Abstract
The two heavy chains of kinesin-1 are dimerized through extensive coiled coil regions and fold into an inactive conformation through interaction of the C-terminal tail domains with the N-terminal motor (head) domains. Although this potentially allows a dimer of tail domains to interact symmetrically with a dimer of head domains, we report here that only one of the two available monomeric tail peptides is sufficient for tight binding and inhibition of a dimer of head domains. With a dimeric tail construct, the other tail peptide does not make tight contact with the head dimer and can bind a second head dimer to form a complex containing one tail dimer and two head dimers. The IAK domain and neighboring positively charged region of the tail is sufficient for tight half-site interaction with a dimer of heads. The interaction of tails with monomeric heads is weak, but a head dimer produced by the dimerization of the neck coil is not required because an artificial dimer of head domains also binds monomeric tail peptides with half-site stoichiometry in the complete absence of the native neck coil. The binding of tail peptides to head dimers is fast and readily reversible as determined by FRET between mant-ADP bound to the head dimer and a tail labeled with GFP. The association and dissociation rates are 81 µM−1s−1 and 32 s−1 respectively. This half-site interaction suggests that the second tail peptide in a folded kinesin-1 might be available to bind other molecules while kinesin-1 remained folded.
Conventional kinesin-1 (hereafter called kinesin) is a molecular motor protein that moves a large range of vesicular and nonvesicular cargoes towards the plus ends of MTs (1). The core is a dimer of heavy chains that contains N-terminal motor domains (head domains), a long region with multiple coiled coils followed by a C-terminal region (tail domains) that is not coiled coil. Previous work has established that this molecule is folded at physiological ionic strength into a compact conformation that is formed by direct interaction of the tail region with the head/neck regions as determined by cosedimentation of head and tail peptides (2), column binding assays (2–4), FRET (5) and cross-linking (4). Both the basal and microtubule-stimulated ATPase of this folded conformation are inhibited because binding of tail domains inhibits the rate limiting step of ADP release (6;7). Native kinesin also contains light chains that bind some of the cargoes and that weaken the interaction of the tail and head when present (2;8).
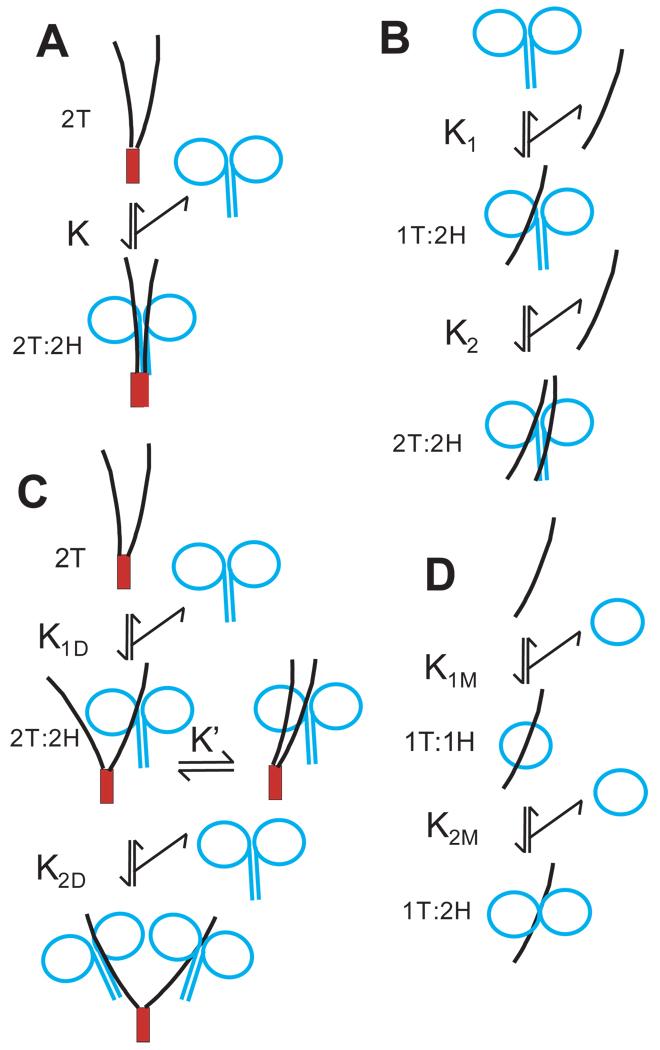
Isolated kinesin head domains are monomeric but stable dimers can be produced by inclusion of the neck coil that can form a coiled coil (9). In native kinesin, the tails are present in a dimeric context because the heavy chains are also dimerized through the additional extensive coiled coil regions of the stalk. Thus the head-tail interaction that produces folding in full length kinesin involves binding of a dimeric tail region to a dimeric head region with each tail peptide potentially interacting with one or both heads and each head potentially interacting with one or both tails, as indicated schematically in Scheme 1A, to form a complex with 2 tail domains and 2 head domains that is designated 2T:2H. A complex with this stoichiometry could have multiple detailed conformations. For example, one proposed model (2) was a 4-helix bundle formed by the neck coiled coil and coiled coil regions of the tail as in Scheme 1A. This was suggested by the failure of tail domains to bind tightly to monomeric head domains that lack the neck coil (2), implying that the neck coil played an important role and that it constituted part of the binding site for the tail domains. In this context, binding of a monomeric tail peptide to a dimer of heads, as indicated in reaction B of Scheme 1, should be highly disfavored relative to a dimer of tails. Even if two monomer tails bound to equivalent sites on the heads, the binding of a tail dimer would still have a large entropic advantage over binding of two separate monomer tail domains. Additionally, the binding of two tail monomers would be cooperative and therefore weak at low concentrations of monomer if both tail peptides had to bind simultaneously for full interaction as in a 4-helix bundle.
Scheme 1.
Possible arrangements of head and tail domains in complexes of different stoichiometry. See Fig. 1 for color scheme and location of peptides in kinesin. Interactions are shown for illustration with the specific cases of NF910-952 as monomeric tail region (curved black line); SC899-952 as a dimer of tail regions (SC domain as red box); monomeric and dimeric head domains as blue circles. The stoichiometries of tail (T) and head (H) peptides in the complexes are indicated.
We report now, however, that monomer tail peptides bind to dimers of head domains with approximately the same high affinity as dimers of tail domains. The binding occurs without indication of cooperative interacts between tail monomers and results in greater than 50% inhibition. Furthermore, titration experiments indicate that the stoichiometry of binding is one tail peptide per dimer of heads with the second tail peptide of a dimeric tail construct adding little additional binding interaction. Thus binding of the first tail peptide in step 1 of Scheme 1B is much tighter than binding of the second tail peptide (half-site negatively cooperativity) and binding of the first peptide is able to inhibit the activity of both heads. This half-site binding of one tail peptide to a head dimer is sufficient to produce the same degree of inhibition as observed in full length folded kinesin heavy chains indicating that half-site binding also occurs in native folded kinesin and that the other tail peptide could be available to bind to other proteins. The kinetics and structural requirements of this half-site binding are reported here.
