Abstract
Aim
To identify and study targets of microRNA biomarkers of glioblastoma survival across events (death and recurrence) and phases (life expectancy or post-diagnostic) using functional and network analyses.
Materials and Methods
microRNAs associated with glioblastoma survival within and across race, gender, recurrence, and therapy cohorts were identified using 253 individuals, 534 microRNAs, Cox survival model, cross-validation, discriminant analyses, and cross-study comparison.
Results
All 45 microRNAs revealed were confirmed in independent cancer studies and 25 in glioblastoma studies. Thirty-nine and six microRNAs (including hsa-miR-222) were associated with one and multiple glioblastoma survival indicators, respectively. Nineteen and 26 microRNAs exhibited cohort-dependent (including hsa-miR-10b with therapy and hsa-miR-486 with race) and independent associations with glioblastoma, respectively.
Conclusion
Sensory perception and G protein-coupled receptor processes were enriched among microRNA gene targets also associated with survival and network visualization highlighted their relations. These findings can help to improve prognostic tools and personalized treatments.
Keywords: Glioblastoma, microRNA, biomarkers, hazard, clinical cohort, gender, race
Glioblastoma multiforme (World Health Organization glioma grade IV) is a primary and aggressive cancer. Glioblastoma patients have a median survival of less than one year, and the incidence of glioblastoma varies among cohort groups, such as race and gender (1, 2). Some genes and microRNAs, small non-coding RNA molecules that can affect the post-transcriptional regulation of genes, exhibit abnormal expression patterns in glioblastoma (3, 4). Data and methodological limitations have prevented the identification of consistent microRNA biomarkers of glioblastoma survival that could be used to develop effective prognosis and diagnostic tools and therapies. Data limitations mostly encompass small data sets with unknown or restricted representation across cohort groups and consideration of a single glioblastoma survival indicator. Methodological limitations include arbitrary discretization of response (e.g. high and low survival) and explanatory (e.g. high or low expression level) variables (5), single-microRNA analysis (6, 7), pre-selection of microRNAs, and use of approaches that cannot accommodate the multifactorial nature of the disease.
The main objective of this study was to identify microRNAs that are reliable indicators of glioblastoma survival and recurrence using survival analysis. The study also aimed at extending the findings to microRNA target genes, their biological processes, molecular functions, and networks. Another goal was to identify and profile cohort-dependent associations between microRNAs and glioblastoma that can be used in personalized therapies.
Materials and Methods
Survival, cohort, recurrence, and microRNA information from 253 individuals diagnosed with glioblastoma and death and recurrence records between the years 1990 and 2008 was considered. Surgical samples corresponded to newly diagnosed glioblastoma cases, had a minimum of 80% tumor nuclei and a maximum of 50% necrosis (8). The data was obtained from The Cancer Genome Atlas (TCGA) December 2009 data freeze (9). Cohort factors were gender (male or female), race (white Caucasian or not), therapy received (radiation therapy alone, RX; chemotherapy plus radiation and no targeted therapy, CRN; chemotherapy plus radiation and targeted therapy, CRT; and all other therapies including no therapy, OTHER), and the detection of glioblastoma recurrence or progression after the original diagnostic (progression/recurrence or not).
Prognostic microRNA biomarkers for two events (death and recurrence) and two phases (from birth to event or from diagnostic to event) were studied through three complementary glioblastoma survivals: life expectancy (years from birth to death associated with glioblastoma), post-diagnostic glioblastoma survival (months from glioblastoma diagnostic to death), and post-diagnostic glioblastoma recurrence or progression (or post-diagnostic recurrence hazard, encompassing the months from glioblastoma diagnostic to reports of progression or recurrence). The last two indicators are also known as overall survival (OS) and progression-free survival (PFS) and offer complementary information to life expectancy (LE). The models used to describe the three indicators are specified in terms of hazard (instead of survival) and thus, hazard or survival is used where appropriate. Table I summarizes the number and distribution of individuals studied across levels of the covariates considered in the model. The median age at diagnosis was 55.7 years. Expression levels of 534 microRNAs were measured using the Agilent 8×15K Human microRNA platform. The data was quantile-normalized, collapsed within microRNA, and log 2-transformed following the procedures described in Beehive (10).
Table I.
Number and distribution of individuals analyzed for overall and post-diagnostic hazard of glioblastoma death and post-diagnostic hazard of glioblastoma recurrence and level of the cohort factors considered.
| Post-diagnostic survival (overall survival)a | Post-diagnostic recurrence (progression-free survival) | ||||
|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | ||
| Total | 253 | 192 | |||
|
| |||||
| N Censored | 26 | 10% | 17 | 9% | |
| Race | Caucasian | 211 | 83% | 161 | 84% |
| Other | 42 | 17% | 31 | 16% | |
| Gender | Females | 91 | 36% | 68 | 35% |
| Males | 162 | 64% | 124 | 65% | |
| Therapy | RX | 38 | 15% | 32 | 17% |
| CRN | 133 | 53% | 113 | 59% | |
| CRT | 31 | 12% | 28 | 15% | |
| OTHER | 51 | 20% | 19 | 10% | |
| Recurrence | Yes | 192 | 76% | 192 | 100% |
| No | 61 | 24% | 0 | 0% | |
The number of patients analyzed for post-diagnostic survival and life expectancy is the same.
N, Number of patients; RX, radiation therapy alone; CRN, chemotherapy plus radiation and no targeted therapy; CRT, chemotherapy plus radiation and targeted therapy; OTHER, all other therapies including no therapy.
Statistical computing method
A Cox survival model together with leave-one-out cross-validation (LOOCV) and discriminant analyses were used to identify microRNA expression profiles associated with glioblastoma survival. This model accommodates censored data resulting from individuals that are alive or that do not have a recurrence record at the end of the period analyzed. The test of no association between the microRNA or cohort prognostic markers and the hazard ratio between gender, race, therapy, or recurrence groups and the 95% confidence interval limits follow a Chi-square distribution. There was no indication of significant departure from the proportional hazards assumption, also confirmed by the overlap on microRNAs between survival indicators.
