Abstract
A large-scale mapping of the worker-honeybee brain proteome was achieved by MudPIT. We identified 2,742 proteins from forager and nurse honeybee brain samples, 17% of the total proteins were found to be differentially expressed by spectral count sampling statistics and a G-test. Sequences were compared with the EuKaryotic Orthologous Groups (KOG) catalog set using BLASTX, and then categorized into the major KOG categories of most similar sequences. According to this categorization, nurse brain showed increased expression of proteins implicated in translation, ribosomal structure and biogenesis (14.5%) compared with forager (1.8%). Experienced foragers overexpressed proteins involved in energy production and conversion, showing an extensive difference in this set of proteins (17%) in relation to the nurse subcaste (0.6%). Examples of proteins selectively expressed in each subcaste were analyzed. A comparison between these MudPIT experiments and previous 2-DE experiments revealed nine coincident proteins differentially expressed in both methodologies.
Keywords: Apis mellifera, brain proteome, differential expression, honeybee subcastes, MudPIT
1. Introduction
Honeybee (Apis mellifera) has been used as a model system in a variety of ethological and behavioral studies, such as navigation, social organization and learning1–5. Labor division in A. mellifera provides an example of a social behavior evolution that is associated with changes in gene regulation, which influences temporal patterns of gene expression6–7. Despite showing complex social behavior, the honeybee constitutes an easily accessible animal with a small, simple brain. In addition, its sequenced genome8 makes this organism a powerful model for comparative proteomic studies.
Numerous studies of neurophysiology and behavior in the honeybee have used transcriptomic techniques, such as expressed sequence tags and cDNA microarrays analysis, to identify genes differentially regulated during caste and subcaste differentiation9–12. Studies at the genomic and transcriptomic levels associated with deeper proteomic analyses are necessary to develop a complementary understanding of the ontogenetic and behavioral transitions in A. mellifera. Recent proteomic studies on A. mellifera have focused on nurse hypopharyngeal gland secretion13 or royal jelly14, brain neuropeptides15, brain mushroom bodies16, honeybee thorax17, and, recently, differences in the whole-body protein profiles of the nurses and foragers18–19.
In a previous report, our group performed comparative proteomic analysis of A. mellifera brain from nurse and forager worker subcastes using two-dimensional gel electrophoresis (2-DE) within a pH range of 4–7 followed by MALDI-TOF mass spectrometry to identify proteins20. A known drawback of our past study was that certain types of proteins possessing important cellular functions were notably difficult to separate or detect using 2-DE. These proteins include membrane, low copy number, highly basic and very large (>150 kDa) or small (<10 kDa) species.
Large-scale analysis strategies such as Multi-dimensional Protein Identification Technology (MudPIT)21–22 have been increasingly used in proteomic projects, allowing analysis via liquid chromatography coupled to mass spectrometry. It efficiently allows extensive mapping of proteomes as well as quantitative comparisons between samples using label-free methods. Label-free quantitative proteomic analyses can be based on normalized spectral count23, where the total number of tandem mass spectra taken on peptides from a given protein in a LC/LC-MS/MS analysis, is linearly correlated with the protein abundance over a dynamic range of two orders of magnitude. In addition, it was shown that the spectral count has the highest technical reproducibility in comparing with others sampling statistics such as sequence coverage and peptide count24.
Using MudPIT and label free quantitation, we performed large-scale mapping of the honeybee brain proteome, both from nurse and forager subcastes. Comparative analysis using G-test statistics of the MS data showed significant differences between forager and nurse brain proteomes.
2. Materials and methods
2.1. Insect Collection and Brain Dissection
Apis mellifera adult worker subcastes (forager and nurse) were collected from colonies at Vereda Rosa (Mel&Mel) Apiary (Brasilia, Brazil). To ensure that fully mature foragers were collected, only those carrying pollen were selected. Nurses were removed from the hive. Bees were anaesthetized with chloroform, and brains were dissected in cold lysis buffer (7 mol/L urea, 2 mol/L thiourea, 1% diothiothretol (DTT), 2% Triton X-100, 0.5% ampholytes 3–10 or 4–7) containing a cocktail of protease inhibitors (Complete, Mini, Protease Inhibitor Cocktail Tablets, Roche Diagnostics, Mannheim, Germany). Head glands were removed and discarded. After thorough washing and soaking with cold lysis buffer, brains were immediately immersed in liquid N2 and stored at −80 °C.
2.2. Sample Preparation
Experiments were carried out with samples obtained from ten brains for each subcaste (forager and nurse) group. Brains were lysed using manual homogenization in 200 μL of lysis buffer followed by incubation for 1 h at room temperature. The samples were centrifuged at 15,000 g for 15 min. The resulting supernatant was submitted to protein quantification assay using the 2D Quant kit (GE Healthcare, Uppsala, Sweden) and confirmed by amino acid analysis. Nurse and forager samples were desalted and lyophilized prior to MudPIT separation. Dialysis was carried out at 4 °C against a slow distilled water flow (about 15 L) using a 2 kDa cutoff membrane (D2272, Sigma, St. Louis, MO) prepared in a microdialyzer system (Pierce, Rockford, IL).
2.3. Trypsin Digestion
The lyophilized fractions were dissolved in 100 mmol/L Tris-HCl buffer pH 8.5. The samples were then reduced by adding 20 mmol/L final concentration of DTT and cysteines were alkylated by adding 20 mmol/L final concentration of iodoacetamide. Subsequently, the samples were digested overnight at 37 °C in a final concentration of 2 mol/L urea with 100 mmol/L Tris-HCl, pH 8.5 containing trypsin at an enzyme/substrate ratio of 1:75 (Promega, Madison, WI, USA). The reaction was stopped by addition of 90% formic acid to a final concentration of 4%. Digested samples were stored at −80 °C until mass spectrometry analysis.