MATERIALS AND METHODS
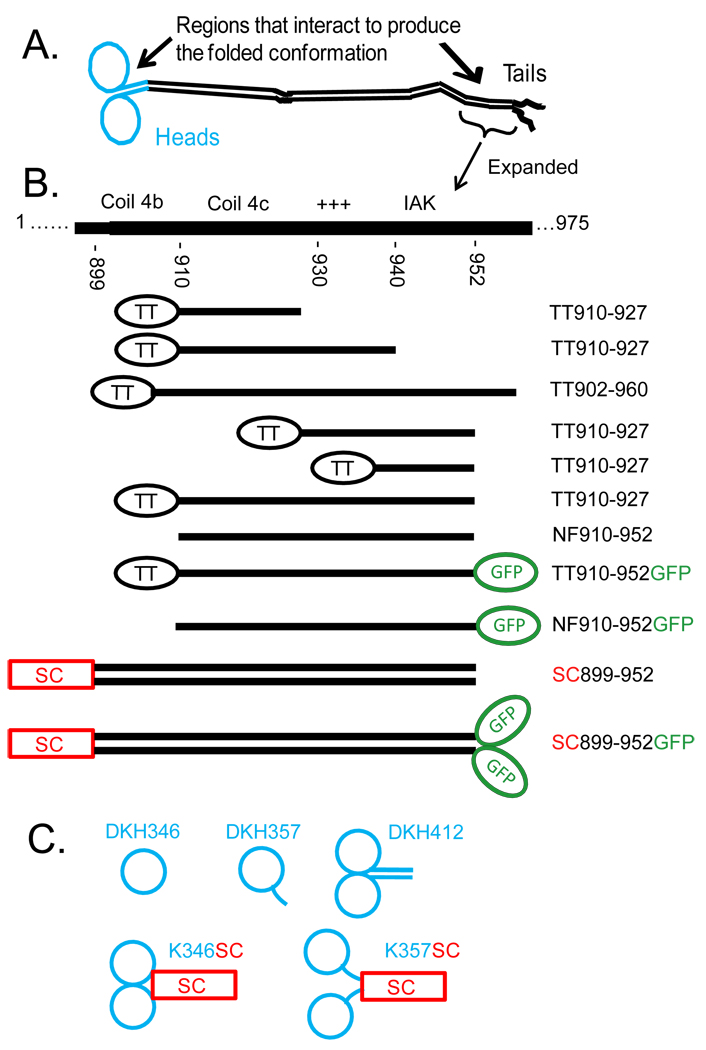
Head constructs DKH346, DKH357 and DKH412 contain residues 1 to 346, 357 and 412 respectively of Drosophila kinesin as previously described (6). DKH412 also contains a HisTag appended to the C-terminal. K346SC is an artificial dimer obtained by fusing DKH346 to the stable coiled coil peptide CGN4-p1 (10) and has TSGG linking the C-terminus of DKH346 to the initial Arg of GCN4-p1. Kinesin tail peptides are indicated by their beginning and ending residue numbers. The prefix TT indicates that they are fused to thioredoxin by a linker that contains a HisTag and a thrombin cleavage site. All tail constructs except ones terminating at residue 960 have a ThrSer appended to the C-terminus. Non-fusion versions were obtained by cleavage with thrombin and are designated by the prefix NF and have a GlySer appended to the N-terminus. mGFP (eGFP containing the A206K mutation to make it monomeric (11)) was fused to the C-terminus where indicated. SC899-952 has the indicated kinesin sequence fused to the stable coiled coil of GCN4 as previously described (6). All constructs were confirmed by DNA sequencing. The tail peptides were purified on NTA columns with elution by imidazole and by ion exchange chromatography on phosphocellulose. Protein concentrations were determined by absorbance at 280 nm in 6 M guanidine-HCl using the extinction coefficients of (12) and a value of 2310 M−1 cm−1 for the ADP bound at the active site of head constructs, except for constructs containing eGFP for which a molar extinction coefficient of 39,000 M−1cm−1 at 490 nm was used (11). Further characterization of the purity and concentrations of the preparations are given in the Supporting Information.
All reactions were conducted at 25 °C in A25 buffer (25 mM ACES/KOH, pH 6.9, 2 mM magnesium acetate, 2 mM potassium EGTA, 0.1 mM potassium EDTA and 1 mM 2-mercaptoethanol) supplemented with KCl where indicated. DKH412•mADP was prepared by equilibration of DKH412•ADP with an excess of mATP for 30 minutes at room temperature. The final ratio of mADP:ADP after hydrolysis and equilibration was 4:1. Release of bound mADP was monitored by FRET from tyrosine to mADP using excitation at 285 nm and a 320 nm long pass emission filter in experiments without GFP, essentially as previously described (6). In the presence of GFP, mant fluorescence was monitored with a mant-selective 417–477 nm band pass filter. Binding of GFP-labeled tail domains to DKH412•mADP was monitored by FRET from mADP to GFP using excitation at 350 nm and a 502–538 nm band pass emission filter. Error bars indicate the standard deviation. ATPase rates were determined from the change in absorbance of NADH at 340 nm when coupled to pyruvate kinase and lactate dehydrogenase as previously described (13). The reaction contained A25 buffer with 2 mM PEP, 0.25 mM NADH, 1 µM ATP and KCl as indicated. In the absence of MTs, this low 1 µM concentration of ATP is sufficient to saturate the basal ATPase of kinesin that has a KM for ATP of ~2.5 nM based on net ATP binding and hydrolysis at ≥4 µM−1s−1 and release as ADP at ~ 0.01 s−1 (6). Use of a low ATP concentration minimizes the contribution of contaminating ATPases that are present in trace amounts in some preparations and have KM values for ATP that are much higher than 1 µM. In some cases a correction of 1.6 times the absorbance change at 420 nm was applied to correct for changes in turbidity. All theoretical fits for mADP release and ATPase measurements were fit to the full quadratic equation for mutual depletion (14).
Sedimentation and diffusion coefficients were determined by sucrose gradient velocity centrifugation and gel filtration essentially as previously described (15) at 4° C in 1000 mM NaCl in A25 buffer supplemented with 0.1 mM ATP when head domains were present. Cosedimentation was performed similarly by sucrose gradient velocity centrifugation in A25 buffer with 25 mM KCl and 0.1 mM ATP.
RESULTS
Isolated tail peptides are monomeric
Fig. 1 gives the sequence of the tail region of Drosophila kinesin heavy chain and the location of the peptides used here. The extreme C-terminal region (953–975) is not required for folding or inhibition of ADP release (6). Coil 4a,b that terminates at approximately residue 910 is the last strongly predicted coiled region (see (16) for a more detailed description of the domains in kinesin). Residues 910–930 (possible Coil 4c) are weakly predicted to be coiled coil. The region 929–938 is highly positively charged with 3 Arg, 1 Lys and 1 His residues and no negatively charged residues. The tail domain binds strongly to MTs and this positively charged region is required (3;7). The following region is highly conserved in kinesin-1s and is designated the IAK region. It has been shown to be necessary for strong inhibition of basal ADP release (6). SC899-952 was previously studied as a dimeric model for the tail region (7). It contains the stable coiled coil of GCN4-p1 (10) fused in the same heptad frame to residue 899 in Coil 4b and extends past the IAK region. Other regions of the tail were expressed as indicated. They were initially purified as fusions with thioredoxin (designated by the prefix TT) and several have been previously described (7). In some cases they were cleaved with thrombin to liberate the tail peptide as a non fusion version designated by the prefix NF. Hydrodynamic analysis in 1000 mM NaCl summarized in Table 1 indicates that TT902-960 and TT910-952-mGFP are monomeric. Thus potential Coil 4c even with part of Coil 4b, in TT902-960 is not sufficient to locally sustain dimerization. As expected from inclusion of the stable GCN4 coiled coil, SC899-952 and SC899-952-mGFP are both dimeric. Essentially identical sedimentations coefficients were obtained in 25 mM KCl (see Fig. 4) and the oligomeric assignments of Table 1 are thus also valid at lower salt concentration.
