Abstract
Circadian clocks synchronise biological processes with the day/night cycle, using molecular mechanisms that include interlocked, transcriptional feedback loops. Recent experiments identified the evening complex (EC) as a repressor that can be essential for gene expression rhythms in plants. Integrating the EC components in this role significantly alters our mechanistic, mathematical model of the clock gene circuit. Negative autoregulation of the EC genes constitutes the clock's evening loop, replacing the hypothetical component Y. The EC explains our earlier conjecture that the morning gene PSEUDO-RESPONSE REGULATOR 9 was repressed by an evening gene, previously identified with TIMING OF CAB EXPRESSION1 (TOC1). Our computational analysis suggests that TOC1 is a repressor of the morning genes LATE ELONGATED HYPOCOTYL and CIRCADIAN CLOCK ASSOCIATED1 rather than an activator as first conceived. This removes the necessity for the unknown component X (or TOC1mod) from previous clock models. As well as matching timeseries and phase-response data, the model provides a new conceptual framework for the plant clock that includes a three-component repressilator circuit in its complex structure.
Keywords: biological clocks, circadian rhythms, gene regulatory networks, mathematical model, systems biology
Introduction
Circadian clocks are found widely among organisms, ranging from cyanobacteria to mammals (Dong and Golden, 2008; Zhang and Kay, 2010). These internal time-keepers generate ∼24 h rhythms of expression of multiple genes even in the absence of any environmental cues, allowing the organism to anticipate each new day. Circadian rhythms can enhance growth and survival (Dodd et al, 2005; Harmer, 2009; Zhang and Kay, 2010). In order to understand the behaviour, mechanisms and properties of the system, we previously built a mathematical model of the plant circadian clock (Pokhilko et al, 2010).
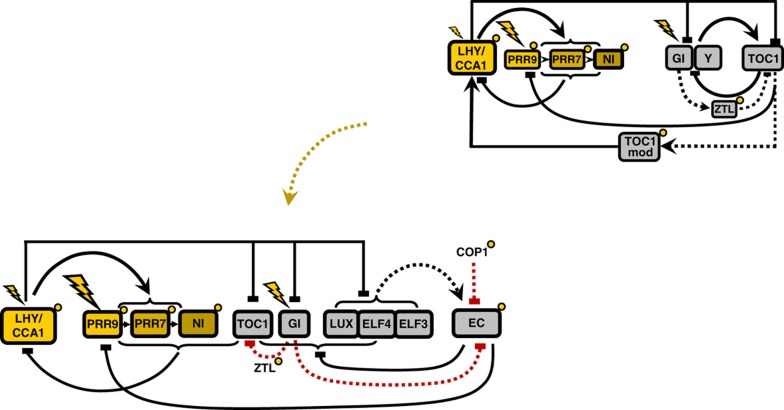
The clock was represented by a three-loop structure of interconnected morning and evening loops (Figure 1, upper right). The morning loop included MYB-related transcription factors LHY (LATE ELONGATED HYPOCOTYL) and CCA1 (CIRCADIAN CLOCK ASSOCIATED 1), which activate the expression of PRR9, PRR7 and PRR5/NI (PSEUDO-RESPONSE REGULATORs 9, 7, 5/night inhibitor) (Farre et al, 2005; Nakamichi et al, 2010). Transcriptional co-regulators PRR9, PRR7 and PRR5 inhibit LHY and CCA1 expression in the model, and in data that showed binding to their promoters (Nakamichi et al, 2010). The evening loop was represented by TIMING OF CAB EXPRESSION1 (TOC1), which inhibited the expression of its unknown activator Y. The hypothetical gene Y was introduced in the model by Locke et al (2005) to describe the observed autonomous oscillations of TOC1 expression in lhy/cca1 double-mutant plants. GIGANTEA (GI), a large plant-specific protein, accelerated the degradation of TOC1 protein through stabilisation of the F box protein ZTL (ZEITLUPE) in the model, as in the data (Kim et al, 2007).
Figure 1.
The revised outline of the Arabidopsis circadian clock. Elements of the morning and evening loops are shown in yellow and grey, respectively. Proteins are shown only for EC, ZTL and COP1 for simplicity. Transcriptional regulation is shown by solid lines. EC protein complex formation is denoted by a dashed black line. Post-translational regulation of TOC1 and the EC by GI, ZTL and COP1 are shown by red dashed lines. Acute light responses in gene transcription are shown by flashes. Post-translational regulation by light is shown by small yellow circles. The previous outline circuit (Pokhilko et al, 2010) is shown on the upper right.
The connections between morning and evening loops were represented in the model by the inhibition of evening gene expression by LHY/CCA1 protein, which was well documented, and by activation of LHY/CCA1 expression by TOC1. Previous models required unknown substances TOC1mod or X to match the observed ∼12 h delay between TOC1 expression and LHY/CCA1 induction (Locke et al, 2005; Pokhilko et al, 2010). Pokhilko et al (2010) introduced an additional connection from the evening loop to the morning loop, based on timeseries data, through inhibition of PRR9 expression by TOC1. This improved the model's description of plant rhythms but left open questions about core parts of the clock mechanism.
Loss-of-function mutants in each of the genes represented in previous clock models remained rhythmic, albeit with varying rhythmic properties. This was problematic, because the model required a hypothetical component Y to explain the rhythms observed in the lhy/cca1 double mutant. GI, the first gene proposed as a candidate for Y, was known not to perform all of the required functions (Locke et al, 2005), so the biological identity of the missing components was unknown. Conversely, three mutants that did cause striking, arrhythmic phenotypes could not be integrated into the model, because the functions of the plant-specific proteins ELF3 (EARLY FLOWERING 3), ELF4 (EARLY FLOWERING 4) and the GARP transcription LUX (LUX ARRHYTHMO) (also known as PCL1) were unclear (Hicks et al, 1996; Covington et al, 2001; Doyle et al, 2002; Hazen et al, 2005).
Recent results demonstrate that ELF3, ELF4 and LUX are the key regulators of clock gene expression at night (Onai and Ishiura, 2005; Kolmos et al, 2009; Dixon et al, 2011; Helfer et al, 2011). ELF3, ELF4 and LUX proteins were shown to form a complex, the EC (evening complex), which binds to the promoters of target genes (Nusinow et al, 2011). Although only LUX protein binds directly to promoters, both ELF3 and ELF4 proteins are important for EC function (Nusinow et al, 2011). The binding of the EC to the promoters of target genes, such as PRR9 and LUX itself, suppresses their expression (Dixon et al, 2011; Helfer et al, 2011). The importance of the ELF3/ELF4/LUX complex for free-running rhythms in constant light, and for entrainment of both wild-type (WT) and the lhy/cca1 double mutant (Hazen et al, 2005; Onai and Ishiura, 2005; Kolmos et al, 2009; Dixon et al, 2011), suggested that ELF3, ELF4 and LUX (the EC genes) are the major elements of the evening loop of the clock. However, the evening loop's structure and integration with the rest of the clock circuit remained unclear.
