phosphoflow cytometric analysis of phosphorylated STAT5 in LMP1-transfected 293T cells. EV, empty vector. D) Western analyses of phosphorylated JAK3 and PD-L1 in the NOR-LCL treated with the JAK3 inhibitor VI (250 nM or 1250 nM) or control compound. Total JAK3 and GAPDH, loading controls. Data are representative of 3 independent experiments.
Previously, we identified a STAT-responsive element within the PD-L1 promoter region that was responsive to JAK2/STAT1 signaling in cHLs with chromosome 9p24.1 amplification (9). In our panel of EBV-transformed LCLs, we observed only low-level STAT1 activity (pSTAT1 expression); however, these cell lines expressed higher levels of the JAK3-associated STAT proteins, pSTAT3 and pSTAT5 (Fig. 4B and S3D). In addition, enforced LMP1 expression increased STAT5 activity (intracellular pSTAT5 levels) (Fig. 4C). Because LMP1 was previously shown to bind JAK3 and activate STAT signaling (35), we asked whether LMP1 also modulated PD-L1 expression via a JAK3-dependent mechanism. We treated the NOR-LCL with a specific JAK3 inhibitor (JAK3 inhibitor VI) and subsequently assessed PD-L1 expression by western blot. In these assays, chemical inhibition of JAK3 decreased PD-L1 abundance (Fig. 4D), indicating that EBV LMP1 likely augments PD-L1 promoter activity via JAK3.
PD-L1 is Frequently Expressed on EBV-positive PTLD
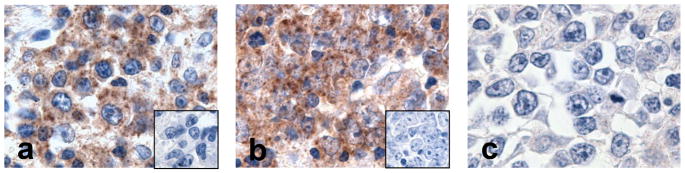
Given the PD-L1 expression in EBV+ cHLs and the EBV LMP1-associated increase in PD-L1 promoter and enhancer activity, we next asked whether additional EBV-associated lymphoproliferative disorders such as post-transplant lymphoproliferative disorders (PTLD) expressed PD-L1. In a series of primary EBV+ PTLDs (5) evaluated by immunohistochemical staining, 73% (19/26 cases) expressed PD-L1 (Fig. 5). These observations extend the spectrum of EBV-associated tumors in which PD-1 signaling may promote tumor immune escape.
Figure 5. PD-L1 expression in primary PTLD tumors.
Immunohistochemical staining for PD-L1 on three representative primary PTLDs. In two of the PTLDs (a and b), PD-L1 expression (brown) is detected in tumor cells with centrocytic/centroblastic morphology which are unstained with isotype control (inset). One tumor shows no staining for PD-L1 (c).
Discussion
Tumor immune evasion is an emerging hallmark of cancer (36). However, the diverse cellular mechanisms for promoting expression of immune evasion molecules in cancers are largely undefined. In cHL, the immunosuppressive microenvironment surrounding the malignant RS cells is suggestive of multiple immune evasion mechanisms (6, 9, 37, 38). Further characterization of the cellular bases for immune evasion will allow emerging targeted immunomodulatory therapies to be directed towards those individuals who may benefit the most. Furthermore, observations made in cHL may be translated to other diseases with shared molecular etiology; such has been the case for AP-1-driven galectin-1 expression in cHL and EBV-positive PTLD (5–7).