Figure 1.
Schematic representation of peptides used in this study. A) Domain arrangement of a full length kinesin heavy chain dimer in an extended conformation. Heads and neck coil in blue with tail region in black (see (16) for a more detailed description). B) Expanded view of the region in the tail that interacts with the heads to produce the folded conformation with indication of the location of the tail constructs. The C-terminal region of kinesin heavy chain contains a region of strongly predicted coiled coil (Coil 4A,B) that is followed by a region that is only weakly predicted to be coiled coil (Coil 4C) and then the remaining C-terminal residues that are not predicted to be coiled coil and are likely unstructured. TT indicates thioredoxin linked to the tail peptide by a linker containing a HisTag and a thrombin cleavage site. NF indicates tail peptides without thioredoxin that have been generated by cleavage with thrombin. SC indicates fusion to the stable coiled coil of GCN4 (10) to force dimerization. C) Head constructs. DKH346 is a complete monomer with full neck linker; DKH357 has part of the neck coil, but not enough to cause dimerization; DKH412 has the full neck coil plus part of the hinge and is dimeric; K346SC and K357SC are DKH346 and DKH357 respectively fused to the stable coiled coil of GCN4 to force dimerization.
Table 1.
Hydrodynamic Characterization
| Construct | s20,wa | D20,wa | Molecular mass (kDa) | Oligomeric | |
|---|---|---|---|---|---|
| (S) | (10−7 cm2/s) | Svedbergb | Calculatedc | State | |
| TT910-952GFP | 3.44 | 7.40 | 41.9 | 46.3 | monomer |
| TT902-960 | 1.96 | 9.07 | 19.5 | 20.6 | monomer |
| SC899-952 | 2.18 | 7.97 | 24.7 | 11.5 | dimer |
| SC899-952GFP | 4.16 | 5.26 | 71.4 | 38.6 | dimer |
Determined in A25 buffer with 1000 mM NaCl. Measurements performed in duplicate or greater with standard deviations < 4%.
Calculated by the Svedberg Equation using a partial volume of 0.73 cm3/g.
Calculated from the amino acid composition.
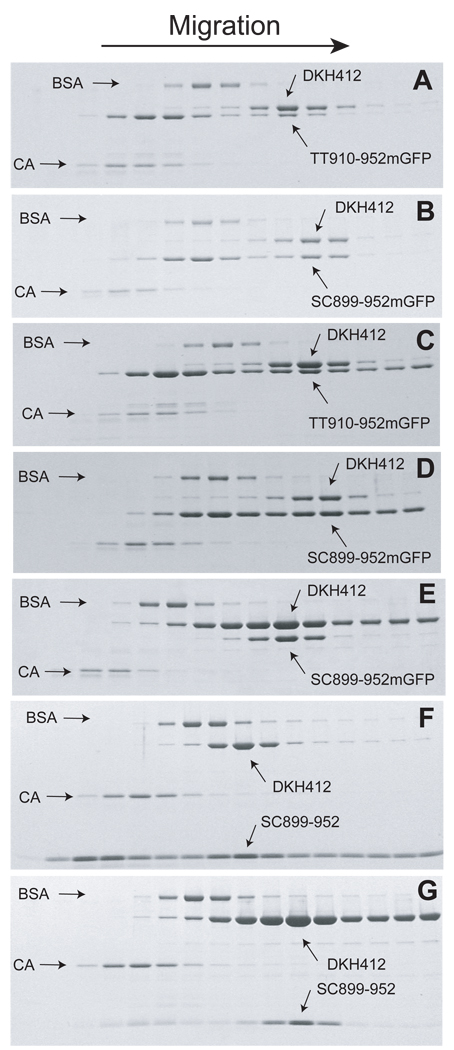
Figure 4.
Cosedimentation of head and tail domains. Bovine serum albumin (BSA) and carbonic anhydrase (CA) were included as internal standards. Following centrifugation, the gradient was fractionated into 15–16 fractions and analyzed by SDS-PAGE. The gel images have been compressed vertically for compact presentation. A. Initial concentrations of 10 and 15 µM for DKH412 and TT910-952mGFP respectively. B. Initial concentrations of 10 and 15 µM DKH412 and SC899-952mGFP respectively. C. Initial concentrations of 30 µM for DKH412 and TT910-952mGFP with 1 µM TT910-952mGFP throughout gradient. D. Initial concentrations of 10 and 15 µM DKH412 and SC899-952mGFP respectively with 1 µM SC899-952mGFP throughout gradient. E. Initial concentrations of 30 and 10 µM DKH412 and SC899-952mGFP with 2 µM SC899-952mGFP throughout gradient. F. Initial concentrations of 30 and 12 µM DKH412 and SC899-952 with 2 µM DKH412 throughout gradient. G. Initial concentrations of 10 and 30 µM DKH412 and SC899-952 with 2 µM SC899-952mGFP throughout gradient.
