Abstract
After 12 years from its first application, microarray technology has become the reference technique to monitor gene expression of thousands of genes in the same experiment. In the past few years an increasing amount of evidence showed the importance of non coding RNA (ncRNA) in different human diseases. The microRNAs (miRNAs) are one of the groups of ncRNA. They are small RNA fragments, 19–25 nucleotides long, with a main regulatory function on both protein coding genes and non-coding RNAs. The application of microarray platforms applied to miRNA profiling determined their deregulation in virtually all human diseases that have been studied. We previously developed a custom miRNA microarray platform, and here we describe the protocol we used to work with it including the oligo design strategy, the microaray printing protocol, the target-probe hybridization and the signal detection.
1. Protein-coding genes versus non-coding RNA
Although 5% of the human genome evolves under purifying selection, only 2% of this set is represented by protein coding RNA (i.e. protein coding genes) (1). The remaining 3% is split into non-genic DNA and non-coding RNA (2, 3). The main differences between protein coding and non-coding RNA (mRNA and ncRNA respectively) is based on the absence of any Open Reading Frame (ORF) for ncRNA and on the difficulty to predict the transcript of ncRNA from its genomic structure (4). The ncRNA family is heterogeneous, based on the different size of the product (from few nucleotides to thousands) and on its function. NcRNA can have a generic function such as ribosomal RNA (rRNA) and transfer RNA (tRNA), both involved in mRNA/protein translation, or small nuclear RNA (snRNA) involved in splicing, and finally small nucleolar RNA (snoRNA) involved in the modification of rRNA. The last members of the ncRNA to be discovered are involved in post-transcriptionally regulating protein expression through small RNA molecules such as small interference RNA (siRNA) (5) and microRNA (miRNA) (6–8).
2. Different approaches to quantify tiny RNA molecules
The mature miRNAs are very small RNAs, 19–25 nucleotides (nt) long and because of this small size there are differences in the approaches to quantify miRNAs and mRNAs. For example small RNAs are less efficiently precipitated in ethanol and for this reason during the isolation by standard Trizol protocol of the RNA, resuspension in ethanol should be avoided. On the other hand, in our experience the miRNAs seems to be more stable than longer RNAs, for example in degraded samples it is still possible to obtain readable miRNA expression data. Moreover other groups reported a higher stability of miRNAs compared to mRNA in samples obtained from formalin-fixed paraffin-embedded tissues (9–11).
When the first miRNA was described (6, 7, 12), Northern blotting was used to detect these small RNAs. The protocol for Northern Blotting was modified to detect such a small RNA fragments, a hundred-fold less than the average coding RNA, by using high percentage (%) of urea-acrylamide gels instead of usual agarose gels.
We can classify three main techniques to detect and quantify miRNA in tissue samples. They are the cloning of miRNA, the PCR- based detection, and finally the hybridization with selective probes. The cloning of miRNA was the main tool used to identify miRNAs (7, 13, 14). Cloning offers the advantage to discover new miRNAs not predicted from bioinformatics and to sequence the miRNAs. On the other hand, the quantification ability is smaller compared to other approaches. The PCR-based technique is able to detect low copy number with high sensitivity and specificity on both the precursor (15, 16) and the mature form of miRNAs (17). It is cheap and it can be used extensively for clinical samples with minute amounts of available RNA. The hybridization techniques comprise Northern blotting, in situ hybridization (17, 18), bead-based flow-cytometry (19) and microarray (Liu et al, 2004). Northern Blotting using radioactive probes, is very sensitive, but it is very time-consuming. Northern blotting is not practical in large clinical studies to detect the expression of hundreds of miRNAs. Northern blotting also requires large amounts (5 to 25 μg) of total RNA from each sample. Since the discovery of miRNAs in C. elegans, the number of miRNA quickly increased and were found in all the eukaryotic species analyzed (12, 20–22). In August 2007, the number of miRNAs has passed 500 [http://miRNA.sanger.ac.uk/sequences/], and almost a thousand have been predicted in the human genome. Intronic miRNA precursors that bypass Drosha processing have been recently described in Drosophila melanogaster (D. melanogaster) (23, 24). These recent discoveries could also potentially expand the number of miRNAs The increasing number of miRNAs and the analysis of large cohort of patients require a technique able to process multiple miRNAs at the same time and use a small amount of RNA obtained from patients.