Here, we observed detectable PD-L1 expression in 100% of primary cHLs with diploid 9p24.1, prompting us to define additional mechanisms of PD-L1 induction in this disease. Our comprehensive PD-L1 regulatory element analysis revealed a candidate enhancer element with tandem AP-1 binding sites. These results were of particular interest because AP-1 is constitutively active in cHL and RS cells express high levels of both cJUN and JUN-B (7, 39). We found that the AP-1 components, cJUN and JUN-B, bound the predicted PD-L1 AP-1-responsive enhancer element and the enhancer augmented PD-L1 promoter-driven luciferase activity in an AP-1 dependent manner. Furthermore, specific inhibition of cJUN activity with a dominant-negative construct decreased PD-L1 expression. Together, these results define an additional AP-1-dependent mechanism for PD-L1 expression in cHL. Recent studies also indicate that PD-L1 is infrequently altered by genetic translocation (38).
We observed a mutually exclusive relationship between EBV infection and 9p24.1 amplification in this series of primary cHLs, suggesting that these may represent alternative mechanisms of PD-L1 induction. In support of this hypothesis, we found strong expression of PD-L1 on EBV-transformed B-cells (LCLs) and activity of both the PD-L1 promoter and enhancer in these cells. Furthermore, the EBV-encoded antigen, LMP1, increased PD-L1 promoter-driven luciferase expression and over 70% of primary EBV-driven PTLDs expressed PD-L1 on the malignant B cells. These findings are of additional interest given the recently reported expression of the PD-1 receptor on EBV-specific CD8+ T cells (40). In previous array comparative genomic hybridization analyses, 9p24.1 amplification was not identified in EBV+ PTLDs (41, 42), further supporting the notion that EBV infection and 9p24.1 copy gain represent alternative mechanisms of PD-L1 induction.
We previously found that the 9p24.1 amplicon increased both the gene dosage of PD-1 ligands and their induction by JAK2 (9). Given the known role of JAK/STAT signaling in the induction of PD-L1, we asked whether EBV LMP-1 modulated JAK/STAT activity and PD-L1 expression in EBV-transformed LCLs. Of interest, EBV-LCLs expressed higher levels of the JAK-3-associated STAT proteins and chemical JAK3 inhibition decreased PD-L1 expression in LCLs. These findings highlight alternative mechanisms of JAK/STAT-mediated PD-L1 induction that may be operative in EBV+ cHLs. Of note, additional molecular bases of JAK activation have been described in cHL including mutations of SOCS-1 (43), downregulation of miR-135a (44) and rare rearrangements of JAK2 (45).
The additional mechanisms of PD-L1 induction, AP-1 activation and EBV infection, expand the group of cHLs that may be amenable to PD-1 blockade and identify EBV-PTLD as another lymphoid tumor in which to consider this treatment strategy. The data also provide a likely mechanism for the observed expression of PD-L1 in other malignancies with constitutive AP-1 activity such as anaplastic large cell lymphoma (7, 46, 47) and melanoma (48) and bases for further analyses of the PD-1 pathway in additional EBV-associated tumors (2, 49).
Furthermore, our studies demonstrate that AP-1 activation and EBV infection induce the expression of two immunoregulatory molecules, galectin-1 and PD-L1, which limit anti-tumor T-cell responses by different and potentially complementary mechanisms (5–7). These data highlight the existence of synergistic AP-1 dependent mechanisms of tumor immune evasion and suggest additional approaches to combined targeted immunotherapy.
Supplementary Material
Statement of Translational Relevance.
Tumor immune escape is an emerging hallmark of cancer that is becoming increasingly targetable with neutralizing antibodies. However, in order for these therapies to be effective, they must be directed toward tumors that are amenable to targeted immunomodulation. We recently found that in cHL, 9p24.1 amplification increases the expression of the immune evasion molecule, PD-L1, by two complementary mechanisms. Here, we show that cHLs with normal 9p24.1 copy numbers also express detectable PD-L1. In these tumors, PD-L1 expression is supported by an AP-1-responsive enhancer element in the PD-L1 gene in cHLs which have constitutive AP-1 activity. Furthermore, we found that EBV induces PD-L1 expression via AP-1 and JAK/STAT pathways and EBV+ cHLs with diploid 9p24.1 and the majority of EBV+ PTLDs express detectable PD-L1. Therefore, AP-1- and EBV-driven malignancies with normal 9p24.1 copy numbers, including cHLs and most PTLDs, may also be amenable to PD-1 blockade.