Inhibition of head domains by monomeric tail domains
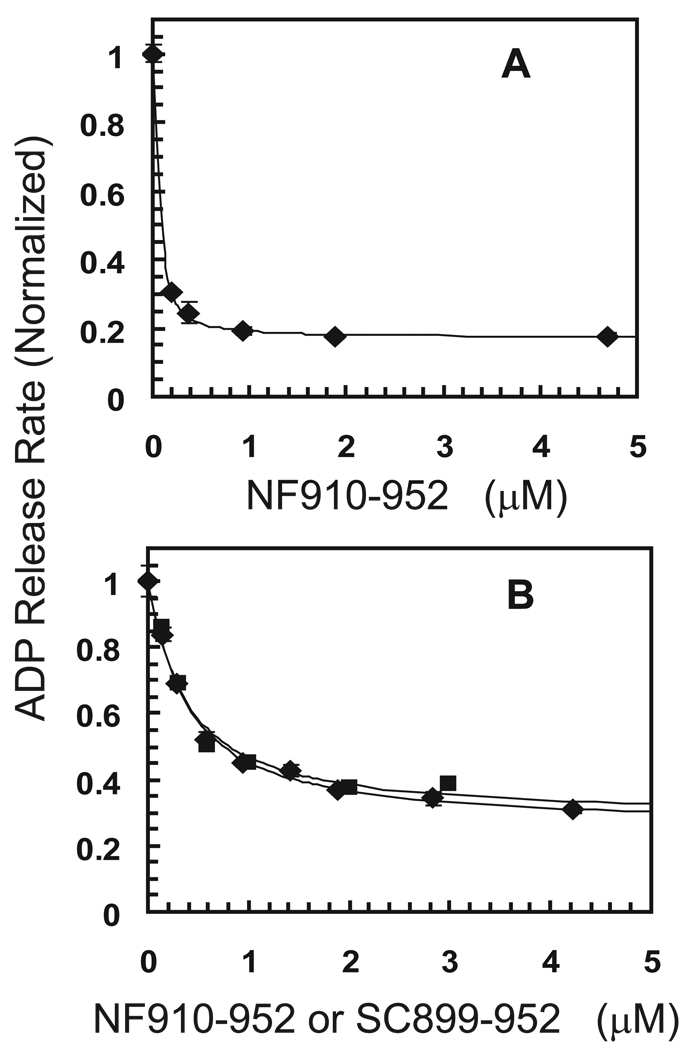
As indicated in Fig. 2A, monomeric tail peptide NF910-952 is a potent inhibitor of mADP release from dimeric DKH412 with tight binding and over 80% maximum inhibition in the absence of added KCl. The apparent Kd obtained from the fit is 0.021 µM, but is not highly accurate because the concentration of DKH412 was 0.1 µM (as dimer) and calculation of the Kd required a large correction for mutual depletion. This tight affinity and high maximum inhibition by monomeric NF910-952 is essentially identical to that observed previously with dimeric SC899-952 (6). The binding of tail domains in 50 mM KCl (Fig. 2B) is weaker and the concentration dependence of the inhibition by monomeric NF910-952 is consistent with simple noncooperative binding with a Kd of 0.33 µM and a maximum extent of inhibition of 75%. Binding of dimeric SC899-952 in 50 KCl (Fig. 2B) is also approximately noncooperative with a Kd of 0.16 µM (per SC899-952 dimer). Thus pre-dimerization of the tail domains is not required for tight binding to a dimer of heads and inhibition of ADP release.
Figure 2.
Inhibition of mADP release by tail domains. A. DKH412•mADP was mixed in the stopped flow in the absence of added KCl with excess unlabeled ADP and monomeric tail construct NF910-952 and the normalized rate of initial ADP release was determined as previously described (6). Final concentrations were 0.2 µM DKH412, 200 µM ADP and NF910-952 as indicated. See text for description of theoretical lines. B. Inhibition of mADP release in 50 mM KCl with 0.2 µM DKH412 and NF910-952 (◊) or SC899-952 (□). All concentrations as monomers.
Stoichiometry with monomeric tails
Inhibition studies at high concentration of heads and tails allow the apparent stoichiometry of the interaction to be determined as indicated in Fig. 3A. In this experiment, the concentration of DKH412 was fixed at 7 µM monomer peptide (3.5 µM DKH412 dimer) as indicated by the vertical lines at 3.5 and 7 µM. The steady state ATPase rate was determined over a range of concentrations of monomeric NF910-952. Because ADP release is the rate limiting step in basal ATP hydrolysis (17), tails are expected to inhibit both ADP release and steady state ATPase similarly. The upper theoretical line is the linear titration that would result if NF910-952 had infinitely tight binding to DKH412 to form a 2T:2H complex with an equivalence point at 7 µM NF910-952 for fixed 7 µM DKH412 (monomer concentration). The lower theoretical line is for linear titration to form the 1T:2H complex with an equivalence point at 3.5 µM (the concentration of DKH412 dimers). Strikingly, maximum inhibition occurs at the stoichiometry of 1 tail peptide per two head peptides. The theoretical line through the points for DKH412 in Fig. 3A is the best fit for an assumed stoichiometry of 3.5 µM binding sites and yields a maximum extent of inhibition of 86% and a Kd of 0.023 µM. Although this estimate of Kd is uncertain due to a large correction for mutual depletion, it confirms the tight binding seen in Fig. 2A by inhibition of ADP release in the absence of added KCl and is consistent with a Kd < 0.1 µM. The protein concentrations were determined by absorbance at 280 nm in 6 M guanidine hydrochloride (12). This is likely to be accurate for DKH412 with two tryptophan and twelve tyrosine residues, but less accurate for NF910-952 because it contains only two tyrosine and no tryptophan residues. However, essentially identical titrations with tight binding and half-site stoichiometry were also observed with monomeric TT927-952 as shown below (fig. 3b) and with TT902-960 and TT910-960 (not shown). These constructs are fused to thioredoxin which contains an additional two tryptophan and two tyrosine residues and thus have large extinction coefficients at 280 nm. Furthermore, highly absorbing contaminants if present in NF910-952 cannot be the cause of halfsite stoichiometry because they would cause overestimation of the equivalence point, not the reduction to half-site that is observed.
Figure 3.
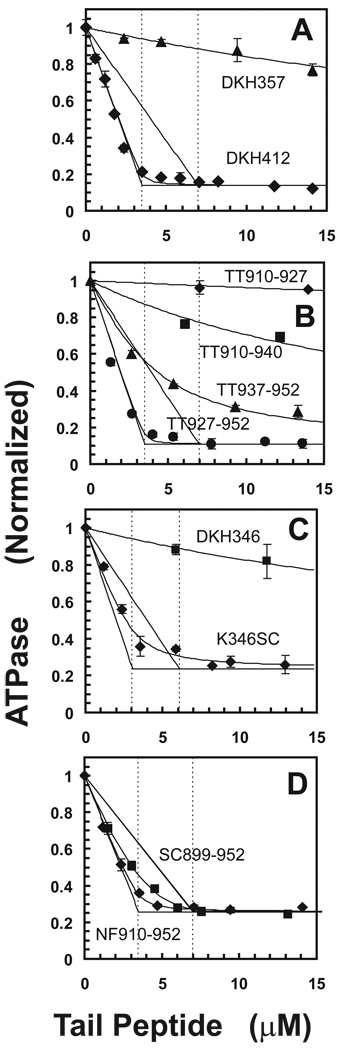
Inhibition of basal ATPase rate by tail domains. A. Influence of NF910-952 on normalized ATPase rate of 7 µM DKH412 monomer (◊) or 13.3 µM DKH357 (△). Vertical lines are at 3.5 and 7 µM for the concentration of DKH412 dimer and monomer respectively. See text for description of theoretical lines. B. Inhibition of 7 µM DKH412 monomer by TT910-927 (◊), TT910-940 (□), TT937-952 (△) and TT927-952 (○). C. Inhibition of 6.8 µM DKH346 (□) or 6 µM K346SC monomer (◊) by NF910-952. D. Inhibition of 7 µM DKH412 by NF910-952 (◊) or SC899-952 (□) in 25 mM KCl.
Monomeric NF910-952, however, only weakly inhibits monomeric DKH357 head domains even at high concentration as also indicated in Fig. 3A. This is consistent with the previously observed failure of tail domains to bind tightly to monomeric head domains (2). Monomeric DKH357 does not show sufficient enough inhibition to allow meaningful calculation of the best fit for both the Kd and the maximum extent of inhibition, but a Kd of 39 µM was determined with assumption of the same 86% maximum extent of inhibition observed with DKH412. In contrast, the Kd value with dimeric DKH412 above is less than 0.1 µM or over 400 fold tighter. This weak binding to monomeric DKH357 may in part represent binding to form a weak 1T:1H complex with DKH357 as in the first step of Scheme 2D or may be the initial lag phase of cooperative binding (combined K1M and K2M) to form a tight 1T:2H complex that would be generated more easily at higher concentrations of DKH357. Dietrich et al. (4) also observed cross-linking of a tail peptide to both monomer and dimer tail domains, but could not establish from these qualitative results that the affinity of the tail peptide for the monomer head domains was orders of magnitude weaker than for the dimer head domains as shown here.