3. Microarray technology on miRNAs
The microarray technology was developed in 1995 (25). It is based on the ability to perform multiple hybridizations in parallel using a glass or quartz support where, depending on the platform used, the probes have been spotted or synthesized by photochemical synthesis, (26–28). The ability to increase the density of the spots on the array resulted in a higher number of genes that are able to be analyzed simultaneously (28, 29). Three different technologies classically exist to detect nucleic acids (DNA or RNA) on an array platform. The first, commonly used for custom arrays, uses glass slides (poly-lysine coated common microscope slides) and is based on the spotting of unmodified oligonucleotides over the slide (30). The second uses glass slides too, and it is also based on the deposition of the probes on the slide. The distinction is that the 5′ terminus of the probe is cross-linked to the matrix on the glass. The number of probes present on these slides can be much higher compared to the former. In the last technology, the probes are photochemically synthesized directly on the surface which must be made of quartz. In the last case, the number of probes rises to millions per area the size of a thumbnail (27). Usually, but not always, the first two are based on the comparison of two samples for each glass (one used as reference) stained with different colors. The third uses a single color hybridization where each slide is hybridized with only one sample.
The short length and the small abundance of miRNAs determine a more difficult design of the probes and require a higher sensitivity. Being miRNAs 19–25nt long, the design of the probe is almost exclusively determined by the sequence of the miRNA itself, which determines a different temperature of annealing for each probe::miRNA interaction.
Since the first time microarray technology was applied to miRNA studies (31, 32), we have identified at least 18 publications describing different approaches for miRNA quantification on microarray platforms (Table 1). Most of them use DNA oligo spotting, some of them use locked nucleic acid (LNA) with the intent of increasing the affinity between the probes and the miRNAs, and reaching more uniform conditions of hybridization among the different probes (33). The ideal technique should be able to detect the miRNAs without any kind of manipulation of the samples, such as enrichment of the low molecular weight species of RNA, retro-transcription of the miRNA, amplification of the miRNA, and in the meantime it should be able to discriminate the two predominant forms of miRNAs (mature and precursor as it occurs with the array initially developed by us). The differences in the expression between mature and precursor forms can represent a significant feature of miRNA biogenesis as it has been proven for two examples, the cluster of mir-143-145 and the cluster of mir-15a-16. In the first example the two miRNAs showed a different expression of the mature form between colon cancer samples and normal colon samples, while no differences were found for the precursor forms (34); while in the second case a mutation of the precursor described in two patients is able to determine a decreased maturation of the miRNA, with a consequent decrease of the mature form (35).
Table 1.
list of all published microarray platforms for microRNA expression studies. (August 2007)
| detection of | |||||||
|---|---|---|---|---|---|---|---|
| Reference | technology | amplificationa | direct microRNA labelling | RetroTranscriptiona | size selectionb | mature form | precursor form |
| (Liu, Calin et al. 2004) | microarray oligo | no | no | yes | no | yes | yes |
| (Babak, Zhang et al. 2004) | microarray oligo | no | no | no | no | yes | yes |
| (Barad, Meiri et al. 2004) | microarray oligo | yes | no | yes | yes | yes | yes |
| (Miska, Alvarez-Saavedra et al. 2004) | microarray oligo | yes | no | yes | yes | yes | no |
| (Nelson, Baldwin et al. 2004) | microarray oligo (RAKE) | no | yes | no | yes | yes | no |
| (Thomson, Parker et al. 2004) | microarray oligo | no | no | no | yes | yes | no |
| (Sun, Koo et al. 2004) | microarray oligo | no | no | yes | no | yes | no |
| (Baskerville and Bartel 2005) | microarray oligo | no | no | yes | yes | yes | no |
| (Goff, Yang et al. 2005) | microarray oligo | no | yes | no | yes | yes | no |
| (Liang, Li et al. 2005) | microarray oligo | no | no | no | yes | yes | no |
| (Shingara, Keiger et al. 2005) Ambion | microarray oligo | no | yes | no | yes | yes | no |
| (O’Donnell, Wentzel et al. 2005) | oligo array | no | yes | no | yes | yes | no |
| (Castoldi, Schmidt et al. 2006) | microarray oligo (LNA) | no | yes | no | no | yes | no |
| (Fang, Lee et al. 2006) | LNA+gold nanoparticles | no | yes | no | no | yes | no |
| (Mattie, Benz et al. 2006) | microarray oligo | no | no | yes | not specified | yes | no |
| (Beuvink, Kolb et al. 2007) | microarray + evanescent resonator | no | yes | no | yes | yes | no |
| (Fan, Chen et al. 2007) | microelectrod- biosensor PNA probes | no | no | no | no | yes | no |
before hybridization
size selection of RNA
4. Significance of microRNA profiling
The miRNAs have been related to the regulation of different biological processes. They have been shown to regulate the Caenorhabditis elegans developmental timing (6, 36), as well as the embryonic and post embryonic differentiation in rodents (37, 38) and in human beings (39–41). MiRNAs have been involved in physiological processes, such as insulin regulation (42, 43), lipid metabolism (44) and synaptic activity, (45).