To create the new clock structure, we first recast the evening loop to include the EC genes, together with post-translational regulation of ELF3 protein by the ubiquitin E3 ligase COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) (Yu et al, 2008) (Figure 1, see Results for further detail). The oscillatory mechanism of the evening loop was analysed using data from the lhy/cca1 double mutant, where only the evening loop sustains rhythmicity. We explored the function of GI in the new circuit, using data from the lhy/cca1/gi triple mutant. Second, we connected the evening loop to the rest of the clock and explored a new mechanism connecting the clock's evening components to the morning genes. In the context of the whole clock circuit, the observed repression of PRR9 by the EC (Dixon et al, 2011; Helfer et al, 2011) creates a three-negative feedback ring structure, termed the repressilator. Another prediction relates to the regulation of LHY and CCA1 expression by TOC1. Although the molecular details remain to be elucidated, our computational analysis revealed that timeseries data on the ztl and prr7/prr9 mutants (Farre et al, 2005; Baudry et al, 2010) are more consistent with TOC1 being an inhibitor instead of an activator of LHY and CCA1 expression. Besides, our new experiments with the toc1 mutant and TOC1-overexpressing (TOC1-ox) plants further supported the negative role of TOC1 in regulation of LHY and CCA1 genes inside the morning loop. The proposed clock circuit integrates both positive and negative connections, including the repressilator, into a complex, multi-loop structure. Our model of this circuit includes significantly more experimental data and explains the clock's responses to multiple genetic and environmental perturbations, now including the canonical response to short light pulses at various times (the phase-response curve (PRC)).
Results
Qualitative analysis leading to revision of the clock gene circuit in Arabidopsis
Figure 1 shows the principal scheme of the new clock model. We justify the new components and circuit structure in outline below, and examine its dynamic behaviour in the following sections. As in all previous models, CCA1 and LHY were treated as a single component (CCA1/LHY). The model consists of 28 ordinary differential equations and 104 parameters. Values of 43 parameters were constrained based on the available data and 61 parameters were fitted to multiple timeseries data sets (see Supplementary Table S1). The value of the six Hill coefficients was set to two. A detailed description of the model is presented in the Supplementary information, together with a discussion of the model's limitations and its robustness to parameter variations (Supplementary Figures S3 and S4).
The evening loop of the clock was fundamentally revised in order to include the ELF3, ELF4 and LUX genes (EC genes). The model includes the formation of the triple ELF3–ELF4–LUX protein complex, the EC (Figure 1), which was shown to be important for clock function (Nusinow et al, 2011). Multiple data show that the EC genes have repressive effects on clock gene expression, so that expression of LUX, ELF4, GI, TOC1 and PRR9 was derepressed in elf3, lux and elf4 mutants (Fowler et al, 1999; Kikis et al, 2005; Kolmos et al, 2009; Dixon et al, 2011; Helfer et al, 2011). The model assumes that the EC suppresses the expression of these five target genes (Figure 1).
To define the minimal structure of the evening loop that remains in the lhy/cca1 mutant, we analysed data on triple mutants lhy/cca1/elf3, lhy/cca1/gi and lhy/cca1/toc1. The data showed that clock entrainment is completely disrupted in the lhy/cca1/elf3 mutant (Dixon et al, 2011), although the remnant circuit is still entrained in lhy/cca1/toc1 (Ding et al, 2007) and lhy/cca1/gi (Locke et al, 2006). This suggested that the EC genes represent a core structure of the evening loop (Figure 1), which drives oscillations in the lhy/cca1 mutant as described below, whereas TOC1 and GI have different roles. Because the remnant circuit in lhy/cca1 is entrained by light signals, we included light-dependent, post-translational regulation of the EC component ELF3 through its observed interactions with COP1 and GI, as detailed below. The new structure of the evening loop allowed us to describe our new data on the lhy/cca1 and lhy/cca1/gi mutants without the hypothetical component Y.
Next, we connected the evening circuit to the rest of the clock. The connection from the morning to the evening loop was described through the suppression of TOC1, LUX, ELF4, ELF3 and GI expression by CCA1 and LHY proteins (Harmer and Kay, 2005; Hazen et al, 2005; Kikis et al, 2005; Locke et al, 2005; Dixon et al, 2011; Li et al, 2011). The connections from the evening loop to the morning loop include the inhibition of PRR9 expression by evening components. Based on indirect observations, we previously suggested the inhibition of PRR9 by TOC1 (Pokhilko et al, 2010). Recent biochemical work demonstrated that PRR9 expression is more likely to be inhibited by the EC (Dixon et al, 2011; Helfer et al, 2011) (Figure 1). Another important connection from the evening to the morning genes is related to the regulation of LHY and CCA1 expression by TOC1 protein (Alabadi et al, 2001; Makino et al, 2002; Mas et al, 2003b; Baudry et al, 2010). Below we changed the sign of TOC1 function in the regulation of LHY and CCA1 expression from an activator to an inhibitor, which improved the model's description of the existing data on the ztl and prr7/prr9 mutants. This revision of TOC1 function removed the need for the hypothetical components X and TOC1mod of our previous models (Locke et al, 2005, 2006; Pokhilko et al, 2010), the most recent of which is hereafter referred to as the P2010 model. To further verify the proposed negative role of TOC1, we measured the level of LHY and CCA1 expression in the toc1 mutant and TOC1-ox plants. Our results showed that LHY and CCA1 mRNA levels were reduced in the TOC1-ox plants and increased in the toc1 mutant, which confirmed our model prediction about the negative regulation of LHY and CCA1 genes by TOC1. Moreover, data published during revision of this manuscript further demonstrated the direct suppressive effect of TOC1 on LHY and CCA1 expression (Gendron et al, 2012). Next, after connecting the loops, we tested the effects of the EC's repressive function on the dynamics of the whole system by comparing the simulated elf3 mutant with WT, and also investigated the sensitivity of the new clock structure to light.