2.4. Multidimensional Protein Identification Technology (MudPIT)
The protein digest was pressure-loaded onto a fused silica capillary desalting column containing 3 cm of 5 μm Polaris C18 material (Metachem, Ventura, CA) packed into a 250 μm i.d. capillary with a 2 μm filtered union (UpChurch Scientific, Oak Harbor, WA). The desalting column was washed with a buffer containing 95% water, 5% acetonitrile, and 0.1% formic acid. After desalting, a 100 μm i.d. capillary with a 5 μm pulled tip packed with 10 cm × 3 μm Aqua C18 material (Phenomenex, Ventura, CA) and by 3 cm × 5 μm Partisphere strong cation exchanger (Whatman, Clifton, NJ) was attached to the filter union and the entire split-column (desalting column–filter union–analytical column) was placed in line with an Agilent 1100 quaternary HPLC (Santa Clara, CA) and analyzed using a modified 12 step separation described previously21. The buffer solutions used contained 5% acetonitrile, and 0.1% formic acid (buffer A), 80% acetonitrile, and 0.1% formic acid (buffer B), and 500 mmol/L ammonium acetate, 5% acetonitrile, and 0.1% formic acid (buffer C). Step 1 consisted of a 100 min gradient from 0–100% buffer B. Steps 2–11 had the following profile: 3 min of 100% buffer A, 2 min of X% buffer C, 10 min gradient from 0–15% buffer B, and 97 min gradient from 15–45% buffer B. The 2 min buffer C percentages (X) were 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, and 60% respectively for the 12-step analysis. The final step, the gradient contained: 3 min of 100% buffer A, 20 min of 100% buffer C, 10 min gradient from 0–15% buffer B, and 107 min gradient from 15–70% buffer B. As peptides eluted from the microcapillary column, they were electrosprayed directly into an LTQ 2D ion trap mass spectrometer (ThermoFisher, San Jose, CA) with the application of a distal 2.5 kV spray voltage. A cycle of one full-scan mass spectrum (400–1400 m/z) followed by 5 data-dependent MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. The application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur datasystem. Each sample was run twice for technical replicates.
2.5. Analysis of Tandem Mass Spectra
Tandem mass spectra were analyzed using the following software analysis protocol. Poor quality spectra were removed from the dataset using an automated spectral quality assessment algorithm25. Tandem mass spectra remaining after filtering were searched with the SEQUEST™ algorithm26 against the NCBI Honeybee protein database (version 4.0 assembly) concatenated to a decoy database in which the sequence for each entry in the original database was reversed22. All searches were parallelized and performed on a Beowulf computer cluster consisting of one hundred 1.2 GHz Athlon CPUs27. No enzyme specificity was considered for any search. SEQUEST results were assembled and filtered using the DTASelect (version 2.0) program28–29. DTASelect 2.0 uses a linear discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false positive rate (5% in this analysis). The false positive rates are estimated by the program from the number and quality of spectral matches to the decoy database.
2.6. Protein Categorization
Protein sequences were compared to the KOG (EuKaryotic Orthologous Groups) catalog set using BLASTX, and then categorized into the major KOG categories of most similar sequence with an E-value cut off inferior to 10e-5, according to their reported function. Categorical redundancy of contigs was not removed when they belonged to multiple KOG categories.
2.7. Assessing differential protein expression using a G-test on spectral counting
First, the spectral counts for each protein were normalized according to total spectral counts for all the proteins in the samples. Let t1 be the total spectral counts for all the proteins in sample 1, t2 be the total spectral counts for all the proteins in sample 2, n1 be the spectral count for the protein in sample 1, n2 be the spectral count for the protein in sample 2, then
where λ is the pseudo spectral count (= 0.5 in this case). The pseudo spectral count is used here to avoid taking logarithm to zero.
The G-value was calculated according to Sokal and Rohlf30:
where f1 and f2 are the normalized spectral counts for the protein in samples 1 and 2, respectively; f1 and f2 are expected spectral counts for the protein in samples 1 and 2, respectively. We assume that the protein is equally expressed, thus f1 = f2 = (f1+ f2)/2.
The calculated G-value was then used to assess whether the protein was differentially expressed according to the chi-square (χ2) distribution table with one degree of freedom. Proteins with G-value larger than 3.841 were considered differentially expressed with P < 0.05.
3. Results and Discussion
3.1. Honeybee brain proteome profile
A proteome profile of the honeybee brain from the nurse and forager stages was obtained from the MudPIT analysis. This is the first large-scale mapping of honeybee-worker brain proteome to our knowledge. A total of 2,987 protein counts (Supplementary table 1) were obtained out of the spectra. Proteins that were detected both in nurse and forager (1619 species) are represented as NF, those found only in nurse (1123 species) are denoted as N, and those presented exclusively in forager (245 species) are assigned as F. After eliminating redundancy, the number of non-redundant proteins comes to 2,742 species, with 1,454 of them being common to both N and F subcastes, 1,063 exclusive to N and 225 exclusive to F. All the non-redundant proteins represent 27.4 % of the 10,157 genes set from initial evaluation of the A. mellifera genome31. Upon classification under GO terms, the figures are 1,376 in NF, 985 in N, 189 in F, making a total of 2,550 classified proteins.
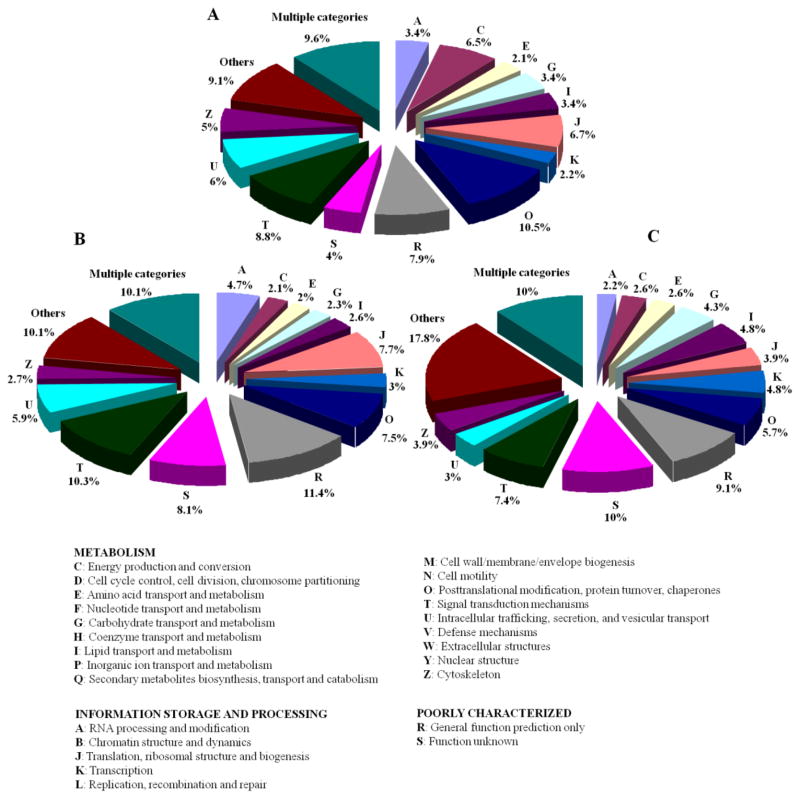
Major protein functions represented in honeybee worker (nurse and forager) brain samples (NF) included energy production and conversion (6.5%), signal transduction mechanisms (8.8%), posttranslational modification, protein turnover, chaperones (10.5%) and translation, ribosomal structure and biogenesis (6.7%) (Figure 1-A). One advantage of MudPIT is that it allows the identification of low copy number proteins. For example, proteins involved in signal transduction processes represented a large fraction in the three groups (NF-8.8%, N-10.3% and F-7.4%, Figure 1). Examples of this group identified were catalytic and regulatory subunits of protein kinases A such as cAMP-dependent protein kinase catalytic subunit (PKA C) isoform 1 and cAMP-dependent protein kinase type II regulatory chain. Previous studies demonstrated that cAMP-dependent kinase (PKA) contributes to the induction of a long-term memory formation in the honeybee Apis mellifera32. Another example was the calcium/calmodulin-dependent protein kinase, isoform B isoform 1. The expression of Ca2+-calmodulin-dependent protein kinase II (CaMKII) in the mushroom bodies of the honeybee brain has been reported33. Key regulators of cellular processes such as transcription factors also typically exist in a very few copies per cell. Examples of identified proteins in this group were nuclear transcription factor Y subunit gamma and C-terminal-binding protein (CtBP protein) (dCtBP) isoform 1.