A multi-step strategy was undertaken to identify and validate microRNA prognostic markers of glioblastoma survival or recurrence. Cohort variables, microRNAs, and interaction terms were included simultaneously in a Cox model, and a combination of forward and stepwise model selection methods were used to identify association for each survival. The associations between all microRNAs in the platform, including 14 microRNA reported to be associated with glioblastoma (hsa-miR-21, hsa-miR-221, hsa-miR-222, hsa-miR-181a, hsa-miR-181b, hsa-miR-7, hsa-miR-128, hsa-miR-124, hsa-miR-137, hsa-miR-451, hsa-miR-10b, hsa-miR-129, hsa-miR-139 and hsa-miR-218) (1) were streamlined using the stepwise method. Following common practice, the resulting microRNA were evaluated using a LOOCV approach (11–13) and classification analyses (14–17). LOOCV is recommended especially for data sets of limited size, providing an almost unbiased estimator and identifying the same best classifiers as other X-fold training-test data partitions (11, 15). For the X-fold validation approach, the specification of suitable training and testing data sets would have required at least 160 patients in each data set (5 patients × 2 races × 2 genders × 4 therapies × 2 recurrence groups) and only 253 patients were available. Use of smaller data sets would have lead to low power and biased findings because of the ill-representation of patients across cohort groups. Patients were classified into high and low survival groups using the median time at the glioblastoma event (death or recurrence) as a cutoff and removing patients with unclear hazard within one unit of the median. Only non-censored records were used to avoid biased classification estimates. Preliminary results from linear and quadratic discriminant, logistic, and k nearest-neighbor analyses were consistent, and quadratic discriminant results are presented.
Validation of the results from the Cox model, LOOCV, and classification analyses on an independent data set was not feasible because no other data set has information on gender, race, therapy, recurrence, and age that would allow testing the cohort-dependent microRNAs identified in this data set. Thus, a two-fold approach was used to offer corroboration of our findings. First, the microRNAs biomarkers identified in this study were searched against the glioblastoma multiforme and cancer literature based on independent data sets. Second, the expression profile of the targets genes of the microRNAs were analyzed (18). The gene targets corresponding to the microRNA associated with glioblastoma survival were obtained from MicroCosm (19, 20). Expression measurements for the target genes were available from the same patients using the Affymetrix HT HG-U133A platform. The normalization and Cox survival models used for the gene targets were the same as described for the microRNA. The target genes subsequently used had a significant association (P-value < 0.001) with either glioblastoma OS, PFS, or LE (18). Functional Gene Ontology (GO) and KEGG Pathway analysis of the significant target genes of the significant microRNAs was undertaken (21, 22). The enrichment of functional categories was evaluated using Fisher’s exact (two-tailed) test and false discovery rate (FDR) multiple test adjustment (23). Network visualization was accomplished by depicting all pair-wise relationships between target genes using the BisoGenet plug-in from the Cytoscape software (24). BisoGenet’s database, SysBiomics, integrates data from multiple public domain datasets such as BIND, HPRD, Mint, DIP, BioGRID or Intact NCBI, UniProt, KEGG, and GO. Based on this information, a global network of relations among microRNA target genes was created and visualized using Cytoscape. The network was inferred using only significant target genes (circular network nodes) of significant microRNAs associated with either glioblastoma survival. Only interactions (network edges) connecting two target genes directly or through an intermediate gene (square gray node) were portrayed to facilitate the visualization of relationships and minimize the incorporation of relationships not relevant to the microRNAs biomarkers detected in this study. Known gene relationships depicted in the network are summarized in the SysBiomics repository (24).
Results
The median length of glioblastoma LE, OS, and PFS was 59 years, 13 months, and 6 months, respectively. Survival length indicators confirm previous reports that most TCGA samples correspond to primary glioblastoma (1, 25). MicroRNAs associated with the three glioblastoma survivals are listed in Tables II to IV, respectively. Hazard ratio estimates >1 indicate an increase in the hazard (decrease in survival probability) per unit increase in the level of microRNA expression, and hazard ratio estimates <1 denote the opposite trend, conditional on all other cohort and microRNA predictors in the model.
Table II.
MicroRNAs associated with life expectancy on a cohort-independent or-dependent manner and supporting independent studies.
| MicroRNA | P-value | Hazard ratio (95% C.I.) | Relevant literature references |
|---|---|---|---|
| hsa-miR-181a* | 0.0537 | RX=0; 0.33 (0.21 to 0.51) RX=1; 1.05 (0.33 to 3.38) |
(3, 4)G |
| hsa-miR-189 | 0.0204 | 0.20 (0.05 to 0.78) | (45)O |
| hsa-miR-19b | 0.0049 | 1.46 (1.11 to 1.90) | (51)G |
| hsa-miR-222 | 0.0258 | 0.83 (0.70 to 0.98) | (6, 37, 38)G |
| hsa-miR-34a | 0.0500 | RX=0; 0.69 (0.57 to 0.85) RX=1; 1.17 (0.72 to 1.89) |
(52, 53)G |
| hsa-miR-550 | <0.0001 | 4.18 (2.31 to 7.56) | (54)O |
| hsa-miR-625 | 0.0119 | 2.48 (1.22 to 5.02) | (55)O |
| kshv-miR-k12-1 | 0.0023 | 2.08 (1.30 to 3.32) | (27)O |
| hsa-miR-10b | <0.0001 | 0.74 (0.64 to 0.85) | (6, 42, 43)G |
| hsa-miR-140 | 0.0130 | 1.57 (1.10 to 2.24) | (38, 39)G |
| hsa-miR-149 | 0.0056 | 0.76 (0.63 to 0.92) | (56)G |
C.I., Confidence Interval; RX=1 denotes radiation therapy alone, RX=0 denotes non-radiation therapy;
glioblastoma multiforme study;
study on any other type of cancer.