The first evidence of the involvement of microRNA in a human disease was set in 2002, when for the first time a deregulation of two miRNAs (mir-15a and mir-16-1) was related to Chronic Lymphocytic Leukemia (CLL) (46). Many other examples followed after that, and many types of tumors were analyzed, ranging from lung cancer (47, 48), breast cancer (49), to colon cancer (34, 50) and nervous tumors such as glioblastoma (51)(for a detailed reference see (52). The majority of the published papers that reported profiling analysis were performed using microarray technology. The creation of lists of miRNAs differentially expressed between tumor and normal tissues or between different tumor types, gives the chance to identify the miRNAs most probably involved in cancer and to identify new diagnostic and prognostic markers (35). Since miRNAs are involved in cell differentiation (53, 54) and in the inflammatory response (55), their deregulation in cancer can be just a consequence of the phenotypic changes in the tumor cells and in the surrounding normal tissues due to the cancer development, and consequently the deregulated miRNAs could not play an active role in the tumor pathogenesis. Consequently, to prove a causative role of a specific miRNA in a tumor pathology, functional studies are mandatory and they have to link the miRNA function with cellular process such as proliferation (56), apoptosis (57, 58), and metastasis/migration (59). A definite proof for the causal link between miRNAs and cancer is the development of B cell malignant proliferation in a miR-155 transgenic mouse model (60)
The leading cause of death in western countries still remains heart diseases (61). Recent data report that mir-1 is involved in the regulation of the cardiogenesis and cardiac conduction in rodents (38). A previous study identified the miRNA signature of cardiac hypertrophy in rodents, and in another study the transgenic mouse model of one of the described upregulated miRNAs (mir-195) was able to reproduce a phenotype close to the disease (62). Among the neurologic diseases, Alzheimer’s disease is the most frequent cause of death in the US (61) and its etiology remains unexplained. The miRNAs are involved in different brain functions too. One of the first examples of miRNA profiling studies showed a specific signature made of 44 miRNAs during the rodent brain development (31). At least two studies correlated the deregulation of miRNAs in the hippocampus of patients affected by Alzheimer’s disease (63, 64). Among the most frequently involved miRNAs in brain development and pathology are mir-9, mir-125b (the_orthologous of lin-4 the first miRNA discovered in C. elegans (6)) and mir-128. Functional studies and mouse models relating miRNA deregulation and brain diseases are still missing, but the profiling studies showed the first evidence of the potential involvement of miRNAs on brain pathologies.
The discovery of miRNAs defined a completely new regulatory system. For this reason these small RNA molecules can help to find some of the hidden etiologic mechanisms for some diseases. Furthermore the better understanding of the etiology and the pathogenesis of a disease can create new diagnostic tools and therapeutic targets. Screening the expression of all miRNAs by cDNA microarray at once, is in our opinion, the first step in the study of the involvement of miRNAs in one specific pathology. The differential expression between different groups becomes the main criteria on microarray analysis and it can suggest that further studies on particular miRNAs that can successfully show a direct involvement in the etiology and or pathogenesis of that disease. In the meantime the use of microarray platforms is probably the best way to study the existence of completely new transcribed genomic elements and to determine their tissue specificity and possible involvement in different diseases such as cancer (65).