Figure 1.
Functional categorization of the honeybee brain proteins. (A) Both honeybee brains, (B) nurse honeybee brain, (C) forager honeybee brain.
Membrane proteins play a central role in cell adhesion, signal transduction, and molecular transportation. Several translocases and other synaptic vesicle membrane proteins were identified with two or more peptides (Supplementary Table 1). Other highly expressed proteins were housekeeping proteins, for example, chaperones (NF-10.5%) and proteins involved in translation, ribosomal structure and biogenesis (NF-6.7%) (Figure 1-A).
The functions of about 10% of the proteins could not be assigned in the KOG catalog in the groups NF and N, and 18% in F. About 4%, 8%, and 10% of identified proteins had unknown functions in NF, N and F groups, respectively. These can be explained by the fact that there may be specific proteins of A. mellifera species, while the search through KOG catalog is based on sequence similarity to other species.
3.2. Quantitative differences between nurse and forager brain
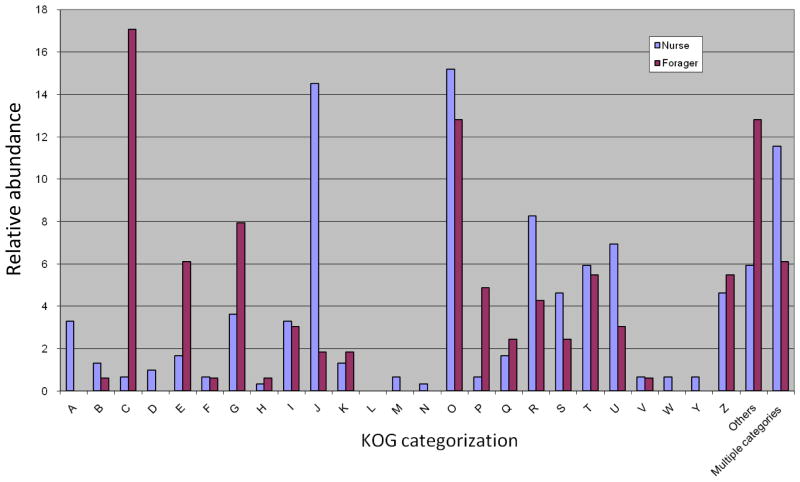
After applying spectral count normalization and G-test, 467 proteins (17%) were found to be differentially expressed. Among these, 164 proteins are more abundant or specifically expressed in forager, while 303 proteins are nurse-specific or more concentrated (Figures 1-B and 1-C and Supplementary Table 2). Figure 2 show a quantitative comparison of protein sets in nurse and forager brains according to KOG categorization. This comparison revealed that the proteins implicated in translation, ribosomal structure and biogenesis are considerably higher in nurse (14.5%) than in forager (1.8%). Elevated levels of this kind of protein in nurse could reflect more active protein synthesis to develop the protein machinery necessary for the changes in brain structure, which precede ontogenesis to foragers.
Figure 2.
Quantitative comparison of KOG protein sets in nurse and forager brains. A: RNA processing and modification; B: Chromatin structure and dynamics; C: Energy production and conversion; D: Cell cycle control, cell division, chromosome partitioning; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; G: Carbohydrate transport and metabolism; H: Coenzyme transport and metabolism; I: Lipid transport and metabolism; J: Translation, ribosomal structure and biogenesis; K: Transcription; L: Replication, recombination and repair; M: Cell wall/membrane/envelope biogenesis; N: Cell motility; O: Posttranslational modification, protein turnover, chaperones; P: Inorganic ion transport and metabolism; Q: Secondary metabolites biosynthesis, transport and catabolism; R: General function prediction only; S: Function unknown; T: Signal transduction mechanisms; U: Intracellular trafficking, secretion, and vesicular transport; V: Defense mechanisms; W: Extracellular structures; Y: Nuclear structure; Z: Cytoskeleton.
In addition, proteins related to posttranslational modification, protein turnover, chaperones, chromatin structure and dynamics, intracellular trafficking, secretion and vesicular transport were also more abundant in nurse brain (Figure 2). Nurse brain also showed a higher expression of proteins associated to RNA processing and modification, cell cycle control, cell division and chromosome partitioning. Higher expression of such protein classes suggests brain differentiation and development in the young workers, as transition from nursing to foraging involves changes in brain structure34. Proteins involved in cell wall, cell membrane, envelope biogenesis and cell motility were identified only in nurse brain.