Localization of the site required for tight binding
Previous work localized the general region in the tail that bound to the heads (2) and more recent studies have refined it further (3;4;6). The different regions of NF910-952 were tested separately for their ability to inhibit DKH412 as indicated in Fig. 3B. TT927-952 contains only the positively charged region and the IAK domain, yet still binds tightly with half-site inhibition. TT937-952 with only the IAK domain still inhibits, but its affinity for DKH412 is reduced with an apparent Kd of ~2 µM versus a Kd < 0.1 µM for TT927-952. As seen previously with SC899-940 (6), some inhibition is still observed on removal of most of the IAK region (TT910-940), but with greatly weakened affinity. Removal of both the IAK and positively charged region (TT910-927) abolishes inhibition. Thus the combined IAK region and adjoining positively charged region is sufficient for tight binding and inhibition with half-site stoichiometry.
Half-site inhibition of an artificial dimer of head domains
If the reason for the weak binding of monomeric DKH357 to tail domains was that tails only bound tightly to dimers of heads, then artificially joining two head domains should provide a tight binding site for monomeric tail domains. To test this, the C-terminus of monomeric DKH357 was fused to the stable coiled coil of GCN4 to produce an artificial dimer of head domains. Although monomeric DKH357 was not strongly inhibited, the artificial dimer (K357SC) was inhibited by NF910-952 with half-site stoichiometry in a manner essentially identically to that observed in Fig. 3A with natively dimeric DKH412 (not shown). However, DKH357 still contains the first part of the neck coil. Even though this partial neck coil is not sufficient to sustain dimerization of the heads on its own, it may still have been needed to provide part of the binding site for the tail peptide. The shorter DKH346 contains only the core motor region and the neck linker in the complete absence of the neck coil. This construct has previously been shown to be a good model for an intact monomeric head because it is short enough to have no detectable tendency to dimerize, but long enough to avoid the abnormal kinetics observed with more severe truncations such as DKH340 (9). As indicated in Fig. 3C, NF910-952 is a potent inhibitor of an artificial DKH346 dimer of K346 (K346SC) with half-site stoichiometry, whereas DKH346 itself is only poorly inhibited. Thus the neck coil is not required for tight binding and half-site inhibition, although the Kd value of 0.33 µM obtained from the fit in Fig. 3C does indicate that binding is not fully as strong as with a native neck coil. It should be noted, however, that K346SC has a flexible spacer with sequence TSGG between the head and the GCN4 coiled coil and this likely entropically disfavors bringing the two head domains together into fixed relative orientation for half-site binding of the tail peptide. Observation of half-site binding with K346SC in spite of the flexible spacer also indicates that only dimerization itself is required for tight half-site binding of tail domains, without requirement for a specific conformation of the heads that might be produced by constrained attachment to the native neck coil.
Stoichiometry with dimeric tails
Titration of a high concentration of DKH412 in the presence of 25 mM KCl with dimeric SC899-952 is indicated in Fig. 3D. Data for monomeric NF910-952 in 25 mM KCl is included for comparison and shows similar half-site stoichiometry as seen in the absence of added KCl. The upper theoretical line is the linear titration that would result if SC899-952 had infinitely tight binding to DKH412 to form the 2T:2H complex with an equivalence point at 7 µM SC899-952 for fixed 7 µM DKH412 as monomers (3.5 µM each as dimers). The lower theoretical line is for linear titration to form the 2T:4H complex with an equivalence point at 3.5 µM SC899-952 for 7 µM DKH412 as monomers (1.75 µM SC899-952 and 3.5 µM DKH412 as dimers). This 2T:4H complex could form if each of the two tail domains of dimeric SC899-952 was able to bind a DKH412 dimer as indicated in Scheme 1C. The observed data with SC899-952 is intermediate between these limits indicating that SC899-952 can bind to DKH412 with less than 1:1 stoichiometry as occurs in the 2T:4H complex. The cosedimentation results below also indicate that the 2T:4H complex should be significantly populated over this concentration range. The theoretical line for inhibition by SC899-952 was fit to the two step binding of Scheme 1C assuming the same maximum extent of inhibition for the 2T:2H and 2T:4H complexes as seen for the 1T:2H complex of NF910-952 and a K1D value equal to that for NF910-952 in 25 mM KCl from the fit for NF910-952 in Fig. 3D (0.066 µM per monomer NF910-952 or 0.033 µM per SC899-952 dimer). Letting only K2D vary, the best fit was obtained at a K2D value of 1 µM. The fit is not highly sensitive to the exact K1D and K2D values, but requires that K1D be tight like the monomer and that K2D be weaker than K1D, but still significant. Essentially identical titrations were obtained with DKH412 and monomeric TT910-952mGFP and dimeric SC899-952-mGFP (not shown) with 66% maximal inhibition and K1D and K2D values of 0.025 and 1.8 µM respectively.
Cosedimentation of head and tail domains
The stoichiometry of the complexes between mono and dimer tail domains and DKH412 was investigated by cosedimentation during sucrose gradient velocity sedimentation as indicated in Fig. 4. The buffer was A25 with 25 mM KCl and 0.1 mM ATP in all cases. When centrifuged alone, DKH412, TT910-952mGFP and SC899-952mGFP have s20,w value of 5.2, 3.2 and 4.3 S respectively under these conditions and migrate slightly ahead of or behind bovine serum albumin. These species were chosen for analysis because they are similar in peptide mass and thus the relative stoichiometries can be more readily estimated from the staining ratios following SDS-PAGE analysis of the gradient fractions. When DKH412 and TT910-952mGFP are loaded together on the gradient in panel A at 10 and 15 µM respectively (all concentrations as monomers so at 5 µM DKH412 dimer and 15 µM TT910-952mGFP), DKH412 now migrates faster as a complex with TT910-952mGFP at approximately 6.0 S. The distribution of TT910-952mGFP is bimodal with most continuing to migrate at the position for free TT910-952mGFP, but with a second peak comigrating with DKH412. In spite of the initial excess of TT910-952mGFP over DKH412, the stoichiometry in the complex is < 0.5 TT910-952mGFP per DKH412 monomer, consistent with a 1T:2H complex. The results in pane B for centrifugation of 10 µM DKH412 with 15 µM SC899-952mGFP is similar except that the complex migrates even faster at 6.7 S and the stoichiometry is close to 1:1 as expected for a 2T:2H complex when dimeric tails are in excess (the molar mass of SC899-952mGFP is 82% of DKH412 and so a 1.0:0.82 staining ratio is expected for a 2T:2H complex as is approximately observed). Note that the association/dissociation of tails and heads is fast (see stopped flow results below) and thus the species are in rapid equilibrium during centrifugation.