Table IV.
MicroRNAs associated with progression-free survival on a cohort-independent or-dependent manner and supporting independent studies.
| MicroRNA | P-value | Hazard ratio (95% C.I.) | Relevant literature references |
|---|---|---|---|
| hsa-miR-181c | 0.0004 | CRN=0: 0.27 (0.16 to 0.47) CRN=1: 0.82 (0.53 to 1.35) |
(6, 38)G |
| hsa-miR-188 | <0.0001 | 2.30 (1.55 to 3.40) | (42)G |
| hsa-miR-222 | 0.0814 | Male: 1.27 (1.02 to 1.58) Female: 1.65 (1.29 to 2.12) |
(6, 37, 38)G |
| hsa-miR-296 | 0.0247 | RX=0: 1.56 (1.14 to 2.14) RX=1: 3.83 (1.82 to 8.07) |
(38, 65)G |
| 0.0633 | CRT=0: 2.04 (1.51 to 2.76) CRT=1: 0.90 (0.39 to 2.10) |
||
| ebv-miR-bart7 | <0.0001 | 0.05 (0.01 to 0.15) | (44)O |
| hsa-miR-486 | 0.0168 | Other: 0.74 (0.44 to 1.25) Caucasian: 1.53 (1.12 to 2.08) |
(42)G |
| hsa-miR-489 | 0.0041 | 0.04 (0.00 to 0.36) | (32)O |
| hsa-miR-512-3p | 0.0257 | Other: 0.00 (0.00 to 0.04) Caucasian: 0.07 (0.02 to 0.28) |
(55, 59)O |
| hcmv-miR-ul70-3p | 0.0004 | Male: 0.43 (0.27 to 0.67) Female: 1.13 (0.71 to 1.79) |
(33)O |
| hsa-miR-552 | 0.0001 | 0.00 (0.00 to 0.01) | (26)G |
| hsa-miR-578 | <0.0001 | 0.00 (0.00 to 0.00) | (66)G |
| hsa-miR-582 | 0.0003 | 5.49 (2.17 to 13.88) | (26)G |
| hsa-miR-584 | 0.0307 | 0.22 (0.05 to 0.87) | (26)G |
| hsa-miR-758 | 0.0029 | CRN=0: 0.77 (0.23 to 2.60) CRN=1: 0.08 (0.03 to 0.21) |
(34)O |
| hsa-miR-93 | 0.0006 | 2.63 (1.51 to 4.85) | (31)O |
| kshv-miR-k12-1 | <0.0001 | 3.19 (1.93 to 5.29) | (27)O |
| kshv-miR-k12-6-5p | <0.0001 | 3.70 (1.93 to 7.10) | (67)O |
| hsa-miR-106b | 0.0014 | RX=0: 0.12 (0.06 to 0.22) RX=1: 0.55 (0.22 to 1.40) |
(43)G |
| hsa-miR-143 | 0.0020 | Other: 0.30 (0.16 to 0.54) Caucasian: 0.83 (0.61 to 1.12) |
(35, 36)O |
C.I., Confidence Interval; CRN=1 denotes chemotherapy plus radiation and no targeted therapy, CRN=0 denotes non-CRN therapy; RX=1 denotes radiation therapy alone, RX=0 denotes non-RX therapy; CRT=1 denotes chemotherapy plus radiation and targeted therapy, CRT=0 denotes non-CRT therapy;
glioblastoma multiforme study;
study on any other type of cancer; n/a, no association with any type of cancer found in literature.
Tables II to IV list previous studies that support the association between the microRNAs and glioblastoma identified in this study. Corroborating our findings, the majority of microRNAs associated with glioblastoma survival (25 out of 45 microRNAs) have also been associated with glioblastoma in independent studies, and the rest (20 microRNAs) have been associated with other types of cancer (Tables II to IV). MicroRNAs in two families (hsa-miR-181 and hsa-miR-34 family) and six microRNAs were associated with multiple survival indicators, while 35 microRNAs were associated with one survival indicator. The same number of positive and negative associations (HR >1 or HR <1) between microRNA expression levels and the three glioblastoma hazards studied were revealed in this study (Tables II to IV). Twenty-six and 19 microRNAs had cohort-independent and-dependent relationships with glioblastoma survival, respectively. The survival plot in Figure 1 depicts the lower post-diagnostic survival probability of females that have a low level of microRNA ebv-miR-bhrf1-1 relative to males with a high expression level. Three microRNAs (hsa-miR-10b, hsa-miR-222, and hsa-miR-140) exhibited different hazard ratio trends across glioblastoma indicator, and the associated confidence interval allowed the identification of the trend best supported by the data.
Figure 1.
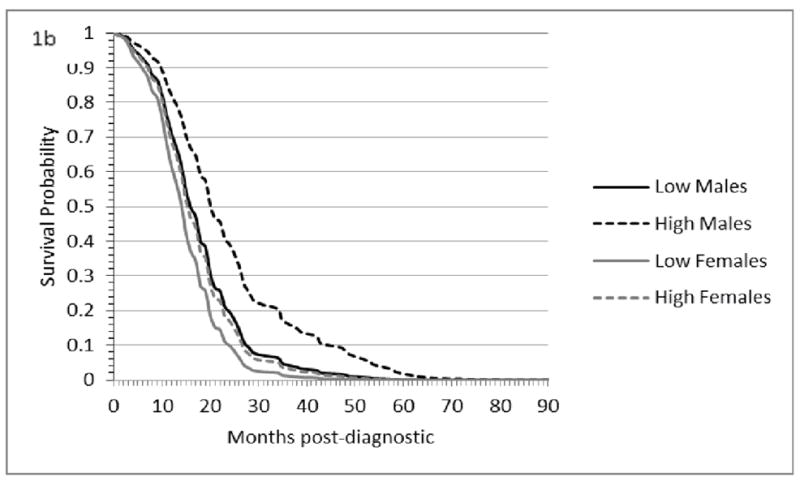
Overall survival plots for males (black lines) and females (gray lines) that have high (dash lines) and low (solid line) levels of ebv-miR-bhrf1-1.