Acknowledgments
Grant Support
The work was supported by funding from the Miller Family Research Fund and the National Institutes of Health (RO1 CA161026).
References
- 1.Kuppers R. The biology of Hodgkin’s lymphoma. Nat Rev Cancer. 2009;9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 2.Young L, Rickinson A. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 3.Portis T, Dyck P, Longnecker R. Epstein-Barr Virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2002;102(12):4166–78. doi: 10.1182/blood-2003-04-1018. [DOI] [PubMed] [Google Scholar]
- 4.Vockerodt M, Morgan S, Kuo M, Wei W, Chukwuma M, Arrand J, et al. The Epstein-Barr virus oncoprotein, latent membrane protein-1 reprograms germinal centre B cells towards a Hodgkin’s Reed-Sternberg-like phenotype. J Pathol. 2008;216(1):83–92. doi: 10.1002/path.2384. [DOI] [PubMed] [Google Scholar]
- 5.Ouyang J, Juszczynski P, Rodig SJ, Green MR, O’Donnell E, Currie T, et al. Viral induction and targeted inhibition of galectin-1 in EBV+ post-transplant lymphoproliferative disorders. Blood. 117:4315–22. doi: 10.1182/blood-2010-11-320481. accompanying editorial, Blood 2011; 4317:4165–6, cited in SciBx 4314(4319); doi:101038/scibx2011251 2012011. [DOI] [PubMed] [Google Scholar]
- 6.Juszczynski P, Ouyang J, Monti S, Rodig SJ, Takeyama K, Abramson J, et al. The AP1-dependent secretion of galectin-1 by Reed Sternberg cells fosters immune privilege in classical Hodgkin lymphoma. Proc Natl Acad Sci USA. 2007;104(32):13134–9. doi: 10.1073/pnas.0706017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodig SJ, Ouyang J, Juszczynski P, Currie T, Law K, Neuberg D, et al. AP1-dependent galectin-1 expression delineates classical Hodgkin and anaplastic large cell lymphomas from other lymphoid malignancies with shared molecular features. Clin Cancer Res. 2008;14(11):3338–44. doi: 10.1158/1078-0432.CCR-07-4709. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto R, Nishikori M, Kitawaki T, Sakai T, Hishizawa M, Tashima M, et al. PD-1-PD-1 ligand interaction contributes to immunosuppressive microenvironment of Hodgkin lymphoma. Blood. 2008;111:3220–4. doi: 10.1182/blood-2007-05-085159. [DOI] [PubMed] [Google Scholar]
- 9.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116(17):3268–77. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keir M, Butte M, Freeman GJ, Sharpe A. PD-1 and its ligands in tolerance and immunity. Ann Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–22. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–29. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber DI, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–7. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 14.Day CI, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–4. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 15.Radzewicz H, Ibegbu CC, Fernandez MI, Workowski KA, Obideen K, Wehbi M, et al. Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J Virol. 2007;81:2545–53. doi: 10.1128/JVI.02021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trautmann I, Janbazian I, Chomont N, Said EA, Gimmig S, Wilkinson P, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversable immune dysfunction. Nat Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Zhang JY, Wherry EJ, Jin B, Xu B, Zou ZS, et al. Dynamic programmed death 1 expression by virus-specific CD8 T cells correlates with the outcome of acute hepatitis B. Gastroenterol. 2008;134:1938–49. doi: 10.1053/j.gastro.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 18.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci. 2002;99:12293–7. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hino R, Kabashima K, Kato Y, Yagi H, Nakamura M, Honjo T, et al. Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer. 2010;116(7):1757–66. doi: 10.1002/cncr.24899. [DOI] [PubMed] [Google Scholar]
