Abstract
Malignant pleural mesothelioma (MPM) is highly intractable and readily spreads throughout the surface of the pleural cavity, and these cells have been shown to express urokinase-type plasminogen activator receptor (uPAR). We here examined the potential of our new and powerful recombinant Sendai virus (rSeV), which shows uPAR-specific cell-to-cell fusion activity (rSeV/dMFct14 (uPA2), named “BioKnife”), for tumor cell killing in two independent orthotopic xenograft models of human. Multicycle treatment using BioKnife resulted in the efficient rescue of these models, in association with tumor-specific fusion and apoptosis. Such an effect was also seen on both MSTO-211H and H226 cells in vitro; however, we confirmed that the latter expressed uPAR but not uPA. Of interest, infection with BioKnife strongly facilitated the uPA release from H226 cells, and this effect was completely abolished by use of either pyrrolidine dithiocarbamate (PDTC) or BioKnife expressing the C-terminus–deleted dominant negative inhibitor for retinoic acid-inducible gene-I (RIG-IC), indicating that BioKnife-dependent expression of uPA was mediated by the RIG-I/nuclear factor-κB (NF-κB) axis, detecting RNA viral genome replication. Therefore, these results suggest a proof of concept that the tumor cell-killing mechanism via BioKnife may have significant potential to treat patients with MPM that is characterized by frequent uPAR expression in a clinical setting.
Introduction
Several decades ago, asbestos was widely used due to its industrial and economic advantages, nonconductiveness, thermal insulation property, and sound absorbability. Epidemiological and pathological studies, however, revealed its tumorigenic properties,1,2,3 and as a result the use of asbestos was prohibited in almost all developed countries. Because asbestos-induced tumors occur several decades after asbestos exposure, the rapidly increasing numbers of asbestos-related malignancies has become a serious issue. Malignant pleural mesothelioma (MPM) is one of these asbestos-related malignancies arising from the pleural cavity, and is highly intractable and resistant to the current standard therapeutics. In fact, MPM has a median overall survival rate of less than 30 months even when multimodality therapy is used.4,5 Therefore, novel therapeutics for this condition are urgently needed.
Urokinase-type plasminogen activator (uPA) is a member of the trypsin-like serin protease family, and is synthesized and secreted as pro-uPA, having less proteolytic activity itself.6 Pro-uPA binds to the uPA receptor (uPAR), a 55–60 kDa glycoprotein that is anchored on the cell surface by a glycosyl-phosphatidylinositol linkage,7,8 and converts pro-uPA to active-uPA, resulting in localized proteolytic activity around the cell surface.9 A number of studies have demonstrated that the active uPA plays a crucial role in tumor invasion and metastasis via extracellular matrix degradation,10 and there is a consensus that a variety of cancers overexpress uPAR and that such expression is closely associated with poor patient prognosis.11,12,13,14,15 In keeping with these findings, uPAR has been shown to be highly overexpressed in tissue samples from patients with MPM,16,17 and to be rather rarely expressed in nonmalignant tissues except under unusual circumstances such as inflammation of foci;13,14 therefore, uPAR may be a potential target molecule for the treatment of MPM.
We recently developed novel oncolytic viruses based on a recombinant Sendai virus (rSeV) that shows uPA-specific cell killing activity via cell–cell fusion.18 These viruses were designated “oncolytic rSeVs,” and had the following genetic modifications: (i) deletion of the gene encoding matrix (M) protein, which resulted in the loss of budding of secondary viral particles and accumulation of HN (hemagglutinin/neuraminidase) and F (fusion) proteins on the infected cell surface; (ii) replacement of trypsin-susceptible amino acid residues of the F-gene with protease-specific ones; and (iii) truncation of the cytoplasmic domain of the F-gene. As a result, our recent study demonstrated that uPA-dependent oncolytic rSeV (rSeV/dMFct14 (uPA2), named “BioKnife”) showed optimal performance and was applicable to various types of human malignancies,18 including highly malignant glioblastoma multiforme.19
Based on these findings, we here examined the therapeutic efficacy of BioKnife for the treatment of orthotopic xenograft models of human MPM showing extensive pleural spread. Of interest, we found that uPA is not always required for the therapeutic efficacy of BioKnife, because infection of rSeV into MPM cells facilitates tumor expression via a cytoplasmic sensor and signal transduction pathway for RNA viruses, retinoic acid-inducible gene-I (RIG-I),20 thereby facilitating BioKnife-mediated fusion-dependent apoptosis.
Results
Therapeutic potentials of BioKnife for orthotopic mouse models of human MPM
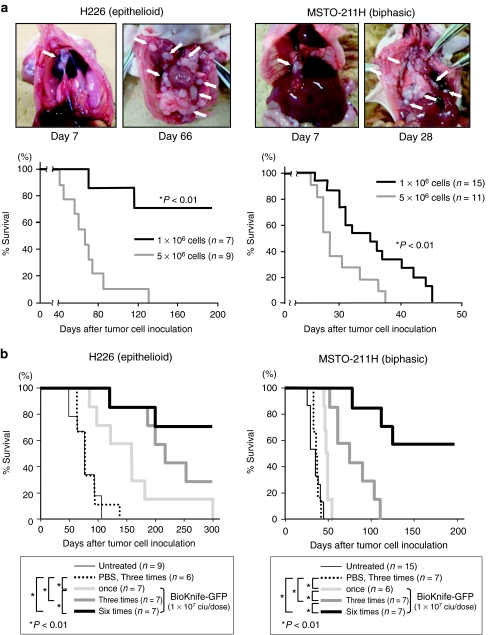
In the initial stage of this study, we established orthotopic xenograft models of MPM by injecting the human MPM cell lines H226 (epithelioid subtype) and MSTO-211H (biphasic subtype) into the right thoracic cavity at two different doses (1 × 106 and 5 × 106 cells/head). As shown in Figure 1a, mice that were intrapleurally administered either H226 or MSTO-211H cells grossly demonstrated some tumor nodules, and these nodules later increased in number in both pleural cavities (white arrows, four upper photographs). In both cases, 5 × 106 cells was considered an appropriate dose, since all the mice administered this dose were killed (within 128 days for H226 and within 38 days for MSTO-211H). Therefore, the following experiments were performed using a dose of 5 × 106 cells.
Figure 1.