Integration of Cox survival model, LOOCV, and discriminant analysis supported the correct classification of 98% and 93% of the patients into the low and high post-diagnostic survival or OS groups, respectively, and the area under the receiver operator characteristic (ROC) was 94%. Likewise, 100% and 91% of the individuals in the high and low PFS groups were correctly classified, and the area under the ROC was 97%. Finally, 86% and 75% of the patients in the high and low LE groups were correctly classified, and the area under the ROC was 85%. Another indicator of the reliability of the integrated approach is that all microRNAs detected in this study have been associated with cancer and the majority with glioblastoma in independent studies. An additional indicator supporting the microRNAs identified is that 239, 418, and 336 gene targets of the microRNAs were significantly associated with LE, OS, and DFS, respectively.
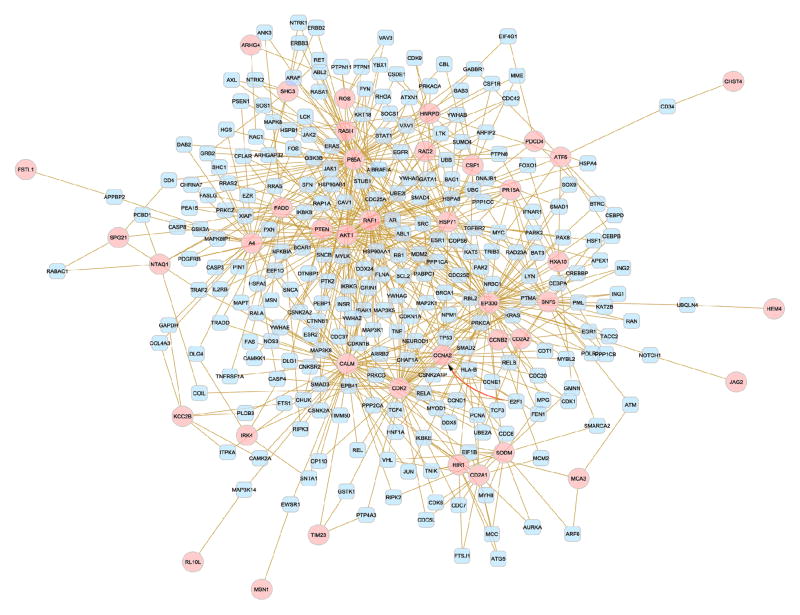
Several GO categories were enriched (FDR-adjusted P-value < 0.05) among the target genes significantly associated with multiple survival indicators. Tables V to VII summarize these findings, with the latter table including an FDR-adjusted P-value <0.01 and a minimum of six genes due to space limitations. Categories are sorted by GO theme, followed by level and P-value. The GO categories enriched across all three survival indicators included sensory perception (of chemical stimulus and smell), neurological process, olfactory receptor activity, rhodopsin-like receptor activity, and transmembrane receptor activity. All GO categories enriched in the post-diagnostic death or OS were also identified in either or both of the remainder indicators. Figure 2 portrays the network including target genes (denoted in pink) of significant miRNAs that also themselves have a significant association with either glioblastoma OS or PFS, and have a minimum of one relationship and at most one indirect relationship with other target genes.
Table V.
Gene Ontology categories enriched (FDR-adjusted P-value <0.05) among the target genes of microRNAs associated with life expectancy
| Gene Ontology | Level | Term | FDR P-value | No. of genes |
|---|---|---|---|---|
| Biological process | 3 | Neurological process (GO:0050877) | 0.0248 | 219 |
| Biological process | 4 | Sensory perception (GO:0007600) | 0.0111 | 151 |
| Biological process | 5 | Sensory perception of chemical stimulus (GO:0007606) | <0.0001 | 70 |
| Biological process | 6 | Sensory perception of smell (GO:0007608) | <0.0001 | 63 |
| Molecular function | 4 | Transmembrane receptor activity (GO:0004888) | 0.0146 | 239 |
| Molecular function | 5 | G Protein-coupled receptor activity (GO:0004930) | 0.0039 | 160 |
| Molecular function | 6 | Rhodopsin-like receptor activity (GO:0001584) | 0.0023 | 134 |
| Molecular function | 7 | Olfactory receptor activity (GO:0004984) | <0.0001 | 60 |
Table VII.
Gene Ontology categories enriched (FDR-adjusted P-value <0.01, number genes >6) among the target genes of microRNAs associated with progression-free survival.
| Gene Ontology | Level | Term | FDR P-value | No. of genes |
|---|---|---|---|---|
| Biological process | 3 | Cell communication (GO:0007154) | <0.0001 | 975 |
| Biological process | 3 | Multicellular development (GO:0007275) | <0.0001 | 507 |
| Biological process | 3 | Neurological process (GO:0050877) | <0.0001 | 248 |
| Biological process | 3 | Anatomical structure development (GO:0048856) | <0.0001 | 483 |
| Biological process | 3 | Cellular organization & biogenesis (GO:0016043) | 0.0008 | 639 |
| Biological process | 3 | Cellular metabolic process (GO:0044237) | 0.0014 | 2290 |
| Biological process | 3 | Cellular developmental process (GO:0048869) | 0.0066 | 551 |
| Biological process | 4 | Signal transduction (GO:0007165) | <0.0001 | 889 |
| Biological process | 4 | Sensory perception (GO:0007600) | <0.0001 | 172 |
| Biological process | 4 | System development (GO:0048731) | <0.0001 | 386 |
| Biological process | 5 | Sensory perception of chemical stimulus (GO:0007606) | <0.0001 | 86 |
| Biological process | 5 | Cell surface receptor linked signal transduction (GO:0007166) | <0.0001 | 433 |
| Biological process | 5 | Organ development (GO:0048513) | 0.0085 | 285 |
| Biological process | 5 | + Regulation of metabolic process (GO:0009893) | 0.0087 | 84 |
| Biological process | 5 | Carboxylic acid metabolic process (GO:0019752) | 0.0095 | 185 |
| Biological process | 5 | + Regulation of cellular process (GO:0048522) | 0.0095 | 214 |
| Biological process | 6 | Organ morphogenesis (GO:0009887) | <0.0001 | 57 |
| Biological process | 6 | Sensory perception of smell (GO:0007608) | <0.0001 | 80 |
| Biological process | 6 | G Protein-coupled receptor protein signaling pathway (GO:0007186) | 0.0099 | 266 |
| Molecular function | 3 | Protein binding (GO:0005515) | <0.0001 | 1601 |
| Molecular function | 4 | Transmembrane receptor activity (GO:0004888) | 0.0017 | 305 |
| Molecular function | 6 | Rhodopsin-like receptor activity (GO:0001584) | 0.0101 | 179 |
| Molecular function | 7 | Olfactory receptor activity (GO:0004984) | 0.0002 | 79 |
Figure 2.