Although binding is tight in 25 mM KCl, some trailing of tail domains behind the complex is expected during migration and this would lower the observed ratio of tail:head domains in the complex. To minimize net dissociation, centrifugation in panel C was performed with a higher initial concentration of 30 µM for both TT910-952mGFP and DKH412 and with a uniform concentration of 1 µM TT910-952mGFP throughout the starting sucrose gradient. Thus the complex was always migrating through a region with at least 1 µM free TT910-952mGFP and the complex should stay largely saturated. The observed s20,w value for the complex in Panel C is unchanged at 6 S and the stoichiometry is still approximately 1T:2H. Inclusion of 1 µM SC899-952mGFP throughout the initial gradient (Panel D) also did not result in a change in s20,w value for its complex or change in the stoichiometry from approximately 2T:2H.
In contrast to the above cosedimentation as the 2T:2H complex when dimeric tails are in excess over dimeric heads, the 2T:4H complex forms if dimeric heads are in excess over dimeric tails. When SC899-952mGFP and DKH412 are cosedimentated at 10 and 30 µM initial concentrations respectively in a gradient containing a uniform level of 2 µM DKH412 throughout (panel E), all of the SC899-952mGFP comigrates with a peak of DKH412 at a larger s20,w value for 7.5 S and with lower stoichiometry, consistent with at least partial conversion to a 2T:4H complex. Because the K2D for binding a second head dimer of 1–2 µM (Fig. 3D) is not much less than the free DKH412 concentration of 2 µM, complete conversion to the 2T:4H complex would not be expected. Exact calculation is complicated because the concentrations in the peak for the complex changes during migration due to radial dilution and spreading by diffusion. For illustration of the expected range, if the concentrations in the peak are approximated as 3 and 8 µM for SC899-952mGFP and DKH412 respectively, then the K1D and K2D values of 0.025 and 1.8 µM estimated above for SC899-952mGFP result in relative stoichiometries at equilibrium of 0.37:0.63 for the 2T:2H and 2T:4H complexes respectively. In panels F and G, SC899-952 without GFP was centrifuged with excess SC899-952 or excess DKH412 respectively. In this case, the lower mass of SC899-952 relative to SC899-952mGFP produces a smaller change in s20,w from 5.2 for free DKH412 to 5.5 and 6.3 S in panels F and G for excess SC899-952 or excess DKH412 respectively. Again, an excess of DKH412 shifts the population to a larger s20,w value as expected for an increase in the relative amount of the more rapidly sedimenting 2T:4H complex. The exact s20,w values expected for the different complexes are not known because the frictional coefficients would be highly dependent on how the components of the complexes were arranged. For example, the mass of the 2T:4H is 1.8-fold greater than that of the 2T:2H complex with SC899-952, yet the observed s20,w value only changes by 1.145-fold. This is in part due to incomplete conversion to the 2T:4H complex under these conditions, but another factor is that the tail region is likely to be unfolded and flexible. Thus the two DKH412 dimers in a 2T:4H complex might be separated enough to be partially hydrodynamically independent. If fully independent in the freely draining limit, the second DKH412 dimer would increase both the mass and frictional coefficient by the same amount with no net change in s20,w value (18).
Kinetics of binding of monomeric tail domains to dimeric head domains
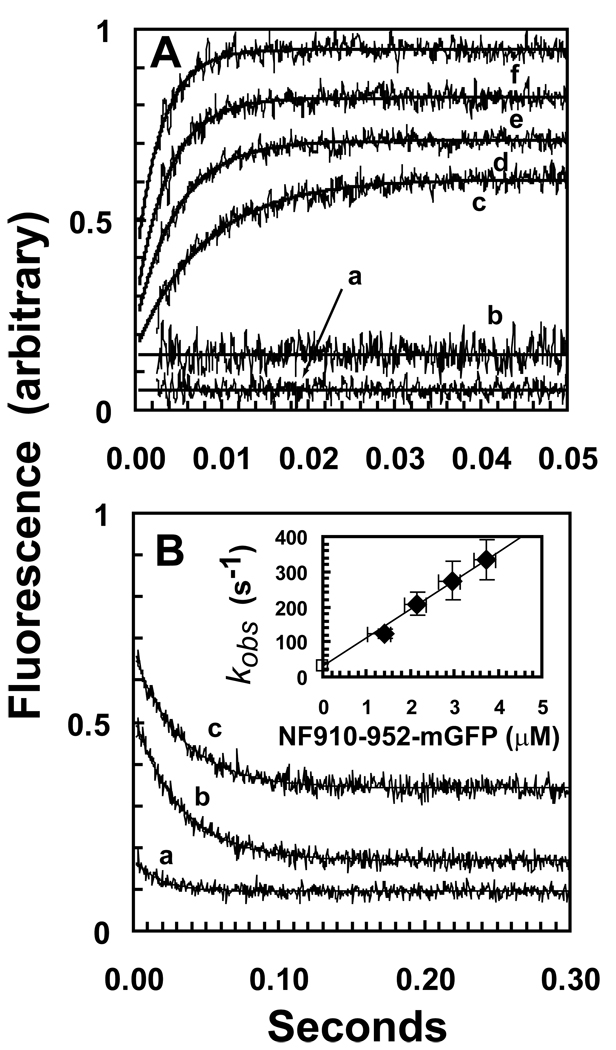
GFP and the mant group of mADP are a potential FRET pair because mADP fluoresces with a maximum at approximately 445 nm when excited at 350, whereas GFP is absorbs maximally at 488. Binding of tails labeled with GFP to the complex of DKH412 with bound mADP should produce an increase in GFP fluorescence when excited at 350 nm if GFP and mADP are close enough in the complex for FRET to occur. Fig. 5A indicates that such an increase is observed and this provides a means to determine the kinetics of binding. At a fixed DKH412 concentration of 0.5 µM dimer, the rate of the transient binding phase increases with the concentration of monomeric NF910-952mGFP (traces c-f) in 50 mM KCl. The amplitudes show only a modest increase with increasing concentration because a Kd of ~0.33 µM (Fig. 2A) produces 75% saturation at even the lowest concentration of 1.4 µM NF910-952mGFP in Fig. 6A (with allowance for mutual depletion). Neither mixing NF910-952 that is not GFP-tagged with DKH412•mADP (trace b) nor mixing DKH412•ADP (not with mADP) with NF910-952mGFP (trace a) produces a transient, even though binding is occurring in both cases. This supports FRET as the basis for the transient because the increase only occurs when a complex can form that contains both mADP and GFP. No significant change in the extent of mADP binding to DKH412 is expected following 2-fold dilution in the stopped flow because the high concentration of free (mADP+ADP) of ~5 µM after mixing is sufficient to maintain saturation given the low nM Kd value of kinesin head domains for ADP and even lower Kd for ADP of the complex with tail domains (6).
Figure 5.