Establishment of orthotopic models of two independent malignant pleural mesotheliomas (H226, epithelioid subtype; and MSTO-211H, biphasic subtype) and the therapeutic potential of uPA activity-dependent rSeV/dMFct14 (uPA2), namely, BioKnife. *P < 0.01. (a) Establishment of orthotopic models of two independent malignant pleural mesotheliomas. Note that 7 days after tumor cell inoculation, tumor nodules were already established, and thereafter the sizes and numbers of nodules increased. The bottom two graphs indicate the dose-dependent survival of H226 (left) and MSTO-211H (right). In both cases, administration of 5 × 106 cells resulted in the death of all the animals. (b) Survival curves indicating the therapeutic effect of multicycle administration of BioKnife-GFP (1 × 107 ciu/dose). Seven days after tumor cell inoculation, when the tumor nodules were almost established, BioKnife-GFP was administered via an intrathoracic route once (day 7), three times (days 7, 9, and 11) or six times (days 7, 9, 11, 13, 15, and 17). The more times administration of BioKnife significantly prolonged the survival of tumor-bearing mice in both tumor cells. The survival curves were determined using the Kaplan–Meier's method, and the log-rank test was used for comparison. ciu, cell infectious unit; GFP, green fluorescent protein; PBS, phosphate-buffered saline; rSeV, recombinant Sendai virus; uPA, urokinase-type plasminogen activator.
Next, to assess the therapeutic effect of the BioKnife, we first examined the dose–response relationship on the tumor-bearing mice via a single intrapleural administration of BioKnife expressing green fluorescent protein (GFP) on day 7 (BioKnife-GFP: 4 × 105, 2 × 106, and 1 × 107 cell-infectious units (ciu)/dose), when the tumors were established. As a result, significant survival prolongation was observed in both tumor models when a dose of 1 × 107 ciu/dose was used (data not shown); therefore, this dose was used for the following experiments.
Subsequently, single and multicycle administrations of BioKnife-GFP (once, on day 7 after tumor cell inoculation only; three times, on days 7, 9, and 11; or six times, on days 7, 9, 11, 13, 15, and 17) were examined. As shown in Figure 1b, an optimized therapeutic effect was achieved by the use of six injections of BioKnife-GFP in both models, as expected.
Evidence of BioKnife-mediated elimination of MPM tumor in vivo
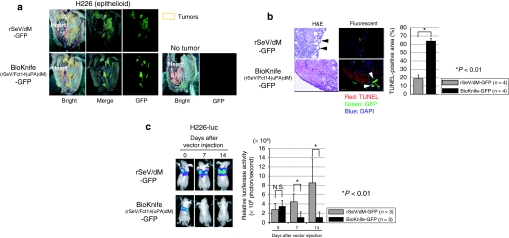
Next, we confirmed the time course of BioKnife-mediated tumor death using H226 as a model. Here we used rSeV lacking the M-gene without F-gene modification (rSeV/dM-GFP)21 as a control virus. Two days after intrapleural injection of BioKnife-GFP (1 × 107 ciu/dose) into the tumor-bearing mice (7 days after tumor cell inoculation), the bilateral thoracic cavities were opened and subjected to a fluorescent dissecting microscope. As shown in Figure 2a, fluorescence was observed and extended to almost all of tumor nodules (surrounded by brown lines). The thoracic cavities without tumor inoculation demonstrated only tiny and scattered dots on the surface, suggesting that MPM tumors were susceptible to both rSeV/dM-GFP and BioKnife-GFP. Similar findings were also observed in the case of MSTO-211H tumors (data not shown).
Figure 2.
Tumor nodule-specific infection and spread of BioKnife-GFP resulted in tumor elimination in the orthotopic MPM model in vivo. *P < 0.01. (a) Representative findings of the pleural surface of orthotopic H226-bearing mice as assessed by using a dissecting fluorescent microscope. Seven days after H226 inoculation, the control virus (rSeV/dM-GFP) or BioKnife-GFP was administrated. Two days later, both thoracic cavities were opened and observed under a dissecting fluorescent microscope. Note that both tumors, but not the adjacent mesothelial cells, exhibited extensive GFP signals, suggesting that the H226 tumors were susceptible to the rSeV infection. Similar findings were also seen when MSTO-211H cells were used (data not shown). n = 4/group. (b) Representative findings of immunohistochemical triple staining (GFP, green; TUNEL, red; and PI for nuclei, dark blue) followed by H&E staining, for the tumor sections obtained as described in a. The right graph is the image analyzer-assisted quantitative analysis of the TUNEL-positive area. Note that the tumors treated by rSeV/dM-GFP showed GFP signals only on the tumor surface, and that a large portion of the tumor cells were histologically viable. There were four mice in each group, and four tumors were excised from each mouse. Error bars in a panel represent the means ± S.D. (c) Time course of tumor reduction. An IVIS in vivo imaging system was employed to assess luciferase bioluminescence using H226 tumor cells that were stably transfected with simian immunodeficiency virus vector expressing firefly luciferase (H226-luc). Note that the in vivo luciferase activity of mice with orthotopic tumors that received rSeV/dM-GFP increased gradually, while that of the mice that received BioKnife-GFP was reduced, as shown in the quantitative analysis of the right graph, indicating that BioKnife-GFP could eliminate tumor cells in a time-dependent manner. Error bars in a panel represent the means ± S.D. n = 4/group. DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; H&E, hematoxylin and eosin; MPM, malignant pleural mesothelioma; NS, not significant; PI, propidium iodide; rSeV, recombinant Sendai virus; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
In contrast to the similar gross findings of GFP fluorescence, histological and immunohistochemical examinations of these tumors demonstrated markedly different features. A large portion of the tumors from the mice administered the control virus (rSeV/dM-GFP) was viable, and GFP expression was seen just on the surface of the tumors (Figure 2b; upper two photomicrographs, black and white arrows). On the other hand, tumors from mice administered BioKnife-GFP showed an extensive dead area, which was positive for terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining (red nuclei), adjacent to the GFP-positive tumor cells (Figure 2b; lower two photomicrographs, black and white arrows). The image analyzer exhibited a significantly larger TUNEL-positive area in BioKnife-GFP treated tumors than in the tumors treated with control virus (Figure 2b, graph).
To assess the time course of tumor elimination, we next employed an IVIS in vivo imaging system (Caliper Life Sciences, Hopkinton, MA) to assess luciferase bioluminescence using H226 tumor cells that were stably transfected with simian immunodeficiency virus vector expressing firefly luciferase (H226-luc). As shown in Figure 2c, the in vivo luciferase activity of mice with orthotopic tumors that received rSeV/dM-GFP increased gradually, while that of mice injected with BioKnife-GFP was reduced, indicating that BioKnife-GFP could eliminate this activity in a time-dependent manner.