Network of target genes of glioblastoma microRNAs.
Footnote. Circular pink nodes denote target genes of microRNAs associated with glioblastoma survival that also have a significant association with survival themselves. Square gray nodes denote a maximum of one intermediate gene between target genes. Edges denote known relationship between genes from several databases and summarized in the SysBiomics repository.
Discussion
All microRNAs associated with glioblastoma survival detected in this study have been confirmed in previous independent studies (25 microRNA) or have been associated with other cancer types (20 microRNAs). This extensive confirmation, the large number of target genes also significantly associated with glioblastoma survival, and the correct classification of patients into survival groups further supports the robustness of our findings. The equal number of positive and negative associations between microRNA expression levels and survival and the fact that 17% of the microRNAs exhibited associations with multiple glioblastoma survival indicators confirm the paradigm that glioblastoma initiation and recurrence are impacted by microRNAs targeting a wide range of oncogenes, tumor suppressor genes, and pathways at different stages of tumor genesis and growth (26). Most microRNAs (64%) exhibited a broad, cohort-independent relationship with glioblastoma survival. This indicates that mainstream and general practices to treat glioblastoma on the basis of microRNA profiles alone are promising. The identification of sex-, race-, and therapy-dependent microRNA biomarkers indicates that general practices can be effectively complemented with personalized practices. The following discussion of the microRNA biomarkers focuses on novel and high impact discoveries, and relevant supporting references for all other microRNAs are listed in Tables II to IV.
Higher levels of Kaposi’s sarcoma-associated herpes virus (kshv) miR-k12-1 were associated with all three glioblastoma survival indicators (Tables II to IV) in agreement with associations between this microRNA and two B-cell-derived cancer types (27). MicroRNAs ebv-miR-bhrf1-1, hsa-miR-565, hsa-miR-137, and hsa-miR-512-3p had gender-, race-, and recurrence-dependent associations with OS and PFS (Tables III and IV). For these four microRNAs, cohort-independent trends in the same direction were reported respectively for Burkitt’s lymphoma, ovarian cancer, chemoradiation-treated rectal cancer, and for both metastatic pancreatic ductal adenocarcinoma cell lines and hepatocellular carcinoma cells linked to the inhibition of the tumorgenesis factor c-FLIP (28–30). Likewise, the gender-, therapy- and race-dependent associations between hsa-miR-93, hsa-miR-489, human cytomegalovirus (hcmv) miR-ul70-3p, hsa-miR-758, hsa-miR-143, and PFS (Table IV) have been confirmed at a cohort-independent level for T-cell leukemia, breast-cancer MCF-7 cells resistant to tamoxifen, tumors from various tissues (e.g. breast, colon, liver), multidrug-resistant variant of a human gastric adenocarcinoma cell line, and for both B-cell chronic lymphocytic leukemia and colorectal cancer cell growth through inhibition of KRAS translation (31–36). The cohort-independent and gender-dependent association of hsa-miR-222 with OS and PFS (Tables III and IV, respectively) confirm the results of Ciafre et al. (37). The therapy-dependent association between glioblastoma and members of the hsa-miR-181 and hsa-miR-34 families (Tables II to IV) are consistent with previous reports (3, 6, 38). High levels of hsa-miR-140 were associated with higher LE and lower and therapy-dependent OS (Tables II and III). The multiple modes of action of hsa-miR-140 are consistent with reports of up-regulation in most glioblastoma cases (38), inhibition of cell proliferation in osteosarcoma and colon cancer cell lines (39), and treatment-dependent action (39). Reanalysis of the association between glioblastoma survival and hsa-miR-140 alone (with and without cohort factors, results not shown) produced trends similar to that in the multi-microRNA models. Thus, our results suggest that the influence of hsa-miR-140 on glioblastoma survival may vary with the glioblastoma phase considered. A gender-dependent association between hsa-miR-26a and OS was uncovered (Table III). The general trend is consistent with the proposed role of hsa-miR-26a promoting glioblastoma cell growth and formation (37, 40), and the gender-dependent model is in agreement with the higher expression of hsa-miR-26a in women than in men diagnosed with hepatocellular carcinoma (41).
Table III.