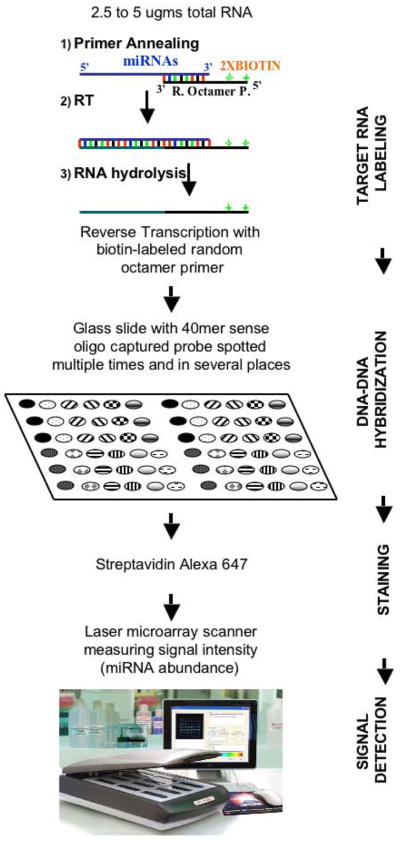
In order to profile miRNA gene expression and to obtain genome wide signatures in diseases such as cancer, we developed a unique miRNA microarray using a CodeLink platform (http://www.osuccc.osu.edu/microarray). We will focus on the equipment, supplies, and protocol that are necessary to develop a microarray platform starting from the oligo design, through the array fabrication and miRNA target preparation, to the hybridization of the samples and the signal detection and data analysis (Figure 1).
Figure 1. Principles of microarray technology used for microRNAs profiling.
A) The microarray based miRNA profiling is presented as described in the majority of profiling studies on primary tumors, initially developed by Liu et al (Liu et al., 2004). Target labeling, hybridization, staining and signal detection are the four main technical steps (presented on the right side). The different replicates of the spots on the glass slide (presented as different types of gray grids) represent different oligonucleotide sequences corresponding to sequences from the precursor miRNA or active miRNA molecule. The main advantage of the microarray based miRNA profiling is the high standardization of the procedure (allowing the processing of tens of samples in parallel), making it relatively easy to be performed. Several technical aspects regarding this technology can be found elsewhere (Liu et al., 2007). Reproduced after Calin GA & Croce CM, Investigation of MicroRNA Alterations in Leukemias and Lymphomas. Methods Enzymol. 2007;427:191–213.
5. Discussion of the equipment (for list of Instruments, Supplies and Reagents see Appendix)
The Ohio State University Comprehensive Cancer Center (OSUCCC) miRNA Expression Bioarrays are transcriptional profiling products designed to monitor the miRNA and other small non-coding RNA levels of multiple genes. The miRNA Expression Bioarrays utilize nucleic acid hybridization of a 5′ biotin-labeled complementary cDNA target with DNA oligonucleotide probes attached to a gel matrix.
The biotin-labeled cDNA targets are prepared by a simple reverse transcription into first strand cDNA. Total RNA is primed for reverse transcription by a random Octomer conjugated with two biotins and a 5′ poly (A) tail. This procedure results in an equal copy number of biotin-cDNA target to the templates of miRNA.
Storage and Handling Conditions
These procedures involve working with RNA; therefore, care must be taken to avoid any potential RNase contamination. All solutions must be RNase-free and pipette tips must be changed before each step. Use commercially prepared nuclease-free H2O (in lieu of H2O treated with diethylpyrocarbonate) for all nucleic acid steps.
Store the cDNA synthesis kits, except for the hybridization buffer, at −20°C (do not store in frost-free freezer).
Store the hybridization buffer components as indicated on the tubes or bottles.
The Invitrogen’s Trizol product is recommended for total RNA isolation. No column procedure should be applied for total RNA isolation or cleaning. miRNA will be lost when a column is applied to total RNA isolation. Any Trizol contamination in total RNA can kill the enzyme in the target labeling reaction.
5.1 MiRNA microarray fabrication
5.1.1 Oligo Design
Two 40 mer oligo probes, one for the mature miRNA and the other for precursor oligo, were designed from the sense strand of both arms of the hairpin structure of the microRNA precursor sequence collected from the Sanger Database (http://microrna.sanger.ac.uk/cgi-bin/sequences/browse.pl). The oligo probes were modified at the 5′ end with Amine-C6 linker and ordered from Integrated DNA technology (IDT) (Coralville, IA, USA) at 50 or 100 μM stock concentration in H2O.
5.1.2 Oligo Probe library for miRNA array fabrication
The working oligo library should be in the concentration of 20 μM in 50 mM Sodium Phosphate buffer pH 8.0 with 2 μM of Sodium-Fluorescein. The oligos were synthesized in a 96 well plate format from the vendor, and we had to assemble in house four 96 well plates into 384 well microtiter plate format by Tecan TeMo liquid handler in order to be able to work with the array fabrication system. For quality control (QC) purpose of chip fabrication, the oligo probes are co-dispensed with Fluorescein (Sigma F-6377).