BioKnife-mediated cell death of MPM is due to uPA- and caspase-dependent apoptosis
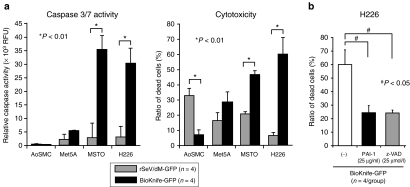
The increase in the TUNEL-positive area of tumors treated with BioKnife-GFP suggested that the BioKnife-mediated death of the tumor cells might have been due to an apoptotic mechanism. We therefore confirmed this in vitro. MPM cells (H226 and MSTO-211H) and control cells (AoSMC: human aortic smooth muscle cells; and Met5A: human normal mesothelial cells) were subjected to a cell proliferation assay (4 days after virus inoculation) and caspase 3/7 activity assay (2 days after virus inoculation), the common pathway of caspase-dependent apoptosis. As shown in Figure 3a, a significant and dramatic increase of caspase 3/7 activity was observed only in MPM cells, but not in control cells (left graph), and corresponding cytotoxicity was also seen in MPM cells (right graph). Interestingly, AoSMCs showed increased cell death when the control virus was used without enhancement of caspase 3/7 activity, suggesting that the death of these cells might be due to the direct cytopathic effect of rSeV/dM-GFP.
Figure 3.
BioKnife-mediated cell death of human MPM cells is regulated by caspase-dependent apoptosis. *P < 0.01 and #P < 0.05. (a) MPM cells (H226 and MSTO-211H) and control cells (AoSMC, human aortic smooth muscle cells; and Met5A, human normal mesothelial cells) were subjected to caspase 3/7 activity assay (left graph, 2 days after virus inoculation), and cell proliferation assay (right graph, 4 days after virus inoculation). Note that significant and dramatic increase of caspase 3/7 activity was observed only in MPM cells, but not in control cells (left graph), and corresponding cytotoxicity was also seen in MPM cells (right graph). AoSMCs showed increased cell death in use of control virus without enhancement of caspase 3/7 activity, suggesting the direct cytopathic effect of rSeV/dM-GFP. Error bars in a panel represent the means ± S.D. n = 4/group. (b) A graph indicates PAI-1 and pan-caspase inhibitor (Z-VAD-FMK, benzyloxy- carbonyl-Val-Ala-Asp-fluoromethylketone) dependent inhibition of H226 cell death via BioKnife-GFP treatment to confirm whether BioKnife-mediated cell death might be dependent on uPA/uPAR system and caspases. Error bars in a panel represent the means ± S.D. n = 4/group. GFP, green fluorescent protein; MPM, malignant pleural mesothelioma; PAI-1, plasminogen activator inhibitor-1; RFU, relative fluorescent unit; rSeV, recombinant Sendai virus; uPAR, urokinase-type plasminogen activator receptor.
To examine whether BioKnife-mediated cell death might be dependent on the uPA/uPAR system and caspases, their inhibitors, human recombinant plasminogen activator inhibitor-1 (PAI-1) and pan-caspase inhibitor (Z-VAD-FMK: benzyloxy-carbonyl-Val-Ala-Asp-fluoromethylketone)22 were used. As shown in Figure 3b, these inhibitors significantly inhibited the BioKnife-GFP-mediated cytotoxicity of H226 cells, indicating that the uPA/uPAR and caspases were essentially involved in the BioKnife-mediated cell death of H226.
Human MPM frequently expresses uPAR
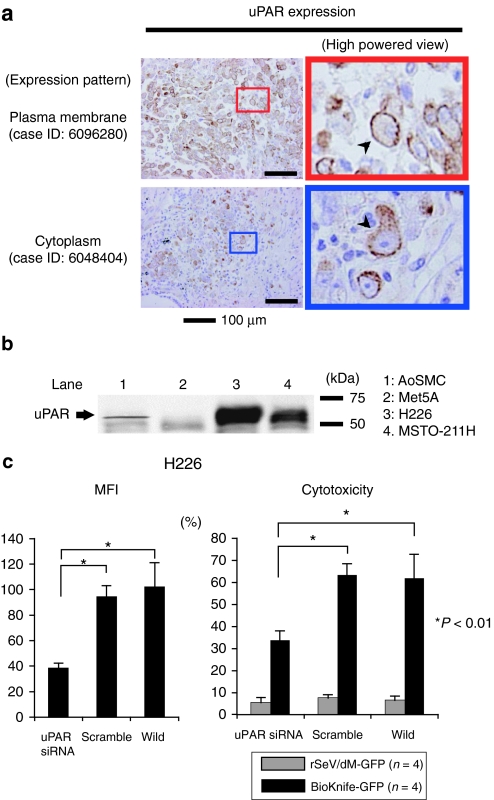
Next, we examined the expression of uPAR in human MPM by immunohistochemistry for surgical specimens (nine cases, summarized in the Supplementary Table S1) and by western blotting for the cell lines. As shown in Figure 4a and the Supplementary Table S1, uPAR expression was found in all cases tested, and, interestingly, staining signals were detected on the plasma membrane (upper two photomicrographs, arrowhead) or cytoplasm (bottom two photomicrographs, arrowhead) or both. There was no staining when isotype-matched murine IgG1 was used as the primary antibody (data not shown). Western blot analysis demonstrated strong expression of the uPAR protein, ~55 kDa, in both the MPM cell lines, H226 and MSTO-211H, as well as a small amount in AoSMCs; there was no uPAR protein in the noncancerous mesothelial cell line, Met5A (Figure 4b).
Figure 4.
uPAR expression in MPM cell lines and surgical specimens. (a) Immunohistochemical detection of uPAR expression in MPM surgical specimens (nine cases, summarized in the Supplementary Table S1). Strong immunoreactivity for uPAR expression was found in all cases tested. Note that staining signals were detected on the plasma membrane (upper two photomicrographs, arrowhead) or cytoplasm (bottom two photomicrographs, arrowhead) or both. There was no staining in use of isotype-matched control antibody (data not shown). (b) Representative western blot analysis demonstrated the strong expression of uPAR protein, ~55 kDa, in both MPM cell lines, H226 and MSTO-211H, and a small amount in AoSMCs, but not in normal mesothelial cell Met5A at all. (c) Graphs indicating the essential role of uPAR on the effect of BioKnife-mediated cytotoxicity via siRNA-mediated knockdown. Left graph shows the siRNA-specific downregulation of uPAR expression on cell surface expressed by mean fluorescent intensity (MFI) assessed by flow cytometry. Right graph indicates the corresponding cytotoxic effect of BioKnife-GFP as well as of control virus. Error bars in panels represent the means ± S.D. n = 4/group. AoSMC, human aortic smooth muscle cells; GFP, green fluorescent protein; Met5A, human normal mesothelial cells; MPM, malignant pleural mesothelioma; siRNA, small interfering RNA; uPAR, urokinase-type plasminogen activator receptor.