Table 1 shows proteins selectively expressed in nurse or forager with the highest normalized spectral counts. One example of protein selectively expressed in nurse brain with high spectral count was isoform A isoform 2 (14-3-3 zeta). According to the information of Gene Ontology (GO) database, 14-3-3 zeta protein is required in Raf-dependent cell proliferation and photoreceptor differentiation during eye development and could be also related with olfactory learning. 14-3-3 zeta was also one of the Apis mellifera sequences that matched with Drosophila melanogaster genes implicated in behavior35. Another protein highly specific for nurse brain was moleskin, partial, assigned in GO as involved in cell proliferation and differentiation (e.g., compound eye cone cell differentiation).
Table 1.
Examples of proteins selectively expressed in nurse or forager honeybee brains with the highest normalized spectral counts.
| Accession No. | Identified protein | Organism/Subcaste | Normalized SpecCount | KOG categories |
|---|---|---|---|---|
| XP_623183.1 | 14-3-3 CG17870-PA, isoform A isoform 2 | Apis mellifera/nurse | 39.5 | Posttranslational modification, protein turnover, chaperones |
| XP_624116.2 | moleskin CG7935-PA, partial | Apis mellifera/nurse | 23.5 | Nuclear structure, Intracellular trafficking, secretion, and vesicular transport |
| XP_392511.2 | eIF5 CG9177-PB, isoform B isoform 1 | Apis mellifera/nurse | 20.5 | Translation, ribosomal structure and biogenesis |
| XP_624598.1 | lethal (1) G0022 CG8231-PA KOG0359, Chaperonin complex component, TCP-1 zeta subunit (CCT6) | Apis mellifera/nurse | 16.5 | Posttranslational modification, protein turnover, chaperones |
| XP_393135.2 | ribosomal protein L23A CG7977-PA KOG1751, 60s ribosomal protein L23 | Apis mellifera/nurse | 14.5 | Translation, ribosomal structure and biogenesis |
| XP_395962.2 | p115 CG1422-PA KOG0946, ER-Golgi vesicle-tethering protein p115 | Apis mellifera/nurse | 13.5 | Intracellular trafficking, secretion, and vesicular transport |
| XP_394518.1 | glutathione S transferase S1 CG8938-PA, isoform A | Apis mellifera/nurse | 12.5 | Posttranslational modification, protein turnover, chaperones |
| XP_001120061.1 | connectin CG7503-PA KOG4194, membrane glycoprotein LIG-1 | Apis mellifera/nurse | 11.5 | Signal transduction mechanisms |
| XP_624589.1 | calmodulin CG8472-PA, isoform A isoform 2 | Apis mellifera/nurse | 9.5 | Signal transduction mechanisms |
| XP_624212.1 | nucleoporin Nup43 (p42) | Apis mellifera/nurse | 9.5 | Nuclear structure |
| XP_623220.1 | tubulin alpha-1 chain | Apis mellifera/forager | 575.6 | Cytoskeleton |
| BAE86928.1 | alpha-glucosidase | Apis mellifera/forager | 367.1 | Carbohydrate transport and metabolism |
| AAP93583.1 | thioredoxin reductase | Apis mellifera ligustica/forager | 44.5 | Posttranslational modification, protein turnover, chaperones |
| XP_623225.1 | Ras-like GTP-binding protein Rho1 isoform 2 | Apis mellifera/forager | 25.1 | General function prediction only |
| AAM20738.1 | alpha-amylase | Apis mellifera mellifera/forager | 19.9 | Carbohydrate transport and metabolism |
| XP_392060.2 | CG6180-PA isoform 1, KOG3346: phosphatidylethanolamine binding protein | Apis mellifera/forager | 19.9 | General function prediction only |
| ABH88169.1 | chemosensory protein 1 | Apis mellifera/forager | 12.2 | No hits found |
| XP_393296.2 | serine/threonine-protein phosphatase PP1-beta catalytic subunit | Apis mellifera/forager | 12.2 | Signal transduction mechanisms, general function prediction only |
| XP_392280.2 | mitochondrial acyl carrier protein 1 CG9160-PB, isoform B isoform 1 | Apis mellifera/forager | 10.9 | Energy production and conversion, lipid transport and metabolism, secondary metabolites biosynthesis, transport and catabolism |
| XP_001123234.1 | catalase, partial | Apis mellifera/forager | 9.6 | Inorganic ion transport and metabolism |
Vitellogenin was also only detected in nurse brain (Suppl. Table 1). It is known that vitellogenin is predominant in hemolymph, with higher titers during the first 2–3 weeks of worker adult life36. The high synthesis rate of this protein in young individuals is one of the key defining characteristics of nurse bees37–41. Vitellogenin has an antioxidant function, which led to the suggestion that it also operates to increase queen lifespan42. Recently, it was evidenced that the silencing of the vitellogenin gene function can drive young bees to become extremely precocious foragers43. The protein is also suggested to prime bees for specialized foraging tasks (nectar/pollen collection) and to influence worker longevity44.
Transcription factor mblk-1 was also only detected in the nurse brain (Suppl. Table 1). A previous report indicated that mblk-1 is a transcription factor that might function in mushroom bodies neural circuits45. Others examples of proteins selectively expressed in nurse brain include proteins related to synthesis processes.
Comparing the fraction of proteins involved in energy production and conversion, experienced foragers showed an extensive difference in this class of proteins (17%) in relation to the nurse subcaste (0.6%) (Figure 2). The large fraction of proteins within this category could be related to higher energetic requirements associated to increased brain activity during learning and memorization processes that are triggered upon foraging. Examples of proteins up-regulated or exclusively detected in forager in this category were α-glucosidase, glucose oxidase, α-amylase, glycerol-3-phosphate dehydrogenase and glucose dehydrogenase. Analysis of gene activity by northern blots and microarrays revealed higher levels of several of these metabolic enzyme RNAs in the brains of experienced honeybees when compared to naive ones11. Moreover, α-glucosidase, glucose oxidase and α-amylase are up-regulated in hypopharyngeal gland of forager subcaste46–47. Proteins implicated in amino acid transport and metabolism as well as inorganic ion transport and metabolism were also up-regulated in forager brain, which reinforces its higher metabolic activity.
Higher expression of guanine nucleotide-binding protein gamma-e subunit precursor (Gamma(e)) was detected in forager in relation to nurse brain (Suppl. Table 1). According to GO information, this subunit is implicated in visual transduction in the compound eye and it is a component of rhabdomere, participating in the phototransduction process. Such a protein might be related to other types of signal transduction in the bee brain. Another up-regulated protein was the serine/threonine-protein phosphatase PP1-beta catalytic subunit. According to GO, PP1 probably participates in the regulation of glycogen metabolism and is involved in regulation of ionic conductance and long-term synaptic plasticity.