- 20.Geng L, Huang D, Liu J, Qian Y, Deng J, Li D, et al. B7-H1 up-regulated expression in human pancreatic carcinoma tissue associates with tumor progression. J Cancer Res Clin Oncol. 2008;134:1021–7. doi: 10.1007/s00432-008-0364-8. [DOI] [PubMed] [Google Scholar]
- 21.Gao Q, Wang XY, Qiu SJ, Yamato I, Sho M, Nakajima Y, et al. Overexpression of PD-L1 significantly associates with tumor agressiveness and postoperative recurrence in human hepatocellular carcinoma. Clin Cancer Res. 2009;15:971–9. doi: 10.1158/1078-0432.CCR-08-1608. [DOI] [PubMed] [Google Scholar]
- 22.Loos M, Giese NA, Kleef J, Giese T, Gaida MM, Bergmann F, et al. Clinical significance and regulation of the costimulatory molecule B7-H1 in pancreatic cancer. Cancer Lett. 2008;268:98–109. doi: 10.1016/j.canlet.2008.03.056. [DOI] [PubMed] [Google Scholar]
- 23.Nomi T, Sho M, Akahori T, Hamada K, Kubo A, Kanehiro H, et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clin Cancer Res. 2007;13:2151–7. doi: 10.1158/1078-0432.CCR-06-2746. [DOI] [PubMed] [Google Scholar]
- 24.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci. 2007;104:3360–5. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutok J, Pinkus G, Dorfman D, Fletcher D. Inflammatory pseudotumor of lymph node and spleen: An entity biologically distinct from inflammatory myofibroblastic tumor. Hum Pathol. 2001;32:1382–7. doi: 10.1053/hupa.2001.29679. [DOI] [PubMed] [Google Scholar]
- 26.Dirks WG, Drexler HG. Online verification of human cell line identity by STR DNA typing. Methods Mol Biol. 2011;731:45–55. doi: 10.1007/978-1-61779-080-5_5. [DOI] [PubMed] [Google Scholar]
- 27.Aerts S, Van Loo P, Thijs D, Mayer H, de Marin R, Moreau Y, et al. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucl Acids Res. 2005;33:W393–6. doi: 10.1093/nar/gki354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmittgen T, Livak K. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 29.Mann K, Staunton D, Thorley-Lawson D. Epstein-Barr Virus-encoded Protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–20. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brune V, Tiacci E, Pfeil I, Doring C, Eckerie S, van Noesel C, et al. Origin and pathogenesis of nodular lymphocyte - predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med. 2008;205:2251–68. doi: 10.1084/jem.20080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozako T, Yoshimitsu M, Fujiwara H, Masamoto I, Horai S, White Y, et al. PD-1/PD-L1 expression in human T-cell leukemia virus type 1 carriers and adult T-cell leukemia/lymphoma patients. Leukemia. 2009;23:375–82. doi: 10.1038/leu.2008.272. [DOI] [PubMed] [Google Scholar]
- 32.Meier A, Bagchi A, Sidhu H, Alter G, Suscovich T, Kavanagh D, et al. Up-regulation of PD-L1 on monocytes and dendritic cells by HIV-1 derived TLR ligands. AIDS. 2008;22:655–8. doi: 10.1097/QAD.0b013e3282f4de23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhlbauer M, Fleck M, Schutz C, Weiss T, Froh M, Blank C, et al. PD-L1 is induced in hepatocytes by viral infection and by interferon-a and -y and mediates T cell apoptosis. J Hepatol. 2006;45:520–8. doi: 10.1016/j.jhep.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. Embo J. 1997;16(21):6478–85. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gires O, Kohlihuber F, Kilger E, Baumann M, Kieser A, Kaiser C, et al. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999;18:3064–73. doi: 10.1093/emboj/18.11.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D. Weinberg R Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Steidl C, Shah S, Woolcock B, Rui L, Kawahara M, Faninha P, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–83. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steidl C, Connors J, Gascoyne R. Molecular pathogenesis of Hodgkin’s lymphoma: increasing evidence of the importance of the microenvironment. J Clin Oncol. 2011;29(14):1812–26. doi: 10.1200/JCO.2010.32.8401. [DOI] [PubMed] [Google Scholar]
- 39.Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F, et al. Aberrantly expressed c-Jun and JunB are hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappaB. EMBO J. 2002;21(15):4104–13. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenough TC, Campellone SC, Brody R, Jain S, Sanchez-Merino V, Somasundaran M, et al. Programmed death-1 expression on Epstein Barr virus specific CD8+ T cells varies by stage of infection, epitope specificity, and T-cell receptor usage. PLoS One. 2010;5(9):1–13. doi: 10.1371/journal.pone.0012926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirel HA, Bernheim A, Schneider A, Meddeb M, Choquet S, Leblond V, et al. Characteristic pattern of chromosomal imbalances in posttransplantation lymphoproliferative disorders: correlation with histopathological subcategories and EBV status. Transplantation. 2006;80:176–84. doi: 10.1097/01.tp.0000163288.98419.0d. [DOI] [PubMed] [Google Scholar]
- 42.Rinaldi A, Kwee I, Poretti G, Mensah A, Pruneri G, Capello D, et al. Comparative genome-wide profiling of post-transplant lymphoproliferative disorders and diffuse large B-cell lymphomas. British J Hematol. 2006;134:27–36. doi: 10.1111/j.1365-2141.2006.06114.x. [DOI] [PubMed] [Google Scholar]
- 43.Weniger M, Melzner I, Menz C, Wegener S, Bucur AJ, Dorsch K, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25:2679–84. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- 44.Navarro A, Diaz T, Martinez A, Gaya A, Pons A, Gel B, et al. Regulation of JAK2 by miR-135a: prognostic impact in classical Hodgkin lymphoma. Blood. 2009;114:2945–51. doi: 10.1182/blood-2009-02-204842. [DOI] [PubMed] [Google Scholar]
- 45.Van Roosbroeck K, Cox L, Tousseyn T, Lahortiga I, Gielen O, Cauwelier B, et al. JAK2 rearrangements, including the novel SEC31A-JAK2 fusion, are recurrent in classical Hodgkin lymphoma. Blood. 2011;117:4056–64. doi: 10.1182/blood-2010-06-291310. [DOI] [PubMed] [Google Scholar]
- 46.Andorsky DJ, Yamada RE, Said J, Pinkus GS, Betting DJ, Timmerman JM. Programmed death ligand 1 is expressed by non-Hodgkin lymphomas and inhibits the activity of tumor-associated T cells. Clin Cancer Res. 2011;17:4232–44. doi: 10.1158/1078-0432.CCR-10-2660. [DOI] [PubMed] [Google Scholar]
- 47.Leventaki V, Drakos E, Medeiros L, Lim M, Elenitoba-Johnson K, Claret F, et al. NPM-ALK oncogenic kinase promotes cell-cycle progression through activation of JNK/cJun signaling in anaplastic large-cell lymphoma. Blood. 2007;110:1621–30. doi: 10.1182/blood-2006-11-059451. [DOI] [PubMed] [Google Scholar]
- 48.Kang D, Motwani M, Fisher P. Role of the transcription factor AP-1 in melanoma differentiation. Int J Oncol. 1998;13:1117–43. doi: 10.3892/ijo.13.6.1117. [DOI] [PubMed] [Google Scholar]
- 49.Hsu MC, Hsiao JR, Chang KC, Wu YH, Su IJ, Jin YT, et al. Increase of programmed death-1-expressing intratumoral CD8 T cells predicts a poor prognosis for nasopharyngeal carcinoma. Mod Pathol. 2010;23(10):1393–1403. doi: 10.1038/modpathol.2010.130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.