MicroRNAs associated with overall survival on a cohort-independent or -dependent manner and supporting independent studies.
| MicroRNA | P-value | Hazard ratio (95% C.I.) | Relevant literature references |
|---|---|---|---|
| hsa-miR-182 | 0.0245 | RX=0: 0.67 (0.57 to 0.77) RX=1: 1.00 (0.71 to 1.38) |
(42)G |
| 0.0027 | CRT=0: 0.66 (0.56 to 0.77) CRT=1: 1.19 (0.83 to 1.69) |
||
| hsa-miR-189 | 0.0316 | 0.12 (0.02 to 0.83) | (45)O |
| hsa-miR-196a | 0.0168 | 1.39 (1.06 to 1.81) | (57)G |
| hsa-miR-221 | 0.0298 | RX=0: 0.67 (0.43 to 1.04) RX=1: 0.41 (0.22 to 0.75) |
(6, 37, 38, 46)G |
| hsa-miR-222 | <0.0001 | 2.14 (1.51 to 3.03) | (6, 37, 38)G |
| hsa-miR-23b | 0.0135 | 1.61 (1.10 to 2.35) | (37)G |
| hsa-miR-26a | 0.0020 | Male: 1.33 (1.02 to 1.71) Female: 2.52 (1.78 to 3.58) |
(37, 40, 41)G |
| hsa-miR-324-5p | <0.0001 | 2.73 (1.80 to 4.14) | (58)G |
| hsa-miR-34c | 0.0106 | 0.62 (0.43 to 0.90) | (52, 53)G |
| ebv-miR-bhrf1-1 | 0.0009 | Other: 0.09 (0.01 to 0.51) Caucasian: 1.83 (1.16 to 2.88) |
(28)O |
| 0.0008 | Male: 0.65 (0.35 to 1.24) Female: 2.77 (1.43 to 5.38) |
||
| hsa-miR-512-3p | 0.0030 | 0.28 (0.12 to 0.65) | (55, 59)O |
| hsa-miR-565 | 0.0996 | Other: 2.97 (1.71 to 5.16) Caucasian: 1.80 (1.41 to 2.30) |
(29)O |
| 0.0003 | Pr/Re=0: 3.80 (2.40 to 6.02) Pr/Re=1: 1.59 (1.27 to 2.00) |
||
| hsa-miR-572 | 0.0691 | 0.76 (0.57 to 1.02) | (60)O |
| hsa-miR-766 | 0.0052 | 1.57 (1.15 to 2.16) | (61)O |
| kshv-miR-k12-1 | <0.0001 | 2.77 (1.78 to 4.31) | (27)O |
| kshv-miR-k12-6-3p | 0.0608 | 1.54 (0.98 to 2.43) | (62)O |
| hsa-miR-101 | 0.0065 | 1.63 (1.15 to 2.32) | (26)G |
| hsa-miR-10b | 0.0146 | RX=0: 1.16 (0.97 to 1.38) RX=1: 0.74 (0.52 to 1.04) |
(6, 42, 43)G |
| hsa-miR-134 | 0.0007 | 2.11 (1.37 to 3.25) | (43)G |
| hsa-miR-137 | 0.0010 | CRN=0: 2.11 (1.45 to 3.05) CRN=1: 0.94 (0.67 to 1.32) |
(30)O |
| hsa-miR-140 | 0.0010 | CRN=0: 0.21 (0.12 to 0.37) CRN=1: 0.65 (0.37 to 1.15) |
(38, 39)G |
| hsa-miR-148a | <0.0001 | 1.65 (1.35 to 2.02) | (63)O |
| hsa-miR-409-3p | 0.0001 | 0.43 (0.28 to 0.66) | (64)O |
C.I., Confidence Interval; RX=1 denotes radiation therapy alone, RX=0 denotes non-RX therapy; CRT=1 denotes chemotherapy plus radiation and targeted therapy, CRT=0 denotes non-CRT therapy; Pr/Re=1 denotes glioblastoma recurrence or progression report, Pr/Re=0 denotes no recurrence or progression report; CRN=1 denotes chemotherapy plus radiation and no targeted therapy, CRN=0 denotes non-CRN therapy;
glioblastoma multiforme study;
study on any other type of cancer.
Additional analyses resolved the apparent inconsistencies in the trends between previous reports and our study for seven microRNAs hsa-miR-182 (42), hsa-miR-106b (43), ebv-miR-bart7 (44), hsa-miR-189 (45), hsa-miR-221 (46), hsa-miR-21 (47), and hsa-miR-10b (6, 42, 43). For hsa-miR-182, hsa-miR-106b and hsa-miR-221, the individual microRNA analysis supported the multi-microRNA results. For ebv-miR-bart7, hsa-miR-189 and hsa-miR-10b, the individual analysis did not detect a significant trend. In one case, hsa-miR-21 was not detected when considered simultaneously with other microRNA but was significant when considered alone, in agreement with Chan et al. (47). These results suggest that identification of biomarkers on an individual basis may result in spurious associations and also validate the approach used in this study to identify biomarkers that simultaneously considers multiple microRNAs.
The large number of gene targets of the detected microRNAs that also exhibited significant association with glioblastoma survival further substantiates our findings. Sensory perception, neurological process, olfactory receptor, and transmembrane receptor activity were among the processes and functions consistently over-represented among the target genes of microRNAs associated with all three glioblastoma survival indicators. The neurological and sensory perception processes are consistent with reports of glioblastoma candidates for single nucleotide polymorphisms of sensory perception genes and with reports that individuals with brain tumors lose sensory perception (48). Oncogenes act by mimicking the growth signals transmitted by transmembrane receptors (49). G Protein-coupled receptor (GPCR) activity (e.g. rhodopsin-like gene) regulates cellular motility, growth and differentiation, and gene transcription, three factors central to the biology of cancer (50). The network of gene targets that have significant association with glioblastoma survival displays known relationships, including many in the signaling pathways that involve GPCR, including MAPK, adipocytokine, chemokine, ErBB, FC epsilon RI, mTOR, neurotrophin, notch, p53, phosphatidylinositol, RIG-I-like receptor, T-cell receptor, TGF-beta receptor, toll-like receptor, VEGF, and Wnt signaling pathways (Figure 2).
In summary, this study confirmed 25 microRNAs previously associated with glioblastoma survival and identified 20 other microRNA that have been previously associated with other cancer types. This confirmation and the high correct classification of patients into survival groups suggests that the biomarkers revealed in this study are good leads for empirical confirmation, improved prognostic tools, and personalized treatments of glioblastoma multiforme. Six and 39 microRNAs were identified as biomarkers of multiple or single glioblastoma survival indicators, respectively, suggesting the multifactorial and multifaceted genomic basis of this cancer. Nineteen microRNAs exhibited gender-, race-, therapy-, or recurrence-dependent associations with glioblastoma survival, suggesting that personalized prognostic and treatments that consider individual variation can improve the outcome for glioblastoma patients. Sensory perception and GPCR activities are among the processes of the microRNA target genes associated with survival.