New structure of the evening loop accounts for data on the lhy/cca1 and lhy/cca1/gi mutants
The EC as the main element of the evening loop
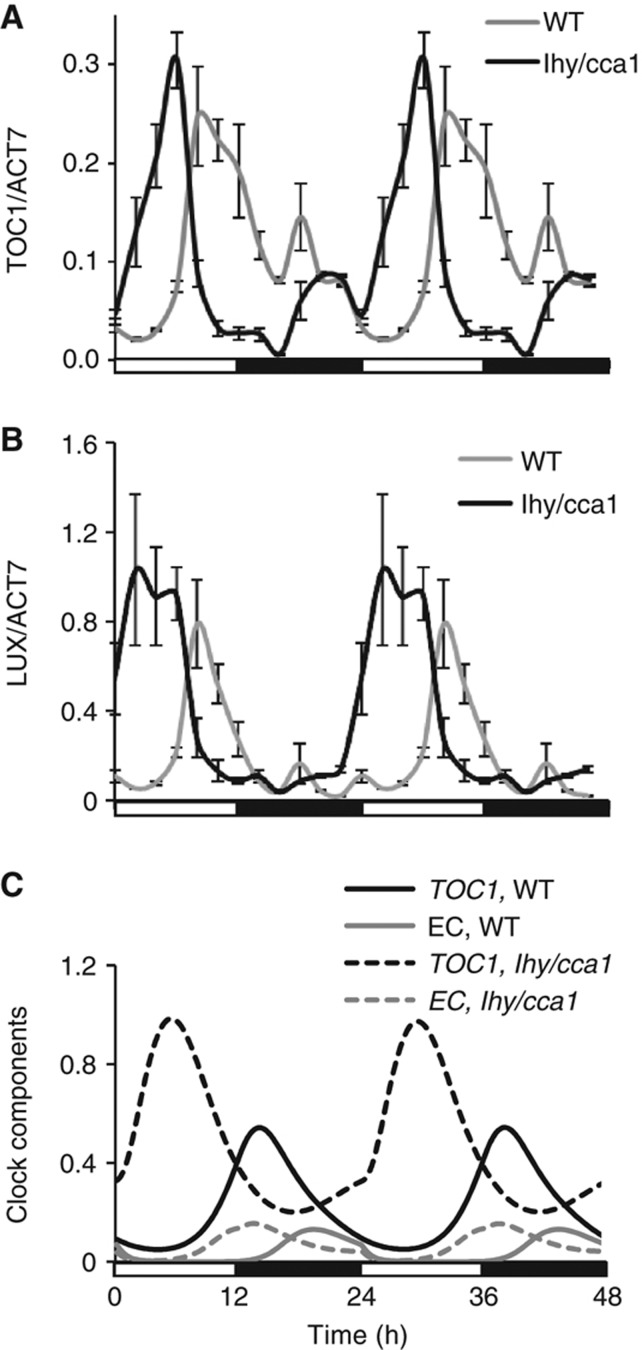
Based on the published data, we revised the structure of the evening loop, which supports oscillations in the lhy/cca1 double mutant. It is represented by the formation of the EC by ELF3, ELF4 and LUX proteins (Nusinow et al, 2011). Inhibition of ELF4 and LUX expression by the EC creates a negative feedback loop (Kikis et al, 2005; Helfer et al, 2011). To verify the new structure, we studied clock gene expression in the lhy/cca1 double mutant computationally and experimentally under various light conditions. Expression peaks of the evening genes TOC1 and GI were first shown to be advanced to a morning phase in RT–qPCR assays of the lhy/cca1 mutant compared with WT plants grown under LD cycles (Figure 2A and B) (Alabadi et al, 2001; Mizoguchi et al, 2002; Locke et al, 2005). Transcriptome data from the lhy/cca1 mutant identified GI as the most LHY/CCA1-responsive, evening-expressed gene (Supplementary Figure S2C). However, ELF3, ELF4 and LUX were shown to be similarly affected (Supplementary Figure S2; Hazen et al, 2005; Kikis et al, 2005; Dixon et al, 2011; Li et al, 2011). We therefore extended the inhibitory action of LHY/CCA1 to all evening genes in our model, and the resulting simulations agreed with these data (Figure 2C): the early expression is caused by the loss of transcriptional inhibition by LHY/CCA1 in the morning. LHY/CCA1 regulation in WT delays the rising phase of evening gene expression, as in previous models.
Figure 2.
Regulation of TOC1 and LUX expression in the evening circuit of the clock. The phase advance of TOC1 (A) and LUX (B) expression in the lhy/cca1 double mutant (black line) compared with WT (grey line) was measured by qRT–PCR assays of plants grown under 12L:12D cycles, as described in Supplementary information. (C) Model simulations demonstrate that in both WT and lhy/cca1 plants, the increase in EC (grey lines) coincides with the time of the fall in the expression of the EC's target genes (such as TOC1, black lines). Data are double-plotted to facilitate comparison to simulations. Light conditions are shown by open and filled bars below the figure.
TOC1 is also repressed by the EC in the model, which is based on the data on the high level of TOC1 expression in the elf3 and elf4 mutants (Kolmos et al, 2009; Dixon et al, 2011) and the presence of two consensus LUX-binding sites, GAT(A/T)CG, in the TOC1 promoter (Helfer et al, 2011). Simulated TOC1 RNA levels fall after dusk as EC levels rise, demonstrating that negative feedback from the EC is an important determinant of the falling phase of the evening genes' expression in the WT (Figure 2C). In the lhy/cca1 mutant, this feedback is the only cause of oscillation in the model, so the profile of TOC1 RNA level almost mirrors the EC profile (Figure 2C). The mutant's observed short period in constant conditions (17–18 h; Locke et al, 2005, 2006) reflects the lack of the additional delays from LHY and CCA1 inhibitor proteins (Supplementary Figure S5), which accelerates the expression of the EC genes. Formation of the EC then leads to autoinhibition of EC gene expression.
Regulation of EC activity by COP1 and GI
The lhy/cca1 mutant retained light entrainment (Alabadi et al, 2001; Mizoguchi et al, 2002; Locke et al, 2005), so light inputs must target at least one component of the evening loop. We therefore included the regulation of EC activity by light through targeted degradation of the EC component ELF3 by the COP1 ubiquitin E3 ligase, which was shown to be important for clock function (Millar et al, 1995b; Yu et al, 2008). To describe the kinetics of COP1 in diel cycles, we used the recent observation that COP1 protein exists in two different forms (Chen et al, 2010). Similarly to Pokhilko et al (2011), we assumed that light/dark transitions switch between the activities of these two, distinguishable E3 ligases, a night-active form (COP1n) and a day-active form (COP1d). Recent data suggested that COP1d might be related with a CULLIN 4 (CUL4) complex with COP1, where COP1 acts as a scaffold for a CUL4-based ubiquitin E3 ligase (Chen et al, 2006, 2010). Here, we assumed that COP1d is more active in the targeted degradation of the EC component ELF3, which thus alters the abundance profile of the EC (Supplementary Figure S6). ELF3 levels peak in the mid-night phase in the simulated WT, as observed (Liu et al, 2001; Dixon et al, 2011), so EC levels have already fallen substantially before dawn. Light regulation of COP1 activity (Pokhilko et al, 2011) then results in ELF3 degradation to a still lower level in the morning (Figure 2C). The further fall in EC levels is predicted to derepress the EC target genes such as TOC1. Its expression increases immediately after dawn in the lhy/cca1 mutant (Figure 2; Supplementary Figures S2 and S6), though the LHY and CCA1 repressors mask this effect in WT plants. Thus, the model predicts that COP1 is important for the timing of evening gene expression in the lhy/cca1 mutant. In the WT, this regulation would most affect genes that are more strongly regulated by LUX than by LHY and CCA1.