We further assessed the role of uPAR expression on the effect of BioKnife-mediated cytotoxicity via small interfering RNA (siRNA)-mediated knockdown for the “loss of function” study. Left graph in Figure 4c shows the siRNA-specific downregulation of uPAR expression on cell surface expressed by mean fluorescent intensity assessed by flow cytometry. Corresponding right graph indicates the cytotoxic effect of BioKnife-GFP but not of control virus, indicating that the expression of uPAR is essential to the biological activity of BioKnife.
BioKnife stimulates uPA expression via a RIG-I- and NF-κB-dependent mechanism
We subsequently confirmed uPA release from MPM cells into the culture medium by casein zymography. Surprisingly, H226 cells that were highly susceptible to the BioKnife treatment did not express uPA at all (Figure 5a). A similar result was found by uPA-specific enzyme-linked immunosorbent assay (data not shown). We therefore hypothesized that the infection with BioKnife might stimulate uPA expression. This hypothesis was based in part on the previous demonstration that genome replication of SeV is recognized by a cytoplasmic sensor for RNA viruses, the DExD/H-box RNA helicase known as RIG-I and thereafter activates nuclear factor-κB (NF-κB).20 In addition, it has been reported that NF-κB/Rel was essential for the constitutive expression of uPA in some cancer cells;23 therefore, such a signal transduction pathway might be involved.
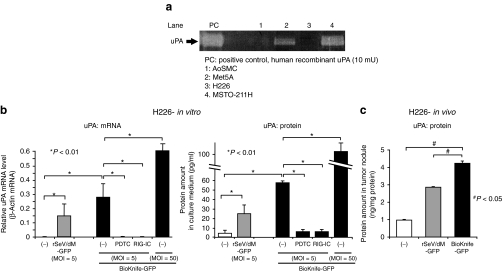
Figure 5.
BioKnife infection stimulates uPA-negative H226 cells to express uPA through RIG-I and NF-κB dependent signal transduction pathway. (a) Representative casein zymography demonstrated the strong expression of active uPA in mesothelial cell line Met5A and MPM cell line MSTO-211H, and very small amount in AoSMCs, but not in MPM cell line H226 at all. (b) Graphs indicating that the infection of rSeV-based vectors, including rSeV/dM-GFP as well as BioKnife-GFP, stimulate uPA in mRNA (left graph) and protein (right graph) levels. Note the dose-dependent upregulation and that addition of PDTC, an inhibitor for NF-κB, to the culture medium and BioKnife-expressing RIG-IC, a C-terminus-deleted dominant negative inhibitor for RIG-I, completely abolished the expression of uPA mediated by rSeV. Error bars in a panel represent the means ± S.D. n = 4/group. (c) The graph indicates that rSeV-based vectors, rSeV/dM-GFP as well as BioKnife-GFP, stimulated uPA expression from uPA-negative H226 tumors in vivo. Seven days after intrapleural tumor inoculation into the nude mice, 1 × 107 ciu/dose of rSeV/dM-GFP or BioKnife-GFP was administered into the thoracic cavity. Two days later, the mice were killed, the tumor nodules were enucleated, and the protein extracts were subjected to human uPA-specific ELISA. The error bars in the panel represent the means ± S.D. n = 4/group. AoSMC, human aortic smooth muscle cells; ciu, cell infectious unit; ELISA, enzyme-linked immunosorbent assay; GFP, green fluorescent protein; Met5A, human normal mesothelial cells; MOI, multiplicity of infection; MPM, malignant pleural mesothelioma; mRNA, messenger RNA; NF-κB, nuclear factor-κB; PDTC, pyrrolidine dithiocarbamate; RIG-I, retinoic acid-inducible gene-I; rSeV, recombinant Sendai virus; uPA, urokinase-type plasminogen activator.
To clarify this hypothesis, we here tested the effect of the NF-κB inhibitor pyrrolibine dithiocarbamate (PDTC) and BioKnife dually expressing both GFP and t he dominant negative mutant RIG-I (RIG-IC)19,24 on the BioKnife-mediated uPA expression from H226 cells. As shown in Figure 5b, both the messenger RNA and protein expressions of uPA were observed when either the control virus rSeV/dM-GFP or BioKnife GFP was used, suggesting that a common mechanism of SeV infection might be crucial to these findings. Importantly, treatment of PDTC as well as use of BioKnife-GFP expressing RIG-IC almost completely abolished the expression of uPA, indicating that a RIG-I- and NF-κB–dependent signal transduction pathway is a key to the inducible expression of uPA from MPM. In addition, use of more BioKnife, multiplicity of infection (MOI) = 50, showed apparent dose–response effect on the stimulation of endogenous uPA expression.
Finally, we confirmed that similar enhancement of uPA expression due to rSeV-based vectors might be representative in vivo in an H226 orthotopic MPM model. Seven days after intrapleural tumor inoculation into the nude mice, a 1 × 107 ciu/dose of rSeV/dM-GFP or BioKnife-GFP was administrated into the thoracic cavity. Two days later, the mice were killed, the tumor nodules were enucleated, and the protein extracts were subjected to a human uPA-specific enzyme-linked immunosorbent assay. As shown in Figure 5c, the tumors treated with control virus showed upregulation of uPA expression, a finding that was significantly enhanced by BioKnife-GFP treatment.
Altogether, these findings strongly suggested that BioKnife would be effective to treat MPM, because this type of tumor frequently expresses uPAR. Importantly, tumor cell-derived uPA, a ligand protease of uPAR, may not be seriously important to the therapeutic effect of BioKnife, because infection with BioKnife itself stimulates uPA expression from MPM via a RIG−/− and NF-κB–dependent pathway for autocrine activation of this type of fusogenic oncolytic virus.
Discussion
We here investigated the therapeutic potential and antitumor mechanism of our uPA activity-dependent oncolytic virus, BioKnife, to treat murine models of MPM. The key observations made in this study were as follows: (i) The repeated administration of BioKnife was effective and significantly prolonged the survival of two independent orthotopic murine models of MPM in vivo; (ii) BioKnife-mediated tumor cell death was mediated by caspase-dependent apoptosis; (iii) human MPM frequently expressed uPAR protein, but expression of uPA was not always seen; and (iv) uPA activity-dependent BioKnife was also effective even on uPA-negative MPM, because infection of BioKnife itself stimulated the expression of uPA from MPM cells through a RIG-I- and NF-κB–mediated signaling pathway. These findings suggest the potential utility of BioKnife to treat MPM in clinical settings.