5.1.3 MiRNA array fabrication
The Codelink™ activated slides (GE Healthcare, USA, PN 300011) are loaded onto an Omnigrid 100 slide holder. The 384 well oligo library plates are loaded sequentially onto a Gene Machine OmniGrid 100 arrayer (Genomic Solution, Ann Arbor, MI, USA) and the miRNA arrays are printed with a designed printing protocol (Reference, Omnigrid user manual website).
5.1.4 MiRNA array post-printing processes
5.1.4.1 Quality Control Scanning
The printed array slides were scanned by Axon Scanner 4200 at 488nm excitation length in order to detect fluorescein, and QC image files were saved.
5.1.4.2 Oligo probe coupling
Under 70% humidity overnight, the dispensed amine-modified oligo probes on the slides couple with NHS ester group of coated polymer gel matrix and are covalently immobilized on the surface of Codelink™ activated slides.
5.1.4.3 miRNA array blocking
The NHS ester groups surrounding the spots of oligo probe dispensed need to be blocked, in a New Brunswick Innova 4080 Shaking incubator, in a solution of 50°C prewarmed 100mM Bicine (Sigma B-8660) and 100mM Taurine (Sigma T8691), pH 9.0 at 50°C for 60 mins. Then the blocked slides are rinsed with deionized H2O and further washed in 50°C pre-warmed 4X SSC/0.1% SDS buffer for 30 mins with 50 rpm agitation. The slides were rinsed with H2O and spun dry. The slides were now ready to hybridize the labeled miRNA cDNA targets and should be stored in a desiccator (Fisher Catalog# 08-642-23C) until use.
5.2 Sample Preparation
5.2.1 Synthesis of biotin labeled First-Strand cDNA targets
5.2.1.1
Prepare each total RNA sample for manual target preparation:
5 μg Total RNA (optimal concentration should be determined for each source of total RNA)
2 μ 0.5 ug/μl primer
X μl Nuclease-free H2O to 12 μl final volume
12 μl Total Volume
5 ug total RNA in 10 ul RNAse free H2O.
2 ul oligo primer (5′ biotin-AAA-AAA-AAA-AAA-(T-biotin)-AAA-AAA-AAA-AAA-NNN-NNN-NN 3′) (0.5ug/ul) (where N stands for random octamer).
5.2.1.2
Incubate 10 minutes in a 70°C water bath then immediately place the tube on ice.
5.2.1.3
Centrifuge for 5 seconds to collect the sample at the bottom of the tube and immediately place the tube on ice.
5.2.1.4
With the tube remaining on ice, add the following reagents to the 12 μl of total RNA/control mRNA/primer mix.
4 μl 5X first-strand buffer
2 μl 0.1 M DTT
1 μl 10 mM dNTP mix
1 μl Superscript™ II RNaseH− reverse transcriptase (200 U/μl)
20 μl Final Volume
5.2.1.5
Incubate 90 mins in a 37°C water bath.
5.2.1.6
Centrifuge for 5 seconds to collect the sample at the bottom of the tube.
5.2.1.7
RNA template degradation. After a 90 min incubation for the first strand synthesis, adding 3.5 ul of 0.5M NaOH/50 mM EDTA into 20 ul reaction mix and incubate at 65°C for 15 min to denature the DNA/RNA hybrids and degrade the RNA template. Then neutralize the reaction with 5 ul of 1M Tris-HCI, pH 7.6 (Sigma). Each labeled target should be in a volume of 28.5 ul. The sample preparation is now done and the samples are stored at −80° C until use.
5.3 miRNA Microarray Hybridization and Data Collection
5.3.1 miRNA Microarray Hybridization
5.3.1.1
Prime all channels of the Tecan HS4800 hybridization stations and load hybridization chambers to HS 4800.
5.3.1.2
Load pre-printed miRNA array face-up to the Tecan HS4800 hybridization station. Close hybridization chambers on HS4800.
5.3.1.3
Run hybridization program for the chip hybridization with labeled cDNA targets:
Prime the chip in the hybridization chamber at 23° C with 6X SSPE with 0.05% Tween 20 for 1 min.
Inject 75 ul prehybridization mix of 6X SSPE/2X Dehardt/30% Formamide and prehybridize the chip at 25° C for 30 mins.
Inject the hybridization mix of labeled biotin-cDNA in 6X SSPE/2X Dehardt/30% Formamide and hybridize at 25° C for 18 hours.
Wash in 0.75X TNT buffer at 23° C for 5 min.
Wash in 0.75X TNT buffer at 37° C for 10 min.
Water rinse on the HS4800 at 23° C for 30 seconds.