Additionally to COP1, GI protein also modulates the kinetics of the evening loop (Locke et al, 2005). Light-dependent stabilisation of ZTL by GI, and hence destabilisation of TOC1 protein (Kim et al, 2007), resulted in the simulated period lengthening in the gi mutant of the P2010 model. The short period of the most of gi mutants suggested another important function of GI in the clock (Martin-Tryon et al, 2007). Here, we added the binding of GI to ELF3 protein (Yu et al, 2008). The binding of GI to F box proteins in the presence of light suggested GI's ability to negatively regulate various protein targets (Kim et al, 2007; Sawa et al, 2007). Thus, we assumed that GI can accelerate the destruction of the EC by bringing F box proteins into its vicinity. Below we simulated computationally the possible outcomes of this role of GI and then tested the model predictions experimentally using the lhy/cca1/gi mutant.
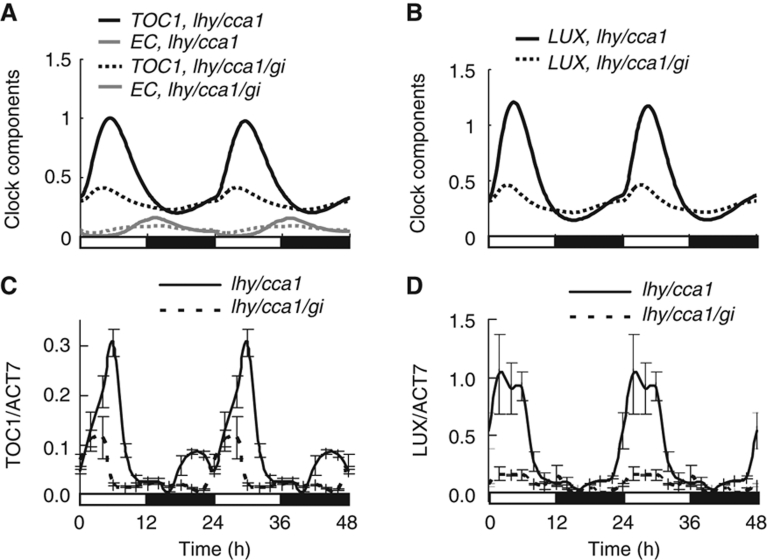
The model predicted that the absence of GI should prevent EC levels from falling to their normal trough and thus reduce the peak levels of all EC-targeted evening genes (LUX, TOC1, GI, ELF4) in the lhy/cca1/gi triple mutant compared with lhy/cca1 double mutant. Figure 3A and B show model simulations of TOC1 and LUX expression, respectively. qRT–PCR measurements of TOC1 and LUX expression confirmed this prediction (Figure 3C and D), demonstrating the indirect positive effect of GI on evening gene expression, consistent with previous reporter gene data (Locke et al, 2006). In our simulations of constant light conditions, this resulted in arrhythmia of the simulated lhy/cca1/gi mutants, in contrast to the short-period oscillations in lhy/cca1 (Supplementary Figure S7), which both are consistent with experimental observations and with previous models (Locke et al, 2006).
Figure 3.
The role of GI in the regulation of TOC1 expression by the evening loop. Model simulations demonstrate lower peak levels of TOC1 (A) and LUX (B) expression (black lines) in lhy/cca1/gi (dotted lines) compared with lhy/cca1 mutants under 12L:12D cycles. This results from increased EC levels (grey lines) during the morning in the lhy/cca1/gi mutant. (C, D) qRT–PCR measurements of TOC1 and LUX expression in lhy/cca1/gi and lhy/cca1 mutants under 12L:12D. Data are double-plotted to facilitate comparison to simulations.
The above simulations showed that COP1 and GI regulate the level of the ELF4–ELF3–LUX complex (EC) in both lhy/cca1 mutants and WT. Despite this post-translational control of the complex, the temporal profiles of the bulk levels of ELF3, ELF4 and LUX proteins in our model mainly reflected the kinetics of the corresponding mRNAs, as shown in Supplementary Figure S8.
Regulation of morning-expressed genes by the evening components of the clock
TOC1—a repressor of the morning loop
The importance of the evening gene TOC1 in the regulation of the morning components LHY and CCA1 has long been assumed, based on multiple experimental observations (Alabadi et al, 2001; Mas et al, 2003b). However, the exact mechanisms of TOC1 action are still unknown. Based on gene expression analysis with mutant plants (Alabadi et al, 2001), TOC1 was previously suggested to play the role of an activator of LHY/CCA1 expression (Alabadi et al, 2001; Locke et al, 2005; Baudry et al, 2010; Pokhilko et al, 2010). However, our analysis of the available data revealed inconsistency between the data and the activator role of TOC1. For example, the increase of TOC1 level in the ztl mutant, caused by the slowing of TOC1 protein degradation rate, leads to a substantial lengthening of the clock period (Mas et al, 2003b; Somers et al, 2004; Kevei et al, 2006). This lengthening was accompanied by a lower amplitude of LHY and CCA1 expression in ztl plants (Baudry et al, 2010). This observation cannot easily be reconciled with the activation of LHY and CCA1 expression by TOC1, which should result in a higher amplitude of LHY and CCA1 in ztl mutants, as revealed by simulation of the P2010 model (Supplementary Figure S9). In addition, recent data showed that the expression of the LHY and CCA1 inhibitors PRR9 and PRR7 is low in the ztl mutant (Baudry et al, 2010). It is hard to explain why LHY and CCA1 mRNA does not rise in the ztl mutant, despite a low level of inhibitors and a higher level of the presumed activator TOC1.
On the contrary, our simulations below show that the results on ztl mutants can easily be described by assuming that TOC1 acts as a repressor of LHY/CCA1 expression. The increase of TOC1 level in the ztl mutant results in a prolonged inhibition of LHY/CCA1, which lengthens the circadian period and reduces the amplitude of LHY/CCA1 expression. As TOC1 (PRR1) belongs to the PRR gene family, the repressive function of TOC1 makes it consistent with the other PRR proteins, such as PRR9, PRR7 and PRR5 (Nakamichi et al, 2010; Pokhilko et al, 2010). Thus, we extended the wave of PRR inhibitors of LHY/CCA1 in the model by including TOC1 (Figure 1), and explored the effect of TOC1 repression on the clock's dynamics.