In general, MPM spreads throughout the pleural cavity, and metastasis of MPM to distant sites is somewhat rare.26 Such characteristics of MPM may be a favorable feature for oncolytic virotherapy using Bioknife. As shown in Figure 2, the tumor nodules infected by BioKnife led to widespread infection among uPAR-expressing cells via cell–cell fusion activity. Although BioKnife theoretically cannot be delivered to the distant metastasis, BioKnife has significant benefit in the eradication of adjacent malignant tumors.
In the present study, we have shown that MPM would be a good target of uPA activity-dependent BioKnife, because the majority of MPM in this series expressed uPAR, as shown in Figure 4 and the Supplementary Table S1, and uPAR plays a key role in tumor cell invasion, migration, angiogenesis, and metastasis.10 This finding of frequent uPAR expression in MPM has been confirmed by other groups.16,17 Importantly, we here demonstrated that the release of uPA from MPM was not a serious issue in terms of the therapeutic efficacy of BioKnife, because infection of BioKnife to MPM cells strongly stimulated uPA expression. Furthermore, we demonstrated that rSeV-mediated induction of uPA was controlled by a RIG-I- and NF-κB–dependent signal transduction pathway (Figure 5). To our knowledge, this is the first study to report the stimulation of uPA expression after SeV infection.
The data obtained in this study confirmed that uPA activated by uPAR is critical on the effect of BioKnife vector. Both expression of uPA and uPAR in tumor cells is important for the effect of BioKnife, and in turn, we here found that the low expression of uPA could be compensated by the signals related to the BioKnife infection. A contradictory finding, however, that somewhat effect of BioKnife on nonmalignant Met5A cells (Figure 3a) showing ~30% cytotoxicity that express uPA but not uPAR at all. This may be explained by an important study that shows uPAR-independent mechanism for activating uPA at cell surface with low affinity and high capacity.25,26 These findings suggest that (i) uPAR expression enhances the virus' cytotoxic effects on tumor cells, however, (ii) cytotoxicity may still occur in tumors without significant uPAR expression. This also implies that the cytotoxic effect of BioKnife would be predicted by uPAR expression level on tumor cells, but uPAR-negative tumor may not always be resistant to BioKnife. These theoretical considerations are clearly important to determine the possible biomarkers predicting the outcome in future research and development of BioKnife in clinical setting. Therefore, more basic research is needed to predict the efficacy of the BioKnife.
Considering the clinical setting, we should emphasize that the current preclinical protocol could be applicable to patients. For instance, video-assisted thoracoscopic surgery27 and a chest tube could be used as an administration route. Importantly, MPM often forms nodular lesions on the pleural surface, which is reflected in the orthotopic MPM murine model in Figure 2. This accessible route could enable us to administer BioKnife repeatedly and safely. Therefore, further preclinical studies using large animals are warranted to examine these techniques in more clinically relevant situations.
In summary, we here demonstrated the antitumoral activity, survival benefit, and applicability of repeated intrathoracic administration of BioKnife, a uPA activity-dependent oncolytic SeV vector, to the MPM in vitro and in vivo. It is important to note that these effects were independent of the release of uPA from tumor cells, because infection of BioKnife could stimulate uPA expression through a RIG-I- and NF-κB–dependent mechanism. Therefore, these findings suggest the potential utility of BioKnife for the treatment of MPM, and further studies are warranted to examine whether this new modality could be effective in a clinical setting as a therapeutic alternative for this intractable disease.
Materials and Methods
Cells and reagents. The human mesothelioma cell lines MSTO-211H and NCI-H226 and the SV40 transformed human mesothelial cell line Met5A (all purchased from American Type Culture Collection, Rockville, MD) were maintained in RPMI-1640 containing 10% fetal bovine serum, 100 IU/ml penicillin, and 100 µg/ml streptomycin. AoSMC (EIDIA, Tokyo, Japan) were maintained in the presence of smooth muscle cell basal medium-2 (EIDIA) with 5% fetal bovine serum, 0.1% GA-1000 (gentamicyn and amphotericin B), 0.1% insulin, 0.2% human basic fibroblastic growth factor (EIDIA), and 0.1% human epidermal growth factor (EIDIA). Cells were used at passage 4–10 for the experiments. The cells were maintained in a humidified atmosphere of 5% CO2 in air at 37 °C.
Z-VAD-FMK, an irreversible, cell permeable, and broad caspase inhibitor, was purchased from Medical & Biological Laboratories (Nagoya, Japan) and used as previously described.22
Establishment of the in vivo orthotopic xenograft model and treatment. The animal experiments were reviewed and approved by the Institutional Committee for Animal Care and Use and by the Biosafety Committee for Recombinant DNA experiments of Kyushu University. These experiments were also done in accordance with the recommendations for the proper care and use of laboratory animals and according to The Law (No. 105) and Notification (No. 6) of the Japanese Government. The MPM in vivo orthotopic xenograft models were established as previously described.28 In brief, 4-week-old male balb/c nu/nu mice (Charles Liver Grade; KBT Oriental, Tosu, Saga, Japan) were kept under humane conditions in the animal care facility. MPM cell lines (H226 and MSTO-211H) in log growth phase were collected. After adequate anesthesia, appropriate numbers of tumor cells in 100 µl Hank's buffered salt solution were injected into the left pleural cavity. When tumor cells were accidentally injected into the subcutaneous space or when mice showed pneumothorax or hemothorax, the animals were killed by over-anesthesia and excluded from further analysis. The day the cells were inoculated into the mice was defined as day 0. Seven days later, when the tumor nodules were established, appropriate vectors (1 × 107 ciu/dose) were administered into the left pleural cavity.
Construction and recovery of vectors. The control vector, rSeV/dM-GFP, and BioKnife-GFP (rSeV/Fct14 (uPA)dM-GFP) were constructed as described previously,18,19,21 and all schematic structures of viruses used in this study are shown in the Supplementary Figure S1. In brief, the parent plasmid pSeV18+/dM-GFP, in which the GFP had been substituted for the deleted M gene,21 was digested with Sal I and Nhe I, and the F gene fragment (9,634 bp) was subcloned into LITMUS 38 (New England Biolabs, Beverly, MA). Site-directed mutagenesis was performed using a Quick-Change Mutagenesis Kit (Stratagene, La Jolla, CA), and the mutated F gene was returned to the pSeV18+/dF-GFP backbone. Recovery and amplification of the SeV vector were performed as follows: briefly, LLC-MK2 cells were transfected with a plasmid mixture containing each plasmid–pSeV+18/Fct14 (uPA2)dM-GFP, pGEM-NP, pGEM-P, and pGEM-L–in 110 µl of Superfect reagent (Qiagen, Tokyo, Japan). The transfected cells were maintained for 3 hours, washed three times and incubated for 60 hours in minimum essential medium containing araC. The cells were collected and lysed by three cycles of freezing and thawing. The lysate solution was incubated on the F/M-expressing LLC-MK2 cells in a 24-well plate. Twenty-four hours later, the cells were washed and incubated in minimum essential medium containing araC and 7.5 µg/ml trypsin plus 10 ng/ml urokinase (Cosmobio, Tokyo, Japan). The virus yield was expressed in ciu, as previously described.17,18
Virus titration. The virus titers were expressed as ciu, which were estimated by infecting confluent LLC-MK2 cells in a 6-well plate with diluted solution as previously described.17,18 In brief, LLC-MK2 cells were inoculated in duplicate with a series of dilutions of virus, then incubated for 1 hour and washed twice with phosphate-buffered saline (PBS). Two days after infection, cells were fixed in methanol, incubated with anti-SeV primary antibodies, and then incubated with FITC-labeled goat anti-rabbit IgG (H+L) (Invitrogen, Carlsbad, CA). Immunofluorescent-positive cells were counted and ciu/ml were calculated.