Table VI.
Gene Ontology categories enriched (FDR-adjusted P-value <0.05) among the target genes of microRNAs associated with overall survival.
| Gene Ontology | Level | Term | FDR P-value | No. of genes |
|---|---|---|---|---|
| Biological process | 3 | Neurological process (GO:0050877) | <0.0001 | 371 |
| Biological process | 3 | Cell communication (GO:0007154) | 0.0001 | 1546 |
| Biological process | 4 | Sensory perception (GO:0007600) | <0.0001 | 255 |
| Biological process | 4 | Signal transduction (GO:0007165) | 0.0006 | 1400 |
| Biological process | 5 | Sensory perception of chem. stimulus (GO:0007606) | <0.0001 | 129 |
| Biological process | 5 | Cell surface receptor linked signal transduction (GO:0007166) | 0.0145 | 684 |
| Biological process | 6 | Sensory perception of smell (GO:0007608) | <0.0001 | 123 |
| Biological process | 6 | G Protein-coupled receptor protein signaling pathway (GO:0007186) | 0.0156 | 413 |
| Molecular function | 3 | Receptor activity (GO:0004872) | 0.0040 | 711 |
| Molecular function | 3 | Antigen binding (GO:0003823) | 0.0422 | 16 |
| Molecular function | 4 | Transmembrane receptor activity (GO:0004888) | 0.0002 | 457 |
| Molecular function | 5 | G Protein-coupled receptor activity (GO:0004930) | <0.0001 | 315 |
| Molecular function | 6 | Rhodopsin-like receptor activity (GO:0001584) | 0.0002 | 274 |
| Molecular function | 7 | Olfactory receptor activity (GO:0004984) | <0.0001 | 120 |
Acknowledgments
The support of NCI (Grant Number: 1R03CA143975) to SRZ and KRD, and NIH/NIDA (Grant Number: R21DA027548 and P30DA018310) to BRS and SRZ and USDA NIFA (Number: ILLU-538-554) to NSV are greatly appreciated.
References
- 1.Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386(1):1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 2.Wrensch M, Minn Y, Chew T, Bondy M, Berger MS. Epidemiology of primary brain tumors: current concepts and review of the literature. Neuro Oncol. 2002;4(4):278–299. doi: 10.1093/neuonc/4.4.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. Hsa-Mir-181a and Hsa-Mir-181b Function as Tumor Suppressors in Human Glioma Cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. MicroRNA–181a sensitizes human malignant glioma U87MG cells to radiation by targeting BCL-2. Oncol Rep. 2010;23(4):997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 5.Gaire RK, Bailey J, Bearfoot J, Campbell IG, Stuckey PJ, Haviv I. MIRAGAA-a methodology for finding coordinated effects of microRNA expression changes and genome aberrations in cancer. Bioinformatics. 2010;26(2):161–167. doi: 10.1093/bioinformatics/btp654. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X, Kang C, Pu P. MicroRNA and brain tumors. Chin J Clin Oncol. 2007;4:355–359. [Google Scholar]
- 7.Cui JG, Zhao Y, Sethi P, Li YY, Mahta A, Culicchia F, Lukiw WJ. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol. 2010;98(3):297–304. doi: 10.1007/s11060-009-0077-0. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.TCGA. Available at: http://cancergenome.nih.gov/dataportal/
- 10.Beehive. Available at: http://stagbeetle.animal.uiuc.edu/Beehive.
- 11.Chuang LY, Yang CS, Li JC, Yang CH. Chaotic genetic algorithm for gene selection and classification problems. OMICS. 2009;13(5):407–420. doi: 10.1089/omi.2009.0007. [DOI] [PubMed] [Google Scholar]
- 12.Chuang LY, Yang CH, Wu KC, Yang CH. A hybrid feature selection method for DNA microarray data. Comput Biol Med. 2011;41(4):228–237. doi: 10.1016/j.compbiomed.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Dumur CI, Ladd AC, Wright HV, Penberthy LT, Wilkinson DS, Powers CN, Garrett CT, DiNardo LJ. Genes involved in radiation therapy response in head and neck cancers. Laryngoscope. 2009;119(1):91–101. doi: 10.1002/lary.20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Tang X. A new classification model with simple decision rule for discovering optimal feature gene pairs. Comput Biol Med. 2007;37(11):1637–1646. doi: 10.1016/j.compbiomed.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Petrausch U, Martus P, Tonnies H, Bechrakis NE, Lenze D, Wansel S, Hummel M, Bornfeld N, Thiel E, Foerster MH, Keilholz U. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye (Lond) 2008;22(8):997–1007. doi: 10.1038/sj.eye.6702779. [DOI] [PubMed] [Google Scholar]
- 16.Oyan AM, Bo TH, Jonassen I, Gjertsen BT, Bruserud O, Kalland KH. cDNA microarray analysis of non-selected cases of acute myeloid leukemia demonstrates distinct clustering independent of cytogenetic aberrations and consistent with morphological signs of differentiation. Int J Oncol. 2006;28(5):1065–1080. [PubMed] [Google Scholar]
- 17.Grutzmann R, Boriss H, Ammerpohl O, Luttges J, Kalthoff H, Schackert HK, Kloppel G, Saeger HD, Pilarsky C. Meta-analysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene. 2005;24(32):5079–5088. doi: 10.1038/sj.onc.1208696. [DOI] [PubMed] [Google Scholar]
- 18.Serão NVL, Delfino KR, Southey BR, Rodriguez-Zas SL. Development of a transcriptomic-based index to prognosticate cancer. Abstract Book of the 6th International Symposium on Bioinformatics Research and Applications; 2010; 2010. [Google Scholar]
- 19.European Bioinformatics Institute. [Accessed June, 2010.];MicroCosm. Available at: http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5.