Glutamine synthetase 2 catalyses the synthesis of glutamine from glutamate, decreasing the glutamate levels in synapses. This amino acid acts as a neurotransmitter performing important functions in mammal brains as motor control, synaptic plasticity, learning, memory, cognition and brain maturation during development48–49. In Drosophila and other arthropods, glutamate also presents substantial signaling roles as a neurotransmitter50. Levels of glutamate and glutamine synthetase transcripts in the Drosophila nervous system were related to the maintanance of flight stability51.
Mitochondrial acyl carrier protein 1 is an accessory and non-catalytic subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (complex I). It has the role of carrying the growing fatty acid chain in fatty acid biosynthesis52–53.
Catalase levels in specific tissues are associated with the antioxidant response. Honeybee flight likely produces high levels of reactive oxygen species in flight muscles. Recently, Williams and co-workers54 showed that catalase and total antioxidant capacity increased in young flight muscle. Surprisingly, such effects were likely due to collecting the young bees soon after orientation flights.
3.3. Differential expression of specific proteins in nurse and forager honeybee brain
In the present work, we showed that several neural-specific proteins are differentially expressed with statistical significance in nurse and forager honeybee brains. An example of an up-regulated neural protein in nurse is synaptojanin, isoform B, which is implicated in the regulation of synapse structure and activity. Nurse also had higher brain expression of failed axon connections, isoform C isoform 1(fax), involved in axonogenesis, which coincides with previous transcriptomic studies in Apis mellifera55. Another neural protein only expressed in nurse brain is contactin. Such a protein is implicated in cell adhesion and is differentially expressed in a number of neuronal tissues during development56. Wrapper is also a nurse subcaste-specific protein, which participates in axonogenesis and gliogenesis57. Those proteins are related to genesis and development of nerve structure, which once again reflects the ongoing cerebral maturation of the nurse subcaste. D2-like dopamine receptor, whose transcripts were more abundant in the oldest workers mushroom bodies58, was detected only in nurse brain according to our results. Such class of receptors may contribute to the differential regulation of distinct neural circuits to behavioral maturation of the bee58.
In the forager subcaste, proteins related to synapses were more abundant. An example of a neural protein up-regulated in forager is calcineurin subunit B isoform 2 (protein phosphatase 2B regulatory subunit), a Ca2+/calmodulin-binding protein probably involved in neurotransmitter secretion and vesicle-mediated transport59–60. Another synaptic vesicle protein up-regulated in forager brain is synaptotagmin, isoform A isoform 1, the major Ca2+ sensor for synaptic vesicle exocytosis61. Forager brain also showed increased expression of neurocalcin homolog (DrosNCa), which inhibits the phosphorylation of rhodopsin, and has high-level expression in the cortical regions of the central brain62–63. A protein synapse-associated protein 47kDa, isoform A was also up-regulated in the forager brain. Neurons release neurotransmitters by calcium-dependent exocytosis of synaptic vesicles64. The high expression of synaptic proteins in honeybee forager brain could be related to the higher cerebral activity required for navigation, learning, memorization and communication of feeding places.
Genes such as period65 and foraging66, that have been proposed to be differentially expressed between nurse and forager brains based on transcriptomic analysis, were not found in the present work. We also did not find members of the nuclear receptor family, which play key roles in embryonic and postembryonic development67, but we could detect a nuclear receptor binding SET domain protein (Supplementary Table 1) expressed only in nurse brain. Similarly, no HR38 receptor, that mediates ecdysteroid signaling, was found among our results. However, an ecdysteroid-regulated gene E74 (AmE74) product was found in forager brain in the present study. Previous report showed that subsets of mushroom body interneurones in the brain of the adult worker bees expressed AmE74, suggesting that it is involved in neural function68. Yamazaki et al.69 showed that HR38 expression was concentrated in a subset of the mushroom body neurons in the forager brain, suggesting that ecdysteroid-signaling in the mushroom bodies might be involved in the division of labor of the workers. We also did not detect the juvenile hormone diol kinase16, a protein assigned by 2-DE as expressed preferentially in the mushroom bodies of the worker brain. On the other hand, we found that the juvenile hormone epoxide hydrolase was expressed in the worker brain, both in nurse and forager (Supplementary Table 1).
3.4. Comparison between 2-DE and MudPIT results
Previous experiments carried out by our group identified proteins differentially expressed in nurse and forager honeybee brain by 2-DE and MALDI-TOF methodologies20. Results obtained by both 2-DE and MudPIT (Table 2) coincide to nine differentially expressed proteins between the subcastes. These nine proteins represent more than 50% of the proteins observed by 2DE as differentially expressed in the previous study. For both methods, similar fold change values were measured in relation to differential expression of proteins. Small differences between fold changes were probably due to the analysis by 2-DE, which took into account individual spots when isoforms for posttranslational modification were present. In the case of MudPIT, all isoforms are jointly considered. Therefore, isoform analysis is an advantage of 2-DE over MudPIT. An example is α-glucosidase, whose study by two-dimensional electrophoresis allowed the detection of various isoforms, while MudPIT did not. One exception was the abundant major royal jelly proteins (MRJPs), which had one of their members (royalactin) recently demonstrated to induce queen differentiation70, showing a large difference between change factors observed in the two approaches. Based on 2-DE results, this protein is the most abundant in both forager and nurse samples. A limitation of the spectral count method is that the MudPIT response to proteins with higher expression levels can saturate. Thus, the spectral count for each protein showed an effect of saturation above 30 spectra by HPLC run71. Therefore, MudPIT may under report extremely abundant proteins.
Table 2.