Kinetics of binding between DKH412•XmADP and NF910-952mGFP monitored by increase in FRET from mADP to GFP. A. Binding kinetics. NF910-952mGFP was mixed with DKH412•mADP in the stopped flow and the transient increase in fluorescence was determined. DKH412•mADP was 1 µM and NF910-952mGFP concentrations were 1.6, 2.4, 3.2 and 4 µM for traces c-f respectively (all concentrations are for monomers after mixing). Each trace in the figure is the average of 5–10 individual transients and the smooth line is the fit to a single exponential. Trace a is 1 µM DKH412 (without mant) mixed with 2 µM NF910-952mGFP. Trace b is 1 µM DKH412•mADP mixed with 1 µM NF910-952 (without GFP). Vertical offsets have been applied to the traces to allow for compact presentation, but the amplitudes of the transients have not been adjusted and are directly comparable between panels A and B. B. Dissociation kinetics. The complex of DKH412•mADP and NF910-952mGFP (at a stoichiometry of 1T:2H) was mixed in the stopped flow with 0, 2.5 and 10 µM NF910-952 for traces a-b respectively. Final concentrations of DKH412•mADP and NF910-952mGFP were 1 µM and 0.5 µM respectively. (INSERT) The observed rate constants for transients c-f in panel A are plotted versus the concentration of NF910-952mGFP. The value of 32 s−1 for dissociation of the complex was used as the limiting value for kobs extrapolated to zero NF910-952mGFP concentration (open square). The slope of the line gives the rate constant for bimolecular binding of 81 µM−1s−1. The vertical error bars are ± the standard deviations of the individual transients that were averaged to give the traces in panel A. Experiments could not be performed under true pseudo first order conditions with a large excess of NF910-952mGFP over DKH412•mADP because the rate of the transient becomes too fast at higher NF910-952mGFP concentration and the DKH412•mADP concentration could not be reduced without losing amplitude. As an approximation, the plotted concentration is the average of the starting and final values (estimated assuming a Kd for binding of 0.4 µM and corrected for mutual depletion). The horizontal error bars are included for reference and indicate the limiting concentrations for zero and 100% binding (depletion of free NF910-952mGFP by 0.5 µM due to complete binding to the 0.5 µM DKH412•mADP dimer).
Dissociation rate constant
When a preformed DKH412•mADP•NF910-952mGFP complex is mixed with excess NF910-952, a decrease in fluorescence of the same amplitude is observed (Fig. 5B) as the GFP-labeled tail dissociates and is replaced by the excess non-GFP labeled tail with accompanying loss of FRET. Similar rate and amplitude values (30.9 and 32.8 s−1 and 0.35 and 0.34 for traces b and c respectively) are obtained with a 5 or 20-fold excess of unlabelled tail domain, indicating that the observed rate of ~32 s−1 represents the rate of initial dissociation of NF910-952mGFP from the complex. In the absence of NF910-952mGFP in the chase, a small drop in amplitude is also observed (trace a). The magnitude of this drop is consistent with a Kd of ~0.4 µM determined below and the 2-fold dilution of the complex on mixing in the stopped flow that will result in a net increase in the degree of dissociation.
Kinetically-determined Kd
The insert to Fig. 5B indicates that the kobs values for binding of NF910-952mGFP to DKH412 increase with increasing concentration of NF910-952mGFP to over 300 s−1 without indication of saturation. Using the dissociation rate of 32 s−1 from Fig. 5B as the limiting value for kobs in the absence of NF910-952mGFP, the data can be fit to a line with a slope of 81 µM−1s−1 for the second order rate constant for binding. The kinetically-determined Kd is koff /kon = 32 s−1 / 81 µM−1s−1 = 0.4 µM. This value in 50 mM KCl is in good agreement with the apparent Ki value of 0.33 µM from the concentration dependence of inhibition of mADP release by NF910-952 without GFP also determined in 50 mM KCl (Fig. 2B).
Detailed analysis of the FRET efficiency is complicated by the 4-fold excess of free mADP over mADP bound to DKH412, but the amplitude of the transient is small (the amplitude of the increase during the transient is only equal to the low level of fluorescence of GFP alone when excited at 350 nm). This suggests that GFP and mADP are not close to each other in the complex. An addition factor is that GFP is linked to Arg-948 of the IAK domain by a long tether of nine amino acids (SGQGATSGG) that is likely to be flexible. Even if the average position of the GFP in the complex was too far from the bound mADP for significant FRET, a low level of FRET could still be observed if some of the conformations of the linker brought the GFP close enough for FRET to occur, or if GFP had a weak binding interaction with the heads. Although FRET should produce a corresponding quenching of mant fluorescence, no decrease was observed with a mant-selective emission filter (not shown). This is likely due to the difficulty in detecting the small expected decrease against the large background signal from a 4-fold excess of free mADP over DKH412•mADP. The small increase in fluorescence at GFP emission wavelengths due to FRET is more readily detected because GFP has only low levels of fluorescence when excited at 350 nm in the absence of FRET. However, regardless of the detailed basis for the increase in fluorescence, the transient observed in Fig. 5 is still a useful monitor of binding kinetics.
DISCUSSION
The results presented here establish that only a single monomeric tail peptide containing the IAK region and the adjoining positively charged region is sufficient for inhibition of dimers of head domains via tight binding with half-site stoichiometry. The second tail domain in an SC899-952 dimer does not appear to make energetically significant interaction with the heads in a 2T:2H complex because the affinity of SC899-952 for head dimers is not greatly increased over that for monomeric tail species. Additionally, the maximum extent of inhibition is over 50% and similar for both the 1T:2H, 2T:2H and 2T:4H complexes indicating that inhibition in all cases arises predominately from interaction of one tail peptide with a dimer of heads and that this asymmetric interaction inhibits both heads without influence of a second tail domain if present. The maximum extent of inhibition of ADP release with folded DKH960 is also greater than 50% and both bound ADP molecules are released in a single first order process (6). This was not unexpected because both of the two tail peptides of DKH960 could in principle bind symmetrically to the two heads of DKH960 to inhibit both heads (Scheme 1A). If the interaction was symmetric, then the ADPs on both heads would be released at the same rate and only a single first order would be observed. The half-site interaction observed here provides an alternative mechanism that allows inhibition of both heads of a dimer by binding of one tail peptide that is either interacting with both heads or binding to one head with induction of a conformational change in the other head that both inhibit ADP release and prevents tight binding of a second tail peptide. Asymmetric interaction in a 1T:2H complex opens the possibility for release of ADP at different rates from the two heads. This is possible, but would not likely be observed in the net kinetics of ADP release because the association/dissociation kinetics for interaction of tails with heads is orders of magnitude faster than ADP release (tail dissociation at 32 s−1 versus inhibited ADP release at < 0.005 s−1). Each cycle of release and rebinding of tail domains provides an opportunity for switching of the asymmetry between heads that could average their properties over the much slower time scale ADP release. It is also possible that the two head domains in a dimer are interacting asymmetrically even in the absence of tail domains, but interconvert between conformations rapidly enough so that release of both ADP molecules is observed in a single first order process. An asymmetric arrangement is in fact observed in the X-ray structure of dimeric kinesin head domains (19).
While one tail domain of dimeric SC899-952 is tightly bound to a dimer of heads, the second tail domain has sufficient freedom for it to bind a second dimer of heads to form the 2T:4H complex, albeit with reduced affinity. A 2T:4H complex is not likely to be physiologically relevant because kinesin in the cell is either attached to cargo or already folded into the inhibited conformation through binding to its own tail region. Observation of a 2T:4H complex though does indicate that the second tail peptide in folded kinesin may not be tightly bound to the head domains and available for interaction with MTs or other interacting molecules such as Unc76 (6;20;21). In particular, the same region of the tail that binds to the heads also binds to MTs at low ionic strength and could drive binding of kinesin to MTs even while still folded if this tail peptide was exposed. However, folded kinesin has negligible affinity for MTs at all but very low ionic strength (7). This masking of the auxiliary MT binding site in the tail could occur if the sites for binding to the heads and binding to MTs were only partially overlapping so that the site for head binding is sufficiently exposed to allow formation of the 2T:4H complex, but the full site needed for MT binding is not as exposed or accessible. Also the steric requirements for binding to MTs may cause interference with some part of the larger full length folded kinesin.