Immunofluorescence staining. The tumors were excised and fixed with 4% paraformaldehyde. Paraffin-embedded samples were cut into sections of 3 µm thickness. Sections were deparaffinized, incubated at 4 °C with anti-GFP (rabbit polyclonal IgG), and subjected to Alexa Fluor 488 conjugate (A-21311, used at 1:300; Invitrogen, Tokyo, Japan). After washing in PBS, the sections were stained by the TUNEL method using an In Situ Cell Death Detection Kit, TMR Red (Roche Diagnostics, Tokyo, Japan) according to the manufacturer's protocol. After staining with 4′,6-diamidino-2-phenylindole (DAPI) (10 µg/ml), the sections were mounted with Vectashield (Vector Laboratories, Burlingame, CA) and analyzed with an all-in-one fluorescent microscope BZ-9000 (Keyence Japan, Osaka, Japan). After immunofluorescent imaging, the sections were re-stained with hematoxylin–eosin. The TUNEL-positive area in tumors was quantified by Image J software from NIH Images. Four tumors were extracted from each of four individual mice.
Luciferase in vivo imaging. H226-luc cells that stably express firefly luciferase were constructed and cloned as follows. Vesicular stomatitis virus G-protein pseudotyped simian immunodeficiency virus vector expressing luciferase (VSV-G-SIV-luciferase) was prepared as previously described.29 H226 cells were transfected with VSV-G-SIV-luciferase at a MOI of 25. Single cells were seeded in a 96-well plate, and then the clone most stably and strongly expressing luciferase was selected using an IVIS Imaging System. Cloned H226-luc cells (1 × 106) were inoculated into the pleural cavity in the manner described above. Seven days later, tumor-bearing mice were treated with a control vector or BioKnife-GFP, and in vivo luciferase imaging was performed using an IVIS 50 instrument at different times (day 0, 7 and 14 after vector injection) after administering the substrate D-luciferin Potassium Salt (Wako, Osaka, Japan) diluted with Dulbecco's PBS (Life Technologies, Tokyo, Japan) at 150 mg/kg by intraperitoneal injection. The mice were fixed on a warmed dark box, 15 minutes after injecting D-luciferin, photons from the bioluminescent tumor in the pleural cavity were detected. The exposure time was 1 minute, the field of view was 10, the binning was medium, and the f-stop was 1. Regions of interest in the tumor image were quantified as photons/second using Living Image software (Caliper LifeSciences, Hopkinton, MA).
Caspase 3/7 activity assay. Caspase-3/7 activities were measured using an Apo-OneTM (superior position) Homogeneous Caspase-3/7 Assay (Promega, Tokyo, Japan). Cells were seeded in a 96-well plate at a density of 5 × 103 cells/well. Twelve hours later, vectors were added to each well at an MOI of 20, and wells with the same volume of PBS added were used as a negative control. Forty-eight hours later, 100 µl of assay reagent (profluorescent substrate Z-DEVD-R110) was added to each well, and then the wells were incubated at room temperature for 30 minutes. The fluorescence of each well was measured at an excitation wavelength of 485 nm and emission wavelength of 535 nm using Tristar LB941 (Berthold Japan, Tokyo, Japan). Caspase-3/7 activity was determined as assay relative fluorescent unit (RFU)–negative control RFU. All experiments were carried out in triplicate.
Cytotoxicity assay. Cytotoxicity was evaluated by Cell Count Reagent SF (Nacalai Tesque, Kyoto, Japan) according to the manufacturer's instructions. Briefly, WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] assay30 is a colorimetric assay based on the ability of viable cells to produce a water-soluble formazan dye by mitochondrial succinate-tetrazolium reductase activity of viable cells. The relative cytotoxicity was calculated as follows: Cytotoxicity (%) = (1−(experimental absorbance − background absorbance)/(absorbance of negative controls − background absorbance)) × 100%. All experiments were performed in quadruplicate.
Immunohistochemistry. A total of nine MPM patients who underwent surgical resection at Kyushu University Hospital between June 2001 and October 2009 were examined. Written informed consent for the comprehensive use of the pathological materials was previously obtained from all the patients, and the study protocol was approved by the institutional ethics review board at Kyushu University. Briefly, 10% formalin-fixed and paraffin-embedded 3-µm–thick sections were deparaffinized, and were incubated in 3% hydrogen peroxidase in ethanol for 30 minutes at room temperature to quench endogenous peroxidase activity. The sections were stained with mouse monoclonal antibody against human uPAR (1:100, clone #3936; American Diagnostica, Stamford, CT) or nonimmunized murine IgG2a (isotype matched) at 4 °C overnight. Then the sections were treated for 60 minutes at room temperature with secondary antibody. Staining for uPAR was completed using EnVision+ Mouse, (Dako Japan, Tokyo, Japan), and then the slides were counterstained with hematoxylin. Evaluation of tissue staining was performed by two independent examiners (Y.M. and K.I.).
Western blotting. Samples were lysed in lysis buffer containing 50 mmol/l Tris HCL (pH 6.8) and 10% sodium dodecyl sulfate (SDS), and the protein concentration for each sample was determined by using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA). Mouse anti-human uPAR monoclonal antibody (American Diagnostica) was used as the primary antibody. Specific bands were visualized using an ECL Plus Western Blotting Detection System (GE Healthcare, Little Chalfont, UK).
uPAR gene silencing by siRNA. Transfection of H226 with siRNA was done using the Lipofectamine RNAiMAX Transfection Reagent (Invitrogen) according to the manufacturer's protocol. Briefly, siRNAs, a siRNA targeting uPAR and scrambled control (sc-36781 and sc-37007, Santa Cruz Biotechnology, Santa Cruz, CA), were used at a final concentration of 10 nmol/l, and transfection reagent was used at the dilution of 1:500 (vol/vol). The knockdown of uPAR (CD87) was assessed by flow cytometry was performed using FACS Calibur (Becton, Dickinson and Company, Franklin Lakes, NJ). Cells (1 × 105) were stained with PE conjugated anti-CD87 monoclonal antibody (clone vim5) or isotype-matched control IgG1κ (BD Biosciences, Franklin Lakes, NJ). The data was analyzed as flow cytometric histograms of ~10,000 events using FlowJo software (Tree Star, Ashland, OR).