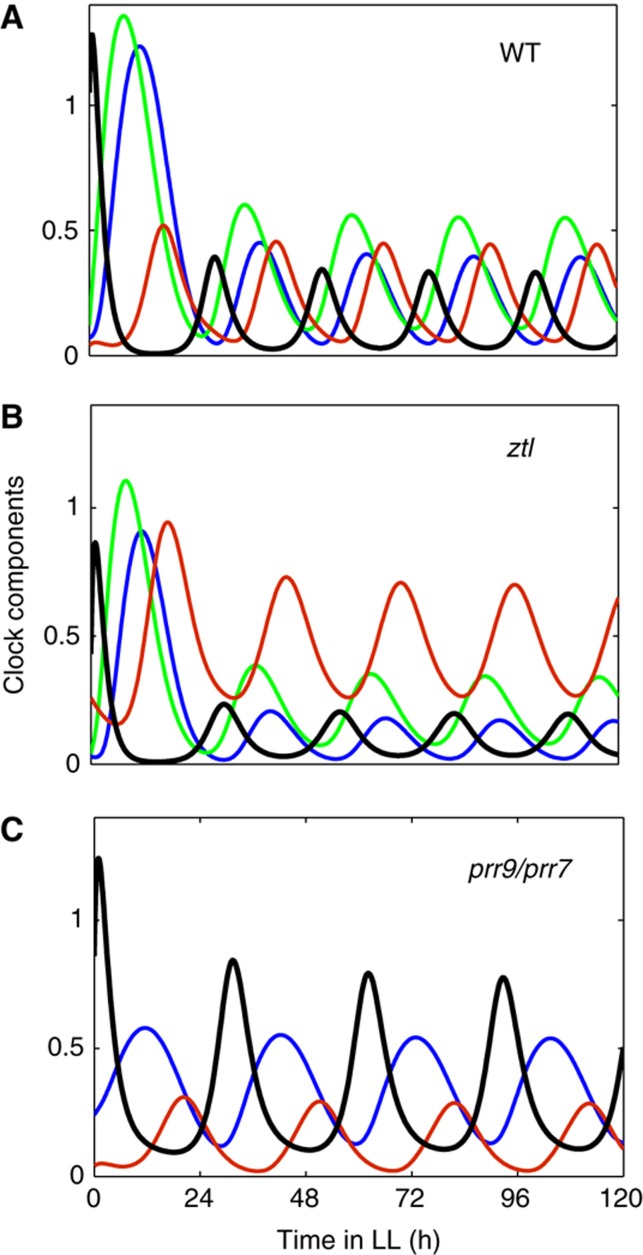
Our simulation of the ztl mutant demonstrated that higher suppression of LHY/CCA1 by TOC1 protein resulted in longer period and lower amplitude of LHY/CCA1 in the ztl mutant compared with WT (Figure 4A and B). These results corresponded to the data (Baudry et al, 2010) and improved the description of ztl compared with the P2010 model (Supplementary Figure S9B). Figure 4A illustrates the participation of TOC1 in the wave of LHY/CCA1 inhibitors in simulations of WT plants in constant light conditions. The absence of TOC1 resulted in a 2.5 h shortening of the period in the toc1 mutant compared with WT (Supplementary Figure S10), which is close to the observed period shortening (Millar et al, 1995a; Mas et al, 2003a). LHY/CCA1 levels were slightly reduced in the simulated toc1 mutant under constant light, but this counter-intuitive result is also consistent with experimental data (Alabadi et al, 2001). The reduction was related to the higher trough level of the remaining LHY/CCA1 inhibitors—the PRR9, PRR7 and NI proteins—in the toc1 mutant (Supplementary Figure S10; please see Supplementary information for detail). Additionally to the correct description of ztl and toc1 mutants, we greatly improved the description of the prr7/prr9 double mutant compared with the P2010 model (Supplementary Figure S9C). The participation of TOC1 in LHY/CCA1 inhibition resulted in robust oscillations in the prr7/prr9 mutant under constant light with a period 30.6 h (Figure 4C), which corresponds to the experimental observations (Farre et al, 2005; Salome and McClung, 2005). In the P2010 model, the simulated period for prr7/prr9 mutants was only 27.5 h, because the only remaining inhibitor (NI) could not provide a long enough delay in LHY/CCA1 expression. Furthermore, the oscillations in the prr7/prr9 mutant simulated by the P2010 model dampened faster than observed in the data (Supplementary Figure S9C). Thus, the introduction of TOC1 repressive function improved the description of multiple mutants in the new model of the clock.
Figure 4.
The improved description of ztl and prr7/prr9 mutants is related to the inhibition of LHY/CCA1 expression by TOC1. The simulated level of LHY/CCA1 mRNA (black) and the repressor proteins PRR7, NI and TOC1 (green, blue and red, respectively) are shown for WT (A), ztl mutant (B) and prr7/prr9 mutant (C) plants. Simulations moved from 12L:12D cycles to constant light at time 0, corresponding to dawn in LD.
To verify the repressive function of TOC1 further, we measured the expression levels of LHY and CCA1 in the toc1 mutant and TOC1-ox plants at the end of the night. TOC1 is predicted to have a larger role than the other, earlier-expressed PRR proteins at this time, when LHY and CCA1 expression starts to rise as they are released from repression. Figure 5A and B show that LHY and CCA1 mRNA levels rise more slowly in the TOC1-ox plants compared with WT, whereas the rise of CCA1 is accelerated in the toc1 mutant. The model simulations of TOC1-ox and toc1 matched our experimental observations (Figure 5C), supporting the proposed repressive function of TOC1 towards LHY and CCA1 expression. This change compared with earlier models affects only the sign of the interaction, not the level of abstraction in the model: the biochemical mechanism of TOC1 action remains to be determined.
Figure 5.
The effect of TOC1 level on the kinetics of LHY and CCA1 expression. qRT–PCR measurements of CCA1 (A) and LHY (B) mRNA levels, and model simulations of LHY/CCA1 (C) expression in TOC1-ox, toc1 and WT plants under 12L:12D cycles were performed as described in the Supplementary information. TOC1-ox was simulated by adding a constant, unregulated activation of TOC1 transcription with a rate constant equal to 0.3 per hour, which correspond the observed expression level of TOC1 in TOC1-ox (Supplementary Figure S1).
The EC controls LHY and CCA1 expression through multiple PRRs
The EC components ELF3, ELF4 and LUX are known to be important for the high-amplitude oscillations of LHY and CCA1 in diel cycles and for rhythmicity of the clock in constant light conditions (Doyle et al, 2002; Hazen et al, 2005; Kolmos et al, 2009; Dixon et al, 2011). We therefore explored the direct and indirect effects of EC action on the clock system, by simulating mutants in the EC genes. Figure 6A demonstrates the reduction of LHY/CCA1 amplitude in the simulated elf3 mutant compared with WT in a 12L:12D cycle, which agrees with experimental observations (Dixon et al, 2011; Helfer et al, 2011). The modelling also showed that this reduction of LHY/CCA1 amplitude prevents rhythmicity in the remnant circuit of elf3 mutants under constant light conditions (not shown), in line with observation (Hicks et al, 1996; Covington et al, 2001; Doyle et al, 2002; Hazen et al, 2005). The model suggested that the effect of the absence of EC on LHY/CCA1 expression is related to the higher level of LHY/CCA1 inhibitors TOC1 and PRR9 in the elf3 mutant (Figure 6B and C), which corresponds to published data (Dixon et al, 2011). The effect of the elf3 mutation on PRR9 is quite subtle compared with its effect on TOC1. To separate the effects of derepressing TOC1 and PRR9 in elf3 mutants, we simulated a hypothetical mutant that lacked only PRR9 inhibition by the EC (Figure 6D). This simulation showed that the amplitude of LHY/CCA1 oscillations falls by 46% in the elf3 mutant and by 24% in the absence of inhibition of PRR9 by EC (Figure 6A and D). Thus, the model predicted that the inhibition of both PRR9 and TOC1 expression by the EC at night is important for robust oscillations of LHY/CCA1 and consequently for the anticipation of dawn by the clock.