- 20.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38(Database issue):D331–335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34(Web Server issue):W472–476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.BisoGenet. Available at: http://bio.cigb.edu.cu/bisogenet-cytoscape/
- 25.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C, Lin J, Hong M, Choudhury Y, Balani P, Leung D, Dang LH, Zhao Y, Zeng J, Wang S. Combinatorial control of suicide gene expression by tissue-specific promoter and microRNA regulation for cancer therapy. Mol Ther. 2009;17(12):2058–2066. doi: 10.1038/mt.2009.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandyopadhyay S, Mitra R, Maulik U, Zhang MQ. Development of the human cancer microRNA network. Silence. 2010;1(1):6. doi: 10.1186/1758-907X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 29.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 30.Svoboda M, Izakovicova Holla L, Sefr R, Vrtkova I, Kocakova I, Tichy B, Dvorak J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33(3):541–547. [PubMed] [Google Scholar]
- 31.Yeung ML, Yasunaga J, Bennasser Y, Dusetti N, Harris D, Ahmad N, Matsuoka M, Jeang KT. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;68(21):8976–8985. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27 Kip1. J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4(11):e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu ZL, Zhao ZR, Zhang Y, Yang YH, Wang ZM, Cui DS, Wang MW, Kleeff J, Kayed H, Yan BY, Sun XF. Expression and significance of FXYD-3 protein in gastric adenocarcinoma. Dis Markers. 2010;28(2):63–69. doi: 10.3233/DMA-2010-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Down-regulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98(12):1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, Zhang CY. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28(10):1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 37.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 38.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20(3):539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28(46):4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23(11):1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, Qin LX, Man K, Lo CM, Lee J, Ng IO, Fan J, Tang ZY, Sun HC, Wang XW. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361(15):1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang L, Mao P, Song L, Wu J, Huang J, Lin C, Yuan J, Qu L, Cheng SY, Li J. miR-182 as a prognostic marker for glioma progression and patient survival. Am J Pathol. 2010;177(1):29–38. doi: 10.2353/ajpath.2010.090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125(6):1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 44.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83(7):3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.George GP, Mittal RD. MicroRNAs: Potential Biomarkers in Cancer. Indian J Clin Biochem. 2010;25:4–14. doi: 10.1007/s12291-010-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C, Wang G, Kang C, Du Y, Pu P. Up-regulation of p27(kip1) by miR-221/222 antisense oligonucleotides enhances the radiosensitivity of U251 glioblastoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26(6):634–638. doi: 10.3760/cma.j.issn.1003-9406.2009.06.006. (Article in Chinese) [DOI] [PubMed] [Google Scholar]
- 47.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 48.Mukand JA, Blackinton DD, Crincoli MG, Lee JJ, Santos BB. Incidence of neurologic deficits and rehabilitation of patients with brain tumors. Am J Phys Med Rehabil. 2001;80(5):346–350. doi: 10.1097/00002060-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 49.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 50.Parker SL, Parker MS, Sah R, Sallee F. Angiogenesis and rhodopsin-like receptors: a role for N-terminal acidic residues? Biochem Biophys Res Commun. 2005;335(4):983–992. doi: 10.1016/j.bbrc.2005.06.158. [DOI] [PubMed] [Google Scholar]
- 51.Chaudhry MA, Sachdeva H, Omaruddin RA. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010;29(9):553–561. doi: 10.1089/dna.2009.0978. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69(19):7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R, Desponts C, Teitell M, Baffa R, Aqeilan R, Iorio MV, Taccioli C, Garzon R, Di Leva G, Fabbri M, Catozzi M, Previati M, Ambs S, Palumbo T, Garofalo M, Veronese A, Bottoni A, Gasparini P, Harris CC, Visone R, Pekarsky Y, de la Chapelle A, Bloomston M, Dillhoff M, Rassenti LZ, Kipps TJ, Huebner K, Pichiorri F, Lenze D, Cairo S, Buendia MA, Pineau P, Dejean A, Zanesi N, Rossi S, Calin GA, Liu CG, Palatini J, Negrini M, Vecchione A, Rosenberg A, Croce CM. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20(5):589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mees ST, Mardin WA, Sielker S, Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M, Haier J. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol. 2009;16(8):2339–2350. doi: 10.1245/s10434-009-0531-4. [DOI] [PubMed] [Google Scholar]
- 56.Dong H, Siu H, Luo L, Fang X, Jin L, Xiong M. Investigating Gene and MicroRNA Expression in Glioblastoma. BMC Genomics. 2010;11(Suppl 3):S16. doi: 10.1186/1471-2164-11-S3-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, Ma X, Hayashi K, Sasaki T. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16(16):4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 58.Stecca B, Ruiz I, Altaba A. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2(2):84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, Chen Z. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2010;23(5):1457–1462. doi: 10.3892/or_00000784. [DOI] [PubMed] [Google Scholar]
- 60.Rauhala HE, Jalava SE, Isotalo J, Bracken H, Lehmusvaara S, Tammela TL, Oja H, Visakorpi T. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int J Cancer. 2010;127(6):1363–1372. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- 61.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4(7):e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samlos MA. PhD thesis. Case Western Reserve University; 2007. Identification and functional analysis of micro-RNAs encoded by Kaposi’s sarcoma-associated herpevirus. [Google Scholar]
- 63.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, García-Foncillas J. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 2009;4(4):e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wurdinger T, Tannous BA. Glioma angiogenesis: Towards novel RNA therapeutics. Cell Adh Migr. 2009;3(2):230–235. doi: 10.4161/cam.3.2.7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mokhtari T. [Accessed June, 2010.];Role of MicroRNAs in the Development of Cancer Stem Cells into Glioblastomas. 2008 Available at: http://realscience.breckschool.org/upper/research/Research2008/Tara.pdf.
- 67.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renee R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81(23):12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]