Casein zymography. Briefly, serum-free supernatants from each cell culture were subjected to 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) containing 0.1% casein plus 4 mg of plasminogen or 0.1% gelatin under a non-reducing condition. The gel was then incubated at room temperature in a 2.5% Triton-X100 solution containing 0.05% sodium azide and 0.05 mol/l Tris (pH 7.5) followed by incubation at 37 °C in 0.1 mol/l Tris-HCl (pH 8.3)/0.5 mol/l sodium chloride solution for 16 hours. The gel was then stained with a 0.25% Coomassie Brilliant Blue, 1.1% acetic acid, 45.5% methanol solution for 40 minutes, followed by destaining in a 25% methanol/10% acetic acid mixture. Enzymatic activity was visualized as clear zones on a blue background.
Quantitative reverse transcription-PCR. Cells were placed in 24-well plates at 5 × 104/well, and each virus was added to the plate at an MOI of 5. To inhibit NF-κB, 50 µmol/l PDTC (final concentration) was added to the well 2 hours before vector infection. Twenty-four hours later, total RNA was extracted and cDNA was synthesized using a SuperscriptIII cells direct cDNA synthesis system (Invitrogen) according to the manufacturer's protocol. Real-time PCR was performed in a Light-Cycler 480 System (Roche Diagnostics) using a QuantiFast SYBR Green PCR Kit (Qiagen) according to the manufacturer's protocol. The specific primer set used in this study was as follows: uPA forward primer (5′-GGAGATGAAGTTTGAGGTGGA-3′), uPA reverse primer (5′-GCAATGTCGTT-GTGGTGAG-3′), β-actin forward primer (5′-CTGGCACCACACCTTCTACAATG-3), β-actin reverse primer (5′-GGCGTACAGGGATAGCACAGC-3). All experiments were carried out in triplicate, and data were analyzed using the Light Cycler 480 software (Roche Diagnostics, Indianapolis, IN). The messenger RNA expression levels of uPA were standardized using β-actin messenger RNA expression levels in each sample.
Enzyme-linked immunosorbent assay. The protein amount of uPA was determined using an IMUBIND uPA kit according to the manufacturer's protocol (American Diagnostica). The experimental conditions were the same as above. The culture medium was used as samples; therefore, receptor-unbound pro-uPA was expressed as the data. Three independent experiments were performed.
Statistical analysis. All data were expressed as the means ± SD. The data were examined statistically using one-way analysis of variance with Scheffe's adjustment. When the number of evaluated groups was small, the data were subjected to the Kruskal–Wallis or the Mann–Whitney U-test. The survival curves were determined using the Kaplan–Meier's method. The log-rank test was used for comparison. A probability value of P < 0.05 was considered statistically significant. Statistical analyses were determined using StatView software (SAS Institute, Cary, NC).
SUPPLEMENTARY MATERIAL Figure S1. Schematic demonstration of the genome structure of vectors used in this study. Table S1. Patients characteristics and immunohistochemical uPAR expression patterns.
Acknowledgments
We thank Drs Takayama and Nakagaki (Department of Respiratory Medicine, Kyushu University) for their generous technical advice on the orthotopic MPM xenograft model, and Kenzaburo Tani (Department of Advanced Molecular and Cell Therapy, Kyushu University Hospital) for his help in the use of the IVIS imaging system. Drs Kitao, Iimori, and Zhao are also gratefully acknowledged for their helpful input. We thank Chie Arimatsu, Aki Furuya, and Ryoko Nakamura for their assistance with the animal experiments, and Drs Tagawa, Kanaya, Ban, and Hironaka for their excellent technical assistance in the vector construction and large-scale production. KN International, Ltd. assisted in the revision of the language in this manuscript. This work was supported in part by a grant from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to Y.M. and Y.Y.). Y.Y. is a member of the Scientific Advisory Board of DNAVEC Corporation. The other authors declared no conflict of interest.
Supplementary Material
Schematic demonstration of the genome structure of vectors used in this study.
Patients characteristics and immunohistochemical uPAR expression patterns.
REFERENCES
- Stanton MF, Layard M, Tegeris A, Miller E, May M, Morgan E.et al. (1981Relation of particle dimension to carcinogenicity in amphibole asbestoses and other fibrous minerals J Natl Cancer Inst 67965–975. [PubMed] [Google Scholar]
- Marinaccio A, Binazzi A, Cauzillo G, Cavone D, Zotti RD, Ferrante P.et al. (2007Analysis of latency time and its determinants in asbestos related malignant mesothelioma cases of the Italian register Eur J Cancer 432722–2728. [DOI] [PubMed] [Google Scholar]
- Murayama T, Takahashi K, Natori Y., and, Kurumatani N. Estimation of future mortality from pleural malignant mesothelioma in Japan based on an age-cohort model. Am J Ind Med. 2006;49:1–7. doi: 10.1002/ajim.20246. [DOI] [PubMed] [Google Scholar]
- Sugarbaker DJ, Flores RM, Jaklitsch MT, Richards WG, Strauss GM, Corson JM.et al. (1999Resection margins, extrapleural nodal status, and cell type determine postoperative long-term survival in trimodality therapy of malignant pleural mesothelioma: results in 183 patients J Thorac Cardiovasc Surg 11754–63; discussion 63–65. [DOI] [PubMed] [Google Scholar]
- Bölükbas S, Manegold C, Eberlein M, Bergmann T, Fisseler-Eckhoff A., and, Schirren J. Survival after trimodality therapy for malignant pleural mesothelioma: Radical Pleurectomy, chemotherapy with Cisplatin/Pemetrexed and radiotherapy. Lung Cancer. 2011;71:75–81. doi: 10.1016/j.lungcan.2009.08.019. [DOI] [PubMed] [Google Scholar]
- Petersen LC, Lund LR, Nielsen LS, Danø K., and, Skriver L. One-chain urokinase-type plasminogen activator from human sarcoma cells is a proenzyme with little or no intrinsic activity. J Biol Chem. 1988;263:11189–11195. [PubMed] [Google Scholar]
- Nielsen LS, Kellerman GM, Behrendt N, Picone R, Danø K., and, Blasi F. A 55,000-60,000 Mr receptor protein for urokinase-type plasminogen activator. Identification in human tumor cell lines and partial purification. J Biol Chem. 1988;263:2358–2363. [PubMed] [Google Scholar]