Figure 6.
Nighttime inhibition of TOC1 and PRR9 expression by the EC is important for the robust oscillation of LHY/CCA1. Model simulations (dashed lines) of the elf3 mutant (A–C) and a hypothetical mutant without inhibition of PRR9 by the EC (D) are shown together with WT simulations (solid lines). The simulations were run under 12L:12D conditions, which are indicated by open (light) and solid (dark) bars.
Improved light sensing by the new clock circuit
Various light input signals are used experimentally to study the mechanisms of light perception by the clock, which results in entrainment of the endogenous oscillator to the environmental day/night cycle. The most obvious manipulation changes the duration of the light interval or day length in an experimental light/dark cycle. Our simulations showed that, similarly to P2010, the new model retains the good match to LHY/CCA1 mRNA data under various photoperiods (Supplementary Figure S11A). In the same time, the new structure of the evening loop provides some delay of evening gene expression in long days and thus provides a better match to the data compared with the P2010 model (Supplementary Figures S11B and S12).
Another way to investigate light sensing by the clock is the so-called PRC, which has a long history in the circadian field. The PRC represents the phase shift of the clock components, after light pulses given at different times to organisms that are kept in darkness (Figure 7A). It is characteristic that the clock sharply changes its response from phase delays to phase advances at a certain time of the subjective night (around 15 h after subjective dawn in Arabidopsis) (Covington et al, 2001). Here, we used our model to investigate the possible mechanisms of this phase shift. Our simulations matched the available data on the PRC of the evening components of the clock (Figure 7A). Although light affects the clock in several places (Figure 1), the PRC in our model is mostly determined by the acute light response in LHY/CCA1 expression. This increase in LHY/CCA1 expression immediately after ‘lights-on’ is caused by a fast transient activation of transcription. In this and all previous models, this is mediated by the yet-unidentified, dark-accumulating activator, protein P (Kim et al, 2003; Locke et al, 2005). A simulated mutant lacking only this response loses its phase response to light (compare Figure 7B and D), indicating that the response is necessary. The most closely related data show that transient, chemical induction of CCA1 expression is sufficient to cause large phase shifts in vivo (Knowles et al, 2008). The increase in the level of LHY/CCA1 protein after the light pulse results in a fast decrease of the expression of LHY/CCA1 target genes, such as LUX (Figure 7B and C) in our model simulations. The resulting shift in LUX phase depends on the phase of LUX expression during the pulse. When the light pulse is given closer to or after the LUX mRNA peak (∼18 h), the increase in LHY/CCA1 level accelerates the fall of LUX mRNA and advances the next peak (Figure 7B). However, earlier pulses delay the rise in LUX mRNA and the next peak (Figure 7C). The phase advance after a pulse at 18 h is lost in a simulated mutant that lacks an acute light response in LHY/CCA1 (Figure 7C). Thus, the model predicted that the acute activation of LHY/CCA1 expression by light is responsible for the observed transition from phase delay to phase advance in the PRC.
Figure 7.
The mechanism of the PRC in plants. (A) A PRC was simulated by monitoring the phase of peaks of LUX expression after light pulses of 1 h duration given on the second day in darkness after 12L:12D entrainment. Data points were taken from Covington et al (2001) for red light pulses. (B–D) The simulated profiles of LUX mRNA (blue) and LHY protein (magenta) with (dashed lines) or without (solid lines) light pulses given at indicated times (arrow)—at 18 h (B), 12 h (C) or at 18 h for a simulated mutant without an acute light response in LHY transcription (D; parameter q1=0). Time 0 refers to the beginning of the second day in darkness.
Discussion
Based on very recent data, we updated the structure of the plant clock and used mathematical modelling to demonstrate a good correspondence of the new model to a wide spectrum of new and older data. The new model better described the clock's response to various genetic and environmental perturbations, and improved our understanding of the clock gene network.
Comparison to earlier models
The most radical change in our model compared with the P2010 model is related to the introduction of the EC genes ELF3, ELF4 and LUX into the clock scheme. The strong phenotypes of the single mutants of these genes suggested that they are very important for the clock. However, the structural relationships between EC genes and the rest of the clock were unknown. Our recent data suggested that the EC is absolutely necessary for the rhythmicity and entrainment of the lhy/cca1 mutant (Dixon et al, 2011). We therefore started building the new structure of the clock from the evening loop, which represents the minimal, EC-containing rhythmic element.
The previous structure of the evening loop was based on the observed, rhythmic TOC1 expression in the lhy/cca1 double mutant (Locke et al, 2005; Pokhilko et al, 2010). In the P2010 model, the evening loop consisted of the hypothetical activator Y of TOC1 expression, which was transcriptionally suppressed by TOC1 protein. In the new model, we replaced Y with the EC genes, which feedback negatively to their promoters, providing oscillations in the lhy/cca1 mutant. The expression profiles of the EC target genes, such as TOC1, are described through the repression from the EC instead of the earlier model's activation by Y. Light regulation is important for the observed entrainment of the evening loop. In previous models, light directly regulated Y expression (Locke et al, 2005). In the new model, light input is provided by the light-dependent degradation of the EC component ELF3, in which COP1 and a related ubiquitin E3 ligase may participate (Yu et al, 2008; Chen et al, 2010). This does not preclude other contributions to light regulation, such as the recently described transcriptional induction of ELF4 (Li et al, 2011).
The next changes in the scheme of the evening loop were related to GI, which was previously proposed as a candidate that accounted for some but not all of Y's functions, on the basis of RNA data and genetic evidence (Locke et al, 2005, 2006). Indeed, GI retains functions in the present model that are consistent with Y but GI appears to be a modulator rather than the major effector. GI still increases TOC1 expression in the model, for example, as observed in data (Figure 3B) (Locke et al, 2006) but the mechanism is by a double inhibition rather than direct activation: GI protein is a negative regulator of the EC, which inhibits TOC1 expression. The introduction of the negative effect of GI on the EC improved the description of the gi mutant compared with the P2010 model: the 2.6 h shortening of circadian period in the simulated gi mutant provides a better match to the data (Park et al, 1999; Gould et al, 2006; Martin-Tryon et al, 2007) than the 2 h lengthening of the gi period simulated in the P2010 model. Thus, we removed the hypothetical gene Y and redrew the structure of the evening circuit by including the important clock components ELF3, ELF4, LUX and COP1, re-connecting them to GI to provide a more realistic structure for the evening loop.