One of the previously puzzling properties of folded DKH960 was that it had greatly increased MT binding at very low ionic strength (see data at 5 versus 50 mM KCl in Fig. 1D of (7)). If unfolding was required for exposure of the tail MT binding site, then MT binding should be even weaker at low ionic strength where the free energy of folding would be greater. However, the affinity of the tail for MTs will also be greatest at low ionic strength and might provide enough interaction energy to overcome the steric or other factors that produce masking and could allow binding to the MT through the tail peptide that is not tightly bound to the heads. The cryoEM reconstruction of a cross-lined head-tail complex (4) indicates that the tail domain can interact with both the head domain and the MT. However, this reconstruction was obtained with a mutant monomeric kinesin head domain that cannot hydrolyze ATP and has greatly elevated MT affinity compared to wild type. It is not known to what extent the altered conformation of the mutant heads in this 1T:1H complex and their forced binding to MTs will perturb the position and conformation of the tail domains in the complex. Wild type folded kinesin has negligible affinity for MTs under these conditions. It will be of interest to also determine the position of the free tail peptide in a 2T:2H complex‥
C-terminal truncations of the kinesin heavy chain at positions 945 or 937 disrupt the IAK region and this results in increased exposure of the auxiliary MT binding site in the tail region with greatly increased net MT affinity and super activated rates of MT-stimulated ATP hydrolysis (7). These truncations also have greatly increased single molecule processivity in motility assays without major inhibition of sliding rate2. The increased MT affinity of these truncations arises from the combined affinity of the heads and tails for MTs. The non-stereospecific electrostatic nature of the binding of the tail domain to MTs likely allows the tail domain to maintain contact with the MT without introducing drag on the moving head domains. Partial or complete unfolding of full length kinesin produced by binding of effectors or phosphorylation could also expose the MT binding site in the tail and result in increased net MT affinity and processivity.
Supplementary Material
Characterization of the protein preparations and Fig. S1. This material is available free of charge via the Internet at http://pubs.acs.org”.
Acknowledgements
We thank Peter Kim for the GCN4-p1 plasmid; Adam Trexler for participation in initial experiments with GFP-labeled tail domains; Chun-Wei Cheh for assistance with ultracentrifucation; and Jessica Woessner for participation in cloning and protein purification.
The abbreviations used are
- MT
microtubule
- mant
N-methylanthraniloyl
- mADP
2’(3’)-O-(N-methylanthraniloyl)adenosine 5’-diphosphate
- GFP
green fluorescent protein
- FRET
Forster resonance energy transfer.
Footnotes
This work was supported in part by National Institutes of Health Grant NS25980 and National Science Foundation Grant 0615549.
Hackney, D.D. 45th Annual American Society for Cell Biology meeting, 2005, abstract 2387.
References
- 1.Adio S, Reth J, Bathe F, Woehlke G. Review: regulation mechanisms of Kinesin-1. J.Muscle Res.Cell Motil. 2006;27:153–160. doi: 10.1007/s10974-005-9054-1. [DOI] [PubMed] [Google Scholar]
- 2.Stock MF, Guerrero J, Cobb B, Eggers CT, Huang T-G, Li X, Hackney DD. Formation of the compact conformer of kinesin requires a C-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J.Biol.Chem. 1999;274:14617–14623. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- 3.Yonekura H, Nomura A, Ozawa H, Tatsu Y, Yumoto N, Uyeda TQ. Mechanism of tail-mediated inhibition of kinesin activities studied using synthetic peptides. Biochem.Biophys.Res.Commun. 2006;343:420–427. doi: 10.1016/j.bbrc.2006.02.169. [DOI] [PubMed] [Google Scholar]
- 4.Dietrich KA, Sindelar CV, Brewer PD, Downing KH, Cremo CR, Rice SE. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc.Natl.Acad.Sci.U.S.A. 2008;105:8938–8943. doi: 10.1073/pnas.0803575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai D, Hoppe AD, Swanson JA, Verhey KJ. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J.Cell Biol. 2007;176:51–63. doi: 10.1083/jcb.200605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackney DD, Stock MF. Kinesin tail domains and Mg2+ directly inhibit release of ADP from head domains in the absence of microtubules. Biochemistry. 2008;47:7770–7778. doi: 10.1021/bi8006687. [DOI] [PubMed] [Google Scholar]
- 7.Hackney DD, Stock MF. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat.Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 8.Verhey KJ, Lizotte DL, Abramson T, Barenboim L, Schnapp BJ, Rapoport TA. Light chain-dependent regulation of Kinesin's interaction with microtubules. J.Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Stock M, Li X, Hackney DD. Influence of the kinesin neck domain on dimerization and ATPase kinetics. J.Biol.Chem. 1997;272:7626–7632. doi: 10.1074/jbc.272.12.7626. [DOI] [PubMed] [Google Scholar]
- 10.Lumb KJ, Carr CM, Kim PS. Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry. 1994;33:7361–7367. doi: 10.1021/bi00189a042. [DOI] [PubMed] [Google Scholar]
- 11.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 12.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal.Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 13.Stock MF, Hackney DD. Assays for kinesin microtubule-stimulated ATPase activity. In: Vernos I, editor. Methods in Molecular Biology-Kinesin Protocols. Totowa: Humana Press; 2001. pp. 65–71. [PubMed] [Google Scholar]
- 14.Griffiths JR. Steady-state enzyme kinetics in mutual depletion systems. Biochem.Soc.Trans. 1979;7:429–439. doi: 10.1042/bst0070429. [DOI] [PubMed] [Google Scholar]
- 15.Stock MF, Chu J, Hackney DD. The kinesin family member BimC contains a second microtubule binding region attached to the N-terminus of the motor domain. J.Biol.Chem. 2003;278:52315–52322. doi: 10.1074/jbc.M309419200. [DOI] [PubMed] [Google Scholar]
- 16.Hackney DD. Jump-starting kinesin. J.Cell Biol. 2007;176:7–9. doi: 10.1083/jcb.200611082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proc.Natl.Acad.Sci.U.S.A. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantor CR, Schimmel PR. Biophysical Chemistry, Part II. New York: W.H. Freeman; 1980. pp. 569–570. [Google Scholar]
- 19.Kozielski F, Sack S, Marx A, Thormahlen M, Schonbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 20.Gindhart JG, Chen J, Faulkner M, Gandhi R, Doerner K, Wisniewski T, Nandlestadt A. The kinesin-associated protein UNC-76 is required for axonal transport in the Drosophila nervous system. Mol.Biol.Cell. 2003;14:3356–3365. doi: 10.1091/mbc.E02-12-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J.Cell Biol. 2007;176:11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the protein preparations and Fig. S1. This material is available free of charge via the Internet at http://pubs.acs.org”.