Coincidence between differentially expressed proteins found by MudPIT and 2DE.
| change factor 2-DE/normalized spectral count | identified protein | organism/protein ID | NSC- N/NSC-F |
|---|---|---|---|
| 9.2/1.7 | major royal jelly protein 1 | Apis mellifera/gi|58585098 | 211.5/124.9 |
| 3.0/3.0 | antdh CG1386-PA | Apis mellifera/gi|66548280 | 55.5/18.6 |
| */14.2 | major royal jelly protein 7 | Apis mellifera/gi|62198227 | 25.5/1.8 |
| */2.3 | major royal jelly protein 2 | Apis mellifera/gi|58585108 | 108.5/47.1 |
| 10.8/32.6 | alpha-glucosidase | Apis mellifera/gi|58585164 | 11.5/374.9 |
| 2.5/2.3 | protein lethal(2)essential for life (Protein Efl21) | Apis mellifera/gi|110750762 | 29.5/67.9 |
| 3.1/6.4 | sarcoplasmic calcium-binding protein 2 CG14904-PA | Apis mellifera/gi|48140312 | 27.5/175.4 |
| 3.9/4.7 | transferrin | Apis mellifera/gi|58585086 | 9.5/44.5 |
| 2.0/1.6 | glutamine synthetase 2 CG1743-PC, isoform C | Apis mellifera/gi|48141383 | 46.5/74.3 |
NSC-N: normalized spectral count nurse subcaste
NSC-F: normalized spectral count forager subcaste
: Stage specific spots in 2-DE
Other proteins that showed quantitative changes by 2-DE also showed differential expression by MudPIT analysis, but with no significance under G-test. It should be noted that both methods agreed for proteins identified as more abundant in nurse brain such as those related to processes of protein synthesis. In forager brain, proteins involved in other metabolic processes, especially energetic metabolism were the most abundant. Differential expression of some of these proteins at a transcriptomic level was reported in previous work. For example, α-glucosidase and protein lethal (2) essential for life (Efl21 protein) genes were found as more expressed in the head of forager bees11.
Concluding remarks
Our study produced a large-scale mapp of the worker honeybee brain proteome. We identified 2,742 proteins from forager and nurse honeybee brain samples, 17% of which were found to be differentially expressed by spectral count sampling statistics and G-test. Some proteins expressed at low copy number per cell and membrane proteins that are difficult to detect by 2-DE were identified by MudPIT. Additionally, neural specific proteins were correlated to putative functions associated to social roles of each honeybee worker subcaste upon ontogenetic development. MudPIT methodology corroborated our previous results in differential expression of nine proteins by 2-DE20, and expanded our findings in an astounding manner. In nurse, proteins involved in protein synthesis and development of cerebral structures were found as more abundant, while, in forager, those related to synaptic activity as well as energy production and conversion were up-regulated. This means that that the nurse brain proteome is deeply committed in building molecular and neural structures, perhaps in preparation for the upcoming forager function, whose brain demands higher energy levels to support a more intense cerebral activity.
Supplementary Material
Acknowledgments
The authors thank Manoel Silva (Apiarios Vereda Rosa, Brasilia, Brasil) for insect collection, and Nuno M. Domingues (Brazilian Center for Protein Research, University of Brasilia) for technical assistance. This work was supported by the travel assistance (grant 453700/2006-3) awarded from CNPq (Brazilian Council for Scientific and Technological Development) to R.B.C., the postgraduate fellowship 190366/2005-2 from TWAS (The Academy of Sciences for the Developing World, Italy) and CNPq to L.G.H, by a computational fellowship BALCH05X5 from CFFT (Cystic Fibrosis Foundation Therapeutics, Inc.) to B.L., by research grants P41 RR011823 from NIH (National Institutes of Health, USA) awarded to J.R.Y., and 307617/2009-2, 477258/2007-7 480333/2004-1 from CNPq to M.V.S.
Footnotes
Supporting Information Available: This material is available free of charge via Internet at http://pubs.acs.org.
References
- 1.Hammer M, Menzel R. Learning and memory in the honeybee. J Neurosci. 1995;15(3 Pt 1):1617–30. doi: 10.1523/JNEUROSCI.15-03-01617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menzel R, Muller U. Learning and memory in honeybees: from behavior to neural substrates. Annu Rev Neurosci. 1996;19:379–404. doi: 10.1146/annurev.ne.19.030196.002115. [DOI] [PubMed] [Google Scholar]
- 3.Giurfa M, Hammer M, Stach S, Stollhoff N, Muller-deisig N, Mizyrycki C. Pattern learning by honeybees: conditioning procedure and recognition strategy. Anim Behav. 1999;57(2):315–324. doi: 10.1006/anbe.1998.0957. [DOI] [PubMed] [Google Scholar]
- 4.Reinhard J, Srinivasan MV, Zhang S. Olfaction: scent-triggered navigation in honeybees. Nature. 2004;427(6973):411. doi: 10.1038/427411a. [DOI] [PubMed] [Google Scholar]
- 5.Dacher M, Gauthier M. Involvement of NO-synthase and nicotinic receptors in learning in the honey bee. Physiol Behav. 2008;95(1–2):200–7. doi: 10.1016/j.physbeh.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 6.Robinson GE, Ben-Shahar Y. Social behavior and comparative genomics: new genes or new gene regulation? Genes Brain Behav. 2002;1(4):197–203. doi: 10.1034/j.1601-183x.2002.10401.x. [DOI] [PubMed] [Google Scholar]
- 7.Toth AL, Robinson GE. Evo-Devo and the Evolution of Social Behavior: Brain Gene Expression Analyses in Social Insects. Cold Spring Harb Symp Quant Biol. 2009 doi: 10.1101/sqb.2009.74.026. [DOI] [PubMed] [Google Scholar]
- 8.Consortium THGS. Insights into social insects from the genome of the honeybee. Apis mellifera Nature. 2006;443(7114):931–49. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee. Apis mellifera Proc Natl Acad Sci USA. 1999;96(10):5575–80. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans JD, Wheeler DE. Expression profiles during honeybee caste determination. Genome Biol. 2001;2(1):RESEARCH0001. doi: 10.1186/gb-2000-2-1-research0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucharski R, Maleszka R. Evaluation of differential gene expression during behavioral development in the honeybee using microarrays and northern blots. Genome Biol. 2002;3(2):RESEARCH0007. doi: 10.1186/gb-2002-3-2-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield CW, Ben-Shahar Y, Brillet C, Leoncini I, Crauser D, Leconte Y, Rodriguez-Zas S, Robinson GE. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103(44):16068–75. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos KS, dos Santos LD, Mendes MA, de Souza BM, Malaspina O, Palma MS. Profiling the proteome complement of the secretion from hypopharyngeal gland of Africanized nurse-honeybees (Apis mellifera L.) Insect Biochem Mol Biol. 2005;35(1):85–91. doi: 10.1016/j.ibmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Furusawa T, Rakwal R, Nam HW, Shibato J, Agrawal GK, Kim YS, Ogawa Y, Yoshida Y, Kouzuma Y, Masuo Y, Yonekura M. Comprehensive royal jelly (RJ) proteomics using one- and two-dimensional proteomics platforms reveals novel RJ proteins and potential phospho/glycoproteins. J Proteome Res. 2008;7(8):3194–3229. doi: 10.1021/pr800061j. [DOI] [PubMed] [Google Scholar]