Unload the chips from the machine for post-hybridization washing.
5.3.2 miRNA Microarray Post-Hybridization
5.3.2.1
Open the hybridization chamber of the HS 4800 and remove the slide chips as quickly as possible and then place them into a slot of the Bioarray Rack, which was placed in the large Reagent Reservoir containing 37° C prewarmed 0.75X TNT. Move the slide into place using the Bioarray Position Tool, tooth-side down. Wash them in 37° C prewarmed 0.75X TNT with agitation in New Brunswick Innova 4080 Shaking incubator at 37° C for 40 mins with 50rpm agitation.
5.3.2.2
Block the chip in TNB blocking buffer (see Appendix) at room temperature for 30 mins.
5.3.2.3
Stain the chips with streptavidAlexa-647 1:500 in TNB buffer at room temperature for 30 mins.
5.3.2.4
Post-stain wash in 1X TNT at room temperature for 40 mins in total with three buffer changes.
5.3.2.5
Rinse the chips with distilled water briefly and spin dry them at 1000 rpm for 1 min.
5.3.3 miRNA Microarray Data Collection
Scan the chip by Axon Scanner at a power setting of Power 100 and PMT 800. The image data may be extracted by GenePix software and saved as a .gpr file for further data analysis
Acknowledgments
Dr. Calin is supported by the CLL Global Research Foundation, and, in part, as a University of Texas System Regents Research Scholar and as a Fellow of The University of Texas M. D. Anderson Research Trust and Dr Croce is supported by Program Project Grants from the National Cancer Institute. We apologize to our colleagues whose work was not cited due to space limitations.
6. Appendixes (Solutions, Equipment, and Supplies)
6.1 MiRNA microarray fabrication (refer to points 5.1.1 through 5.1.4 of Discussion of Equipment)
6.1a Solutions for miRNA array fabrication
6.1.1
100mM Phosphate buffer pH 8.0 (2X): Resuspend 0.69 g of anhydrous monobasic sodium phosphate (Sigma S-3139) and 25.46 g of dibasic sodium phosphate, heptahydrate (Sigma S-9390) in 900 ml of distilled H2O and adjust the solution pH to 8.0 by adding 100ul of 10N NaOH. Add more distilled H2O to 1000 ml volume and filter the solution by 0.22 micron filter unit. The 100 mM (2X) phosphate buffer is stored at 4°C until use.
6.1.2
2uM Fluorescein solution: First prepare a 200 μM solution by dissolving 18.845 mg of Fluorescein (Sigma F-6377) in 250 ml of H2O into as 200 uM solution first. Dilute 5 ml of 200 uM solution into 495 ml of H2O (2 uM Fluorescein solution).
6.1.3
100 mM Bicine and Taurine array blocking solution: Dissolve 48.9 g of Bicine (Sigma B-8660) and 37.5 g of Taurine (Sigma T-8691) in 2400 ml of H2O. Adjust the pH to 9.0 by adding ~40 ml of 10N NaOH. Add more H2O to a volume of 3000 ml.
6.1.4
4.04X SSC/0.1% SDS array washing solution: Make 4.04X SSC from 20X SSC solution (Sigma S-6639) by dilution first. Mix together 2970 ml of 4.04X SSC and 30 ml of 10% SDS.
6.1b Equipment and Materials
Omnigrid 100 Arrayer: Genomic Solution, Inc. 4355 Varsity Dr. Suite E, Ann Arbor MI 48108
Tecan TeMo liquid handler: Tecan TEMO liquid handler, TECAN US Inc. Research Trangle Park, NC27709
Axon Scanner 4200: Molecular Device Corp.1311 Orleans Dr. Sunnyvale CA 94089-1136.