- Ploug M, Kjalke M, Rønne E, Weidle U, Høyer-Hansen G., and, Danø K. Localization of the disulfide bonds in the NH2-terminal domain of the cellular receptor for human urokinase-type plasminogen activator. A domain structure belonging to a novel superfamily of glycolipid-anchored membrane proteins. J Biol Chem. 1993;268:17539–17546. [PubMed] [Google Scholar]
- Vassalli JD, Baccino D., and, Belin D. A cellular binding site for the Mr 55,000 form of the human plasminogen activator, urokinase. J Cell Biol. 1985;100:86–92. doi: 10.1083/jcb.100.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi F., and, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- Alpízar-Alpízar W, Nielsen BS, Sierra R, Illemann M, Ramírez JA, Arias A.et al. (2010Urokinase plasminogen activator receptor is expressed in invasive cells in gastric carcinomas from high- and low-risk countries Int J Cancer 126405–415. [DOI] [PubMed] [Google Scholar]
- Pedersen H, Brünner N, Francis D, Osterlind K, Rønne E, Hansen HH.et al. (1994Prognostic impact of urokinase, urokinase receptor, and type 1 plasminogen activator inhibitor in squamous and large cell lung cancer tissue Cancer Res 544671–4675. [PubMed] [Google Scholar]
- Bianchi E, Cohen RL, Thor AT, Todd RF, 3rd, Mizukami IF, Lawrence DA.et al. (1994The urokinase receptor is expressed in invasive breast cancer but not in normal breast tissue Cancer Res 54861–866. [PubMed] [Google Scholar]
- Li Y., and, Cozzi PJ. Targeting uPA/uPAR in prostate cancer. Cancer Treat Rev. 2007;33:521–527. doi: 10.1016/j.ctrv.2007.06.003. [DOI] [PubMed] [Google Scholar]
- de Bock CE., and, Wang Y. Clinical significance of urokinase-type plasminogen activator receptor (uPAR) expression in cancer. Med Res Rev. 2004;24:13–39. doi: 10.1002/med.10054. [DOI] [PubMed] [Google Scholar]
- Idell S, Pueblitz S, Emri S, Gungen Y, Gray L, Kumar A.et al. (1995Regulation of fibrin deposition by malignant mesothelioma Am J Pathol 1471318–1329. [PMC free article] [PubMed] [Google Scholar]
- Shetty S, Kumar A, Johnson A, Pueblitz S., and, Idell S. Urokinase receptor in human malignant mesothelioma cells: role in tumor cell mitogenesis and proteolysis. Am J Physiol. 1995;268 6 Pt 1:L972–L982. doi: 10.1152/ajplung.1995.268.6.L972. [DOI] [PubMed] [Google Scholar]
- Kinoh H, Inoue M, Komaru A, Ueda Y, Hasegawa M., and, Yonemitsu Y. Generation of optimized and urokinase-targeted oncolytic Sendai virus vectors applicable for various human malignancies. Gene Ther. 2009;16:392–403. doi: 10.1038/gt.2008.167. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Kinoh H, Iwadate Y, Onimaru M, Ueda Y, Harada Y.et al. (2010Urokinase-targeted fusion by oncolytic Sendai virus eradicates orthotopic glioblastomas by pronounced synergy with interferon-ß gene Mol Ther 181778–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M.et al. (2004The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses Nat Immunol 5730–737. [DOI] [PubMed] [Google Scholar]
- Inoue M, Tokusumi Y, Ban H, Kanaya T, Shirakura M, Tokusumi T.et al. (2003A new Sendai virus vector deficient in the matrix gene does not form virus particles and shows extensive cell-to-cell spreading J Virol 776419–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Ikeda Y, Yonemitsu Y, Onimaru M, Nakagawa K, Kohno R.et al. (2008Inhibition of nuclear translocation of apoptosis-inducing factor is an essential mechanism of the neuroprotective activity of pigment epithelium-derived factor in a rat model of retinal degeneration Am J Pathol 1731326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuning U, Guerrini L, Nishiguchi T, Page S, Seibold H, Magdolen V.et al. (1999Rel transcription factors contribute to elevated urokinase expression in human ovarian carcinoma cells Eur J Biochem 259143–148. [DOI] [PubMed] [Google Scholar]
- Okano S, Yonemitsu Y, Shirabe K, Kakeji Y, Maehara Y, Harada M.et al. (2011Provision of continuous maturation signaling to dendritic cells by RIG-I-stimulating cytosolic RNA synthesis of Sendai virus J Immunol 1861828–1839. [DOI] [PubMed] [Google Scholar]
- Brenner J, Sordillo PP, Magill GB., and, Golbey RB. Malignant mesothelioma of the pleura: review of 123 patients. Cancer. 1982;49:2431–2435. doi: 10.1002/1097-0142(19820601)49:11<2431::aid-cncr2820491134>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Longstaff C, Merton RE, Fabregas P., and, Felez J. Characterization of cell-associated plasminogen activation catalyzed by urokinase-type plasminogen activator, but independent of urokinase receptor (uPAR, CD87) Blood. 1999;93:3839–3846. [PubMed] [Google Scholar]
- Yoshino I, Yasunaga T, Hashizume M., and, Maehara Y. A novel endoscope manipulator, Naviot, enables solo-surgery to be performed during video-assisted thoracic surgery. Interact Cardiovasc Thorac Surg. 2005;4:404–405. doi: 10.1510/icvts.2005.108282. [DOI] [PubMed] [Google Scholar]
- Martarelli D, Catalano A, Procopio A, Orecchia S, Libener R., and, Santoni G. Characterization of human malignant mesothelioma cell lines orthotopically implanted in the pleural cavity of immunodeficient mice for their ability to grow and form metastasis. BMC Cancer. 2006;6:130. doi: 10.1186/1471-2407-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Ikeda Y, Yonemitsu Y, Miyazaki M, Inoue M, Hasegawa M.et al. (2010Inhibition of choroidal neovascularization via brief subretinal exposure to a newly developed lentiviral vector pseudotyped with Sendai viral envelope proteins Hum Gene Ther 21199–209. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Min W., and, Lillehoj HS. Lymphocyte proliferation response during Eimeria tenella infection assessed by a new, reliable, nonradioactive colorimetric assay. Avian Dis. 2002;46:10–16. doi: 10.1637/0005-2086(2002)046[0010:LPRDET]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic demonstration of the genome structure of vectors used in this study.
Patients characteristics and immunohistochemical uPAR expression patterns.