- 15.Hummon AB, Richmond TA, Verleyen P, Baggerman G, Huybrechts J, Ewing MA, Vierstraete E, Rodriguez-Zas SL, Schoofs L, Robinson GE, Sweedler JV. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006;314(5799):647–9. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 16.Uno Y, Fujiyuki T, Morioka M, Takeuchi H, Kubo T. Identification of proteins whose expression is up- or down-regulated in the mushroom bodies in the honeybee brain using proteomics. FEBS Lett. 2007;581(1):97–101. doi: 10.1016/j.febslet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Schippers MP, Dukas R, Smith RW, Wang J, Smolen K, McClelland GB. Lifetime performance in foraging honeybees: behaviour and physiology. J Exp Biol. 2006;209(Pt 19):3828–36. doi: 10.1242/jeb.02450. [DOI] [PubMed] [Google Scholar]
- 18.Wolschin F, Amdam GV. Comparative proteomics reveal characteristics of life-history transitions in a social insect. Proteome Sci. 2007;5:10. doi: 10.1186/1477-5956-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolschin F, Amdam GV. Plasticity and robustness of protein patterns during reversible development in the honey bee (Apis mellifera) Anal Bioanal Chem. 2007;389(4):1095–100. doi: 10.1007/s00216-007-1523-5. [DOI] [PubMed] [Google Scholar]
- 20.Garcia L, Garcia CHS, Calabria LK, Cruz GCN, Puentes AS, Bao SN, Fontes W, Ricart CA, Espindola FS, Sousa MV. Proteomic analysis of honey bee brain upon ontogenetic and behavioral development. J Proteome Res. 2009;8(3):1464–1473. doi: 10.1021/pr800823r. [DOI] [PubMed] [Google Scholar]
- 21.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 22.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5(11):2909–18. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 25.Bern M, Goldberg D, McDonald WH, Yates JR., 3rd Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 2004;20(Suppl 1):i49–54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 26.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 27.Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR., 3rd Code developments to improve the efficiency of automated MS/MS spectra interpretation. J Proteome Res. 2002;1(3):211–5. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 28.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cociorva D, Yates JR. DTASelect 2.0: Improving the Confidence of Peptide and Protein Identifications. 54th ASMS Annual Meeting Proceedings; Seattle, WA. May, 2006; Seattle, WA: 2006. [Google Scholar]
- 30.Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 3. W. H. Freeman and Co; New York: 1995. p. 887. [Google Scholar]
- 31.Elsik CG, Mackey AJ, Reese JT, Milshina NV, Roos DS, Weinstock GM. Creating a honey bee consensus gene set. Genome Biol. 2007;8(1):R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiala A, Muller U, Menzel R. Reversible downregulation of protein kinase A during olfactory learning using antisense technique impairs long-term memory formation in the honeybee, Apis mellifera. J Neurosci. 1999;19(22):10125–34. doi: 10.1523/JNEUROSCI.19-22-10125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi H, Kamikouchi A, Kage E, Kubo T. Molecular mechanisms of the highly advanced behaviors in the honeybee. Honeybee Science. 2001;22(3):113–120. [Google Scholar]
- 34.Robinson GE. Genomics and integrative analyses of division of labor in honeybee colonies. Am Nat. 2002;160(Suppl 6):S160–72. doi: 10.1086/342901. [DOI] [PubMed] [Google Scholar]
- 35.Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Liu L, Pardinas JR, Robertson HM, Soares MB, Robinson GE. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002;12(4):555–66. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartfelder K, Engels W. Social insect polymorphism: hormonal regulation of plasticity in development and reproduction in the honeybee. Curr Top Dev Biol. 1998;40:45–77. doi: 10.1016/s0070-2153(08)60364-6. [DOI] [PubMed] [Google Scholar]
- 37.Engels W, Fahrenhorst H. Age-dependent and caste-dependent changes in hemolymph protein patterns of Apis mellifera. Wilhelm Roux Arch Entwickl mech Org. 1974;174:285–296. doi: 10.1007/BF00573233. [DOI] [PubMed] [Google Scholar]
- 38.Excels W. Occurrence and Significance of Vitellogenins in Female Castes of Social Hymenoptera. Integr Comp Biol. 1974;14(4):1229–1237. [Google Scholar]
- 39.Fluri P, Lüscher M, Wille H, Gerig L. Changes in weight of the pharyngeal gland and haemolymph titres of juvenile hormone, protein and vitellogenin in worker honey bees. J Insect Physiol. 1982;28(1):61–68. [Google Scholar]
- 40.Engels W, Kaatz HH, Zilikens A, Simões ZLP, Truve A, Braun R, Dittrich F. Honey bee reproduction: vitellogenin and caste-specific regulation of fertility. In: Hoshi M, Yamashita O, editors. Advances in Invertebrate Reproduction 5. Elsevier Science; Amsterdam: 1990. pp. 495–502. [Google Scholar]
- 41.Pinto LZ, Bitondi MM, Simoes ZL. Inhibition of vitellogenin synthesis in Apis mellifera workers by a juvenile hormone analogue, pyriproxyfen. J Insect Physiol. 2000;46(2):153–160. doi: 10.1016/s0022-1910(99)00111-0. [DOI] [PubMed] [Google Scholar]
- 42.Seehuus S-C, Norberg K, Gimsa U, Krekling T, Amdam GV. From the Cover: Reproductive protein protects functionally sterile honey bee workers from oxidative stress. PNAS. 2006;103(4):962–967. doi: 10.1073/pnas.0502681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marco Antonio DS, Guidugli-Lazzarini KR, do Nascimento AM, Simoes ZL, Hartfelder K. RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften. 2008;95(10):953–61. doi: 10.1007/s00114-008-0413-9. [DOI] [PubMed] [Google Scholar]