CodeLink Activated Slides: GE Healthcare (Amersham-300011), Piscataway, NJ
20X SSC Sigma S6639-1L
5M Sodium Chloride, Sigma S5150-1L
Bicine, Sigma B8660-1KG
Taurine, Sigma T8691-100G
Fluorescein, Sigma F-6377
Anhydrous monobasic sodium phosphate, Sigma S-3139
Dibasic sodium phosphate, heptahydrate, Sigma S-9390
6.2 Sample Preparation (refer to point 5.2 of Discussion of Equipment)
6.2.a Reagents
0.5 pmol/μl 3′ NNNNNNNN- (dA) 12 T (biotin) (dA) 12-Biotin 5′ Oligonucleotide primer
5X First-strand buffer
0.1 M Dithiothreitol (DTT)
10 mM dNTP mix
Superscript™ II RNaseH− reverse transcriptase (200 U/μl) (Invitrogen 18064-014)
10mM dNTP mix (Invitrogen 18427-013)
6.2.b Equipment and Materials Supplied by User
Pipette tips, sterile, RNase-free, and aerosol-resistant
Microcentrifuge tubes, sterile, RNase-free, 1.7 ml
Micropipettes (10, 20, 200, 1000 μl)
Nanodrop UV spectrophotometer
Microcentrifuge, room temperature and 4°C
Water bath (settings 70°C, 65°C, 37°C)
Sterile, Nuclease-Free conical tubes (15 and 50 ml)
Speed-Vac Concentrator
Vortex
Pipet Aid and disposable pipets
6.3 miRNA Microarray Hybridization and Data Collection (refer to point 5.3 of Discussion of Equipment)
6.3.1 miRNA Microarray Hybridization
6.3.1a Required Reagents/Kits Supplied by User
0.5% NEN Blocking Reagent (Perkin Elmer No. FP1020)
Molecular Probes Streptavidin-Alexa Fluor® 647 conjugate (staining solution is a 1:500 dilution in TNB; see Appendix 1) (Molecular Probes No. S-21374)
Nuclease-free H2O (Ambion No. 9915G)
1 X PBS; pH 7.4 (Invitrogen/LTI No. 10010-023)
1M Tris-HCl, pH 7.6 (Sigma No. T-2788)
5M NaCl (Sigma No. S-5150)
Tween®-20 (Sigma No. P-7949)
Formamide (Sigma)
50X Denhardt Solution (Sigma)
6.3.1b Other Equipment and Materials Supplied by User
Tecan HS 4800 Hybridization Station
Axon GenePix 4000B scanner
Computer configured for Axon 4000B Scanner
New Brunswick Innova™ 4080 shaking incubator
Sigma/Qiagen Centrifuge (4°–15°C) (Qiagen No. 81010)
Centrifuge Plate Rotor-2x96 (Qiagen No.81031)
Pipette tips, sterile, RNase-free, and aerosol-resistant
Microcentrifuge tubes, sterile, RNase-free, 1.7 ml
Micropipettes
Powder-free gloves
Microcentrifuge
Microtiter plate lid, Black (Corning, # 3935)
Bioarray Processors (GE, Healthcare)
Bioarray Rack (GE No. 600010)
Small Reagent Reservoir (GE No. 600011)
Large Reagent Reservoir (GE No. 600013)
Bioarray Removal Tool (GE No. 600015)
Bioarray Position Tool (GE No. 600016)
6.3.2 miRNA Microarray Post-Hybridization
6.3.2a Stock Solutions for Post-hybridization array processing
TNT Buffer (20 L)
0.1 M Tris-HCl, pH 7.6
0.15 M NaCl
0.05% Tween-20
Rinse a 25 L Carboy out with 150 ml of isopropanol.
Rinse the Carboy twice with 3 L of deionized H2O and completely drain the carboy.
Add 2 L 1 M Tris-HCl.
Add 600 ml 5 M NaCl.
Add 10 ml Tween-20.
Add 17.39 L deionized H2O.
Mix well by swirling.
Filter TNT through a 0.2 micron filter
This solution can be stored up to 2 weeks at room temperature.
0.75 X TNT Buffer
Add 25 ml of deionized water to 75 ml of TNT buffer (from above) per 100 ml of Buffer required.
TNB Buffer (0.5 L)
0.1 M Tris-HCl, pH 7.6
0.15 M NaCl
0.5% NEN Blocking Reagent (Perkin Elmer cat# FP1020)
Add 435 ml nuclease-free H2O.
Add 50 ml of 1 M Tris-HCl, pH 7.6.
Add 15 ml of 5 M NaCl.
Slowly add 2.5 g of NEN Blocking reagent in 0.5 g increments until all 2.5 g of blocking reagent are dissolved, while warming in a water bath at 60°C
Filter TNB Buffer through a 0.88 micron filter.
Aliquot the TNB buffer to 50 ml tubes and store at −20°C.
This solution can be stored for up to 12 weeks at −20°C. Thaw immediately before use.
6.3.3 miRNA Microarray Data Collection
Ideally, the same raw data should be analyzed by two distinct bioinformatics using two independent methods of analyses. For a detailed description, see reference Calin GA & Croce CM, Investigation of MicroRNA Alterations in Leukemias and Lymphomas. Methods Enzymol. 2007;427:191–213.