After rebuilding the evening loop, we connected it to the rest of the clock circuit and re-examined the connections from the evening genes to the morning loop. Based on genetic data, it was previously assumed that TOC1 activates LHY/CCA1 expression (Alabadi et al, 2001; Locke et al, 2005; Pokhilko et al, 2010). The ∼12 h time delay of peak LHY compared with TOC1 expression required a hypothetical, intermediate clock component, X or TOC1mod, in previous clock models to ensure the required, long-lasting positive effect of TOC1 on LHY/CCA1 expression (Locke et al, 2005; Pokhilko et al, 2010). However, our model analysis demonstrated that the data on ztl and prr7/prr9 mutant plants agreed better with a negative role for TOC1 in LHY and CCA1 expression. In addition to Baudry et al (2010), other published data also show lower levels of LHY and CCA1 mRNA in multiple mutants with increased amounts of TOC1 protein (Makino et al, 2002; Somers et al, 2004; Kim et al, 2011). Our data on LHY and CCA1 expression in the TOC1-ox and toc1 mutants further supported the negative role of TOC1 (Figure 5). This repressive function was consistent with TOC1 protein acting immediately after TOC1 RNA expression, allowing us to further improve the model by removing the hypothetical delaying component. Together, the EC-based evening loop and the change in the sign of TOC1 function suggest that the toc1 mutation merely removes the last component in the wave of PRR repressors, leaving the EC-based evening loop intact. The biochemical mechanisms of TOC1's suppressive action remain the object of further studies: our data inform only the sign of the interaction between genes and cannot exclude greater complexity in the molecular interactions involved.
Additionally to removing X and Y, we further simplified the model by greatly reduced number of transcriptional regulators with Hill kinetics, which imply a complex or multimeric regulation. The remaining Hill coefficients are set to 2, which corresponds to the well-justified dimerisation of plant clock components (Fujiwara et al, 2008; Wang et al, 2010; O'Neill et al, 2011). This provided a more realistic description of the clock compared with the P2010 model.
Mutation of an EC gene removes the evening loop in the model but leaves the potential for oscillatory feedback(s) among LHY, CCA1 and the PRRs. Oscillations and related behaviour have been observed in EC gene mutants in some conditions (Hicks et al, 1996; McWatters et al, 2000; Covington et al, 2001; Hall et al, 2003; Wenden et al, 2011). The new model recapitulates the more severe circadian phenotypes of EC gene mutants under constant light conditions (Reed et al, 2000; Doyle et al, 2002; Hazen et al, 2005), which suggest that the morning loop cannot support self-sustaining oscillations, in contrast to the P2010 model. The new model matches the data for EC gene mutants in diel cycles (Figure 6), indicating that the EC also contributes to high-amplitude oscillations of LHY and CCA1 under entrained conditions. Thus, the new model describes a complex, integrated clock structure, with interdependent dynamics. Weak, damping oscillations from the evening loop alone are stabilised by coupling to the morning loop in the intact system.
Abstraction of regulatory circuits from observed pairwise interactions
The model of the plant circadian clock was modified, based entirely on known components and their interactions. Interestingly, the emerging structure of the clock includes a ring of three sequential negative steps, each representing the inhibition of earlier-expressed clock components by the later ones: inhibition of EC genes by the rise of LHY/CCA1 in the late night, of PRR genes by EC in the early night, and of LHY/CCA1 by PRRs in the day. This new structure allows us to re-interpret several previous observations. First, EC genes were previously suggested to be activators of LHY and CCA1 expression based on genetic studies (Doyle et al, 2002; Hazen et al, 2005; Kikis et al, 2005; Onai and Ishiura, 2005). The new data demonstrated that EC proteins repress the expression of PRR genes (Kolmos et al, 2009; Dixon et al, 2011; Helfer et al, 2011). This, together with the previously known, negative regulation of LHY and CCA1 expression by PRR proteins (Farre et al, 2005; Mizuno and Nakamichi, 2005; Nakamichi et al, 2010) allowed the re-interpretation of the positive genetic interaction from the EC genes to LHY and CCA1 as a double-negative interaction (Dixon et al, 2011; Helfer et al, 2011) (Figure 8A). We demonstrated both computationally and experimentally that mutation of the EC components resulted in the decrease of the LHY/CCA1 amplitude (Figure 6A–C), in agreement with the experimental findings (Doyle et al, 2002; Hazen et al, 2005; Kolmos et al, 2009). Thus, the double-negative feedback from EC to LHY/CCA1 is sufficient to describe the experimental data.
Figure 8.
Core interactions in the clock model form a repressilator circuit. (A–C) The sequential expression of LHY/CCA1 (black), PRR genes (blue) and EC genes (green) are sketched relative to a 12L:12D diel cycle. Their regulatory interactions can be explained by double-negative (solid, blunt arrows) or single-positive (dashed arrow) connections, for (A) LHY/CCA1 activation by the EC genes; (B) PRR gene activation by LHY/CCA1; (C) activation of EC genes by PRRs. (D) The core structure of LHY–PRR–EC interactions in the model is shown to include a repressilator, a three-inhibitor ring oscillator (solid lines). Other interactions between LHY, PRRs and the EC (dotted lines) include the activation of PRRs by LHY/CCA1, which was identified as the morning loop, and the autoinhibition of the EC, which represents the evening loop. For clarity, the light inputs, GI and the post-translational regulators are omitted.
Similarly, the model has a double-negative feedback from LHY/CCA1 to PRR genes, which is also based on experimental data. Indeed, LHY and CCA1 proteins inhibit the expression of the EC genes (Hazen et al, 2005; Kikis et al, 2005; Portoles and Mas, 2010; Dixon et al, 2011; Li et al, 2011). The repression of PRR genes by the EC forms a double-negative connection from LHY and CCA1 to the PRRs. This double-negative connection can also be represented by a positive regulation of PRR expression by LHY/CCA1 (Figure 8B), which was previously suggested based on genetic studies (Farre et al, 2005; Ding et al, 2007). Based on the present data, it is not possible to distinguish between double-negative and direct positive connections from LHY to the PRRs, because either or both of them could lead to the observed decrease of PRR expression in the lhy/cca1 double mutant. However, the observed increase of LUX and ELF4 expression in the lhy/cca1 mutant (Hazen et al, 2005; Kikis et al, 2005) suggests that the double-negative feedback mechanism might underlie the decrease of PRR gene expression in the lhy/cca1 mutant. Additionally to the double-negative connection, our current model retains the direct positive connections from LHY/CCA1 to the PRRs, which is supported by the existence of LHY/CCA1-binding sites in the promoter of PRR9 and PRR7 genes (Farre et al, 2005; Harmer and Kay, 2005) and the observed binding of CCA1 to PRR9 and PRR7 promoters (Portoles and Mas, 2010), which can have high affinity (O'Neill et al, 2011). The positive connections were also found in other circadian systems, such as mamallian and fly clocks (Zhang and Kay, 2010). The functional role of the positive connections might be related with increased robustness of oscillations, as was shown for the various negative feedback networks with an additional positive connection (Tsai et al, 2008). Further biochemical studies are required to dissect the relative impact of the positive and double-negative connections from LHY and CCA1 to the PRRs.