- 44.Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007;5(3):e62. doi: 10.1371/journal.pbio.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JM, Kunieda T, Kubo T. The activity of Mblk-1, a mushroom body-selective transcription factor from the honeybee, is modulated by the ras/MAPK pathway. J Biol Chem. 2003;278(20):18689–94. doi: 10.1074/jbc.M300486200. [DOI] [PubMed] [Google Scholar]
- 46.Kubo T, Sasaki M, Nakamura J, Sasagawa H, Ohashi K, Takeuchi H, Natori S. Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L.) with age and/or role. J Biochem. 1996;119(2):291–5. doi: 10.1093/oxfordjournals.jbchem.a021237. [DOI] [PubMed] [Google Scholar]
- 47.Ohashi K, Natori S, Kubo T. Expression of amylase and glucose oxidase in the hypopharyngeal gland with an age-dependent role change of the worker honeybee (Apis mellifera L.) Eur J Biochem. 1999;265(1):127–33. doi: 10.1046/j.1432-1327.1999.00696.x. [DOI] [PubMed] [Google Scholar]
- 48.Meldrum BS. Glutamate as a neurotransmitter in the brain: review of physiology and pathology. J Nutr. 2000;130(4S Suppl):1007S–15S. doi: 10.1093/jn/130.4.1007S. [DOI] [PubMed] [Google Scholar]
- 49.Mattson MP. Glutamate and neurotrophic factors in neuronal plasticity and disease. Ann N Y Acad Sci. 2008;1144:97–112. doi: 10.1196/annals.1418.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinakevitch I, Grau Y, Strausfeld NJ, Birman S. Dynamics of glutamatergic signaling in the mushroom body of young adult Drosophila. Neural Dev. 2010;5(1):10. doi: 10.1186/1749-8104-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nair S, Agrawal N, Hasan G. Homeostasis of glutamate neurotransmission is altered in Drosophila Inositol 1,4,5-trisphosphate receptor mutants. Invert Neurosci. 2007;7(3):137–47. doi: 10.1007/s10158-007-0048-0. [DOI] [PubMed] [Google Scholar]
- 52.Schulte U. Biogenesis of Respiratory Complex I. J Bioenerg Biomembr. 2001;33(3):205–212. doi: 10.1023/a:1010730919074. [DOI] [PubMed] [Google Scholar]
- 53.Cronan JE, Fearnley IM, Walker JE. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Letters. 2005;579(21):4892–4896. doi: 10.1016/j.febslet.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 54.Williams JB, Roberts SP, Elekonich MM. Age and natural metabolically-intensive behavior affect oxidative stress and antioxidant mechanisms. Exp Gerontol. 2008;43(6):538–549. doi: 10.1016/j.exger.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302(5643):296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 56.Faivre-Sarrailh C, Banerjee S, Li J, Hortsch M, Laval M, Bhat MA. Drosophila contactin, a homolog of vertebrate contactin, is required for septate junction organization and paracellular barrier function. Development. 2004;131(20):4931–42. doi: 10.1242/dev.01372. [DOI] [PubMed] [Google Scholar]
- 57.Wheeler SR, Banerjee S, Blauth K, Rogers SL, Bhat MA, Crews ST. Neurexin IV and Wrapper interactions mediate Drosophila midline glial migration and axonal ensheathment. Development. 2009;136(7):1147–57. doi: 10.1242/dev.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphries MA, Mustard JA, Hunter SJ, Mercer A, Ward V, Ebert PR. Invertebrate D2 type dopamine receptor exhibits age-based plasticity of expression in the mushroom bodies of the honeybee brain. J Neurobiol. 2003;55(3):315–30. doi: 10.1002/neu.10209. [DOI] [PubMed] [Google Scholar]
- 59.Yakel JL. Calcineurin regulation of synaptic function: from ion channels to transmitter release and gene transcription. Trends Pharmacol Sci. 1997;18(4):124–134. doi: 10.1016/s0165-6147(97)01046-8. [DOI] [PubMed] [Google Scholar]
- 60.Mansuy IM, Mayford M, Jacob B, Kandel ER, Bach ME. Restricted and Regulated Overexpression Reveals Calcineurin as a Key Component in the Transition from Short-Term to Long-Term Memory. Cell. 1998;92(1):39–49. doi: 10.1016/s0092-8674(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 61.Yoshihara M, Montana ES. The synaptotagmins: calcium sensors for vesicular trafficking. Neuroscientist. 2004;10(6):566–74. doi: 10.1177/1073858404268770. [DOI] [PubMed] [Google Scholar]
- 62.Teng DH, Chen CK, Hurley JB. A highly conserved homologue of bovine neurocalcin in Drosophila melanogaster is a Ca2+-binding protein expressed in neuronal tissues. J Biol Chem. 1994;269(50):31900–7. [PubMed] [Google Scholar]
- 63.Faurobert E, Chen CK, Hurley JB, Teng DH. Drosophila neurocalcin, a fatty acylated, Ca2+-binding protein that associates with membranes and inhibits in vitro phosphorylation of bovine rhodopsin. J Biol Chem. 1996;271(17):10256–62. doi: 10.1074/jbc.271.17.10256. [DOI] [PubMed] [Google Scholar]
- 64.Brose N, Petrenko A, Sudhof T, Jahn R. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science. 1992;256(5059):1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- 65.Toma DP, Bloch G, Moore D, Robinson GE. Changes in period mRNA levels in the brain and division of labor in honey bee colonies. Proc Natl Acad Sci U S A. 2000;97(12):6914–9. doi: 10.1073/pnas.97.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Influence of gene action across different time scales on behavior. Science. 2002;296(5568):741–4. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- 67.Velarde RA, Robinson GE, Fahrbach SE. Nuclear receptors of the honey bee: annotation and expression in the adult brain. Insect Mol Biol. 2006;15(5):583–95. doi: 10.1111/j.1365-2583.2006.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paul RK, Takeuchi H, Matsuo Y, Kubo T. Gene expression of ecdysteroid-regulated gene E74 of the honeybee in ovary and brain. Insect Mol Biol. 2005;14(1):9–15. doi: 10.1111/j.1365-2583.2004.00524.x. [DOI] [PubMed] [Google Scholar]
- 69.Yamazaki Y, Shirai K, Paul RK, Fujiyuki T, Wakamoto A, Takeuchi H, Kubo T. Differential expression of HR38 in the mushroom bodies of the honeybee brain depends on the caste and division of labor. FEBS Lett. 2006;580(11):2667–70. doi: 10.1016/j.febslet.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 70.Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473(7348):478–83. doi: 10.1038/nature10093. [DOI] [PubMed] [Google Scholar]
- 71.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.