7. Safety Precautions
Standard laboratory safety procedures should be followed when using this product. Safety glasses, a lab coat, and appropriate gloves should be worn at all times when in the laboratory. Additional care should be taken when using Qiagen Buffer RLT, which contains chaotropic salts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Waterston RH, Lander ES, Sulston JE. Proc Natl Acad Sci U S A. 2002;99:3712–6. doi: 10.1073/pnas.042692499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowe CB, Bejerano G, Haussler D. Proc Natl Acad Sci U S A. 2007;104:8005–10. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 4.Costa FF. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Lau NC, Lim LP, Weinstein EG, Bartel DP. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 8.Mabuchi H, Fujii H, Calin G, Alder H, Negrini M, Rassenti L, Kipps TJ, Bullrich F, Croce CM. Cancer Res. 2001;61:2870–7. [PubMed] [Google Scholar]
- 9.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Nat Methods. 2004;1:155–61. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Ach RA, Curry B. Rna. 2007;13:151–9. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tricoli JV, Jacobson JW. Cancer Res. 2007;67:4553–5. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Ambros V. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 13.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Jiang J, Liu Q, Yang L. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Nucleic Acids Res. 2005;33:5394–403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. Nat Methods. 2006;3:27–9. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 20.Jones L. Trends Plant Sci. 2002;7:473–5. doi: 10.1016/s1360-1385(02)02361-0. [DOI] [PubMed] [Google Scholar]
- 21.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 22.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Cell. 2002;110:513–20. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 23.Ruby JG, Jan CH, Bartel DP. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. Cell. 2007 doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schena M, Shalon D, Davis RW, Brown PO. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 26.Kreil DP, Russell RR, Russell S. Methods Enzymol. 2006;410:73–98. doi: 10.1016/S0076-6879(06)10004-X. [DOI] [PubMed] [Google Scholar]
- 27.Wolber PK, Collins PJ, Lucas AB, De Witte A, Shannon KW. Methods Enzymol. 2006;410:28–57. doi: 10.1016/S0076-6879(06)10002-6. [DOI] [PubMed] [Google Scholar]
- 28.Dalma-Weiszhausz DD, Warrington J, Tanimoto EY, Miyada CG. Methods Enzymol. 2006;410:3–28. doi: 10.1016/S0076-6879(06)10001-4. [DOI] [PubMed] [Google Scholar]
- 29.Gershon D. Nature. 2005;437:1195–8. doi: 10.1038/4371195a. [DOI] [PubMed] [Google Scholar]
- 30.Hughes TR, Hiley SL, Saltzman AL, Babak T, Blencowe BJ. Methods Enzymol. 2006;410:300–16. doi: 10.1016/S0076-6879(06)10014-2. [DOI] [PubMed] [Google Scholar]
- 31.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. Rna. 2003;9:1274–81. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. Proc Natl Acad Sci U S A. 2004;101:9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. Rna. 2006;12:913–20. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael MZ, SMOC, van Holst Pellekaan NG, Young GP, James RJ. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 35.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 36.Feinbaum R, Ambros V. Dev Biol. 1999;210:87–95. doi: 10.1006/dbio.1999.9272. [DOI] [PubMed] [Google Scholar]
- 37.Houbaviy HB, Murray MF, Sharp PA. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 39.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. J Biol Chem. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 41.Georgantas RW, 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. Proc Natl Acad Sci U S A. 2007;104:2750–5. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA, Van Obberghen E. J Biol Chem. 2007;282:19575–88. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 43.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 44.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Ashraf SI, Kunes S. Curr Opin Neurobiol. 2006;16:535–9. doi: 10.1016/j.conb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 48.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 49.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 50.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, Croce CM. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 53.Sugatani T, Hruska KA. J Cell Biochem. 2007 doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 54.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. Proc Natl Acad Sci U S A. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 56.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 57.Chan JA, Krichevsky AM, Kosik KS. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 58.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma L, Teruya-Feldstein J, Weinberg RA. Nature. 2007 doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 60.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Proc Natl Acad Sci U S A. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minino AM, Heron MP, Murphy SL, Kochanek KD. Natl Vital Stat Rep. 2007;55:1–119. [PubMed] [Google Scholar]
- 62.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. Proc Natl Acad Sci U S A. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lukiw WJ. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 64.Lukiw WJ, Pogue AI. J Inorg Biochem. 2007 doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Cancer Cell. 2007;12:215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]