Finally, the double-negative feedback from PRRs to the EC genes via LHY and CCA1 is also presented in the model (Figure 8C). Analogously, this follows from the data showing that PRR proteins inhibit LHY and CCA1 expression, which in turn inhibit expression of the evening genes. The double-negative connection could lead to the indirect activation of evening gene expression by PRR genes, which was previously suggested from genetic studies (Mizuno and Nakamichi, 2005; Nakamichi et al, 2005).
A counter-intuitive aspect in each of these connections is that the target gene is expressed before its immediate regulator within the day–night cycle. It was therefore natural to propose that LHY and CCA1 activated PRR gene expression, and the EC activated LHY and CCA1 expression, in line with the genetic results from stable mutant plants or mis-expression lines (dashed arrows in Figure 8A–C). Dynamic manipulations of the circuit and direct biochemical studies were required to demonstrate the double-negative mechanisms (Nakamichi et al, 2010; Portoles and Mas, 2010; Dixon et al, 2011; Helfer et al, 2011; this paper). Our mathematical model suggests that the double-negative connections are consistent with the data. Currently, we have left only one positive connection from LHY/CCA1 to the PRR genes, because it is supported by data on the direct binding of CCA1 protein to PRR promoters. Future experiments are necessary to investigate the functional consequence of this binding on PRR gene expression.
A repressilator, the three-inhibitor ring oscillator, was first constructed as a synthetic circuit in Escherichia coli (Elowitz and Leibler, 2000) and is one of a class of well-studied ring systems (reviewed in Purcell et al, 2010). Here, we show that the repressilator structure is present as an integrated element of the more complex circuit in our current model (Figure 8D). Interestingly, a similar repressilator structure was recently found in the mammalian clock, where it also represents only part of the system (Hogenesch and Ueda, 2011; Ukai-Tadenuma et al, 2011). Importantly, the repressilator structures were discovered in both plant and mammalian networks based directly on experimental data. This suggests that, although the real biological systems are more complicated than the simplified structure of the repressilator, some features of repressilator behaviour might be important for clock function. However, as described in the Results, the whole structure of the plant clock includes such important additional elements as the autoregulation of the EC genes; post-translational regulation of the EC by COP1 and GI; and the wave of multiple PRR inhibitors of LHY/CCA1 expression. Additionally, multiple light inputs affect the kinetics of the plant system. In summary, we propose that the plant clock functions as an integrated multi-feedback system, which maintains robust oscillations and entrainment under multiple perturbations.
Materials and methods
Computational and experimental methods are described in detail in Supplementary information.
Supplementary Material
Supplementary Figures S1–12
Acknowledgments
We are grateful to Gavin Steel for expert technical assistance. This work was supported by the European Commission FP7 Collaborative Project TiMet (project 245143) to AJM and others, and by BBSRC and EPSRC SABR award ROBuST (BB/F005237/1) to KJH and others. SynthSys Edinburgh is a Centre for Integrative and Systems Biology supported by BBSRC and EPSRC award D019621.
Author contributions: AP, APF, KJH, MMS and AJM designed the experiments; APF, MMS and KDE performed PCR and transcriptome experiments, respectively; AP and AJM designed the computational analysis; AP performed the computational analysis; AP and AJM wrote the paper with comments from all the authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, Kay SA, Imaizumi T (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, Deng XW (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, Zheng N, Deng XW (2006) Arabidopsis CULLIN4 forms an E3 Ubiquitin Ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Doyle MR, Amasino RM, Davis SJ (2007) A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 176: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Dong G, Golden SS (2008) How a cyanobacterium tells time. Curr Opin Microbiol 11: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338 [DOI] [PubMed] [Google Scholar]
- Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, Hall A (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, Millar AJ (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Ueda HR (2011) Understanding systems-level properties: timely stories from the study of clocks. Nat Rev Genet 12: 407–416 [DOI] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Hall A, Kozma-Bognar L, Kim WY, Eriksson ME, Toth R, Hanano S, Feher B, Southern MM, Bastow RM, Viczian A, Hibberd V, Davis SJ, Somers DE, Nagy F, Millar AJ (2006) Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol 140: 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH (2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carre IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Kim WY, Fujiwara S, Kim J, Cha JY, Park JH, Lee SY, Somers DE (2011) HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc Natl Acad Sci USA 108: 16843–16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Knowles SM, Lu SX, Tobin EM (2008) Testing time: can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? J Biol Rhythms 23: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, Schoof H, Nagy F, Bujnicki JM, Davis SJ (2009) Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J 3: 350–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, Snoep JL, Hucka M (2006) BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res 34: D689–D691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Siddiqui H, Teng Y, Lin R, Wan XY, Li J, Lau OS, Ouyang X, Dai M, Wan J, Devlin PF, Deng XW, Wang H (2011) Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol 13: 616–622 [DOI] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 2005.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003a) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003b) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA (1995a) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua NH, Kay SA (1995b) The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nakamichi N (2005) Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol 46: 677–685 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T (2005) The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol 46: 609–619 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K, Ishiura M (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972 [DOI] [PubMed] [Google Scholar]
- O'Neill JS, van Ooijen G, Le Bihan T, Millar AJ (2011) Circadian clock parameter measurement: characterization of clock transcription factors using surface plasmon resonance. J Biol Rhythms 26: 91–98 [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW, Mizuno T, Millar AJ (2010) Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol 6: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Ramos JA, Holtan H, Maszle DR, Khanna R, Millar AJ (2011) Ubiquitin ligase switch in plant photomorphogenesis: a hypothesis. J Theor Biol 270: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portoles S, Mas P (2010) The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell O, Savery NJ, Grierson CS, di Bernardo M (2010) A comparative analysis of synthetic genetic oscillators. J R Soc Interface 7: 1503–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Bastow RM, Solomon KS, Dowson-Day MJ, Elumalai RP, Millar AJ (2000) Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol 122: 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, McClung CR (2005) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TY, Choi YS, Ma W, Pomerening JR, Tang C, Ferrell JE Jr (2008) Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science 321: 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR (2011) Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144: 268–281 [DOI] [PubMed] [Google Scholar]
- Wang L, Fujiwara S, Somers DE (2010) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J 29: 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenden B, Kozma-Bognar L, Edwards KD, Hall AJ, Locke JC, Millar AJ (2011) Light inputs shape the Arabidopsis circadian system. Plant J 66: 480–491 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, Zhang Y, Lee I, Xie Q, Paek NC, Deng XW (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA (2010) Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–12