Abstract
Autophagy, a process in which cellular components are engulfed and degraded within double-membrane vesicles termed autophagosomes, has an important role in the response to oxidative damage. Here we identify a novel cascade of phosphorylation events, involving a network of protein and lipid kinases, as crucial components of the signaling pathways that regulate the induction of autophagy under oxidative stress. Our findings show that both the tumor-suppressor death-associated protein kinase (DAPk) and protein kinase D (PKD), which we previously showed to be phosphorylated and consequently activated by DAPk, mediate the induction of autophagy in response to oxidative damage. Furthermore, we map the position of PKD within the autophagic network to Vps34, a lipid kinase whose function is indispensable for autophagy, and demonstrate that PKD is found in the same molecular complex with Vps34. PKD phosphorylates Vps34, leading to activation of Vps34, phosphatydilinositol-3-phosphate (PI(3)P) formation, and autophagosome formation. Consistent with its identification as a novel inducer of the autophagic machinery, we show that PKD is recruited to LC3-positive autophagosomes, where it localizes specifically to the autophagosomal membranes. Taken together, our results describe PKD as a novel Vps34 kinase that functions as an effecter of autophagy under oxidative stress.
Keywords: protein kinase D, DAPk, Vps34, autophagy, oxidative stress
Autophagy is an evolutionary conserved mechanism in which bulk cytoplasmic contents are sequestered by double-membrane vesicles named autophagosomes. Autophagy was initially described as a mechanism of cell survival under nutrient-limiting conditions in yeast, but has since been widely implicated in various cellular processes in mammalian systems, ranging from maintaining homeostasis through degradation of long-lived proteins and damaged organelles to blocking tumorigenesis.1, 2, 3 Two ubiquitin-like conjugation pathways mediate the formation of the autophagic vesicle. One pathway involves the covalent conjugation of autophagy related 5 (Atg5) to Atg12, and in a second pathway, phosphatidyl-ethanolamine (PE) is conjugated to LC3 (the mammalian ortholog of Atg8). Upon maturation, the autophagosome fuses with organelles of the endocytic compartment (early or late endosomes and lysosomes) to form the autolysosome, the degradative autophagic compartment. The lipid kinase activity of the class III phosphoinositide 3 kinase (PI3K), Vps34, which produces phosphatydilinositol-3-phosphate (PI(3)P), is one of the most essential requirements for autophagy.4
The serine/threonine kinase protein kinase D1 (PKD) is a regulator of trafficking from the trans-Golgi network (TGN), and also of various signaling processes such as proliferation, motility, and cell death.5 PKD also functions in the cellular response to oxidative stress. Members of the PKC family induce the activity of PKD under oxidative stress by phosphorylating its activation loop, leading to survival signaling through NF-κB.6, 7 We have previously described the PKC-independent activation of PKD by the tumor-suppressor death-associated protein kinase (DAPk) during oxidative stress.8 DAPk has been shown to activate autophagy, at least in part through its ability to phosphorylate Beclin 1, thus releasing it from the suppressive effects of Bcl-2/BclXL.9 Furthermore, it acts as a mediator of autophagy under ER stress.10 Several studies have described the initiation of autophagic processes in response to oxidative stress.11 Autophagy under these conditions may serve to eliminate oxidized and damaged proteins and organelles and thus maintain cellular viability. Yet, if the oxidative damage is extensive, autophagy can provide a route to cell death.
Here, we demonstrate for the first time that PKD positively regulates autophagy and that ectopic expression of PKD is sufficient to induce autophagosome formation. We studied the effects of PKD and DAPk on autophagy in response to oxidative stress and found that both PKD and DAPk are required for the induction of autophagy during oxidative stress, and that PKD acts as a downstream effecter of DAPk in autophagy, thus revealing another mechanism by which DAPk-induced autophagy is regulated. A direct link between PKD and the autophagic machinery is demonstrated here through the binding and phosphorylation of the major autophagic regulator Vps34. Finally, we show that PKD is recruited to LC3-positive autophagic vesicles and is specifically localized to autophagosomal membranes.
Results
PKD is an inducer of autophagy
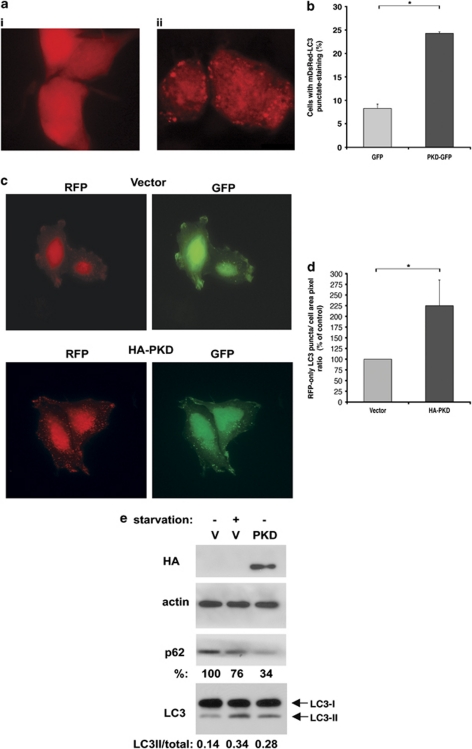
We examined whether PKD could be involved in the regulation of autophagy. Lipidation of LC3, which occurs only under stimulation of autophagy, converts LC3 from its soluble cytoplasmic form (LC3-I) to the membrane-bound, autophagosome-associated form (LC3-II). The recruitment of LC3 to autophagosomes can be detected by fluorescent microscopy as the formation of punctate LC3 dots. To assess whether PKD affects autophagy, cells were transfected with PKD-GFP (or GFP as control) together with monomeric DsRed-LC3 (mDsRed-LC3), and the accumulation of autophagosomes was measured by scoring mDsRed-LC3 punctate staining. Of note, mDsRed-LC3 can potentially serve as a marker for autophagosomes as well as autolysosomes, as unlike the comparable GFP-LC3, the low pKa of mDsRed allows for preservation of its fluorescence within the acidic autolysosome.12 In contrast to cells transfected with GFP, in which mDsRed-LC3 was mostly expressed in a diffused pattern (Figures 1ai and b), ectopic expression of PKD-GFP led to the formation of mDsRed-LC3 puncta (Figures 1aii and b), indicating that PKD induces the accumulation of autophagic vesicles. To determine whether PKD increases autophagic flux, we used a tandem RFP-GFP-LC3 construct.13 This construct allows quantification of both autophagosomes (GFP+RFP-positive puncta) and late-autpohagic structures (RFP-only-positive puncta) in the same cell. HeLa cells were transfected with the tandem RFP-GFP-LC3 construct, together with either empty vector or HA-PKD. Ectopic expression of PKD increased the formation of late-autophagic structures to 225% when compared with control cells (Figures 1c and d), indicating that PKD increases autophagic flux. Notably, similar changes in p62 protein levels and lipidated LC3II levels (calculated as the ratio between LC3II to total LC3 protein levels) were observed upon amino-acid starvation, a canonical way to induce autophagy (Figure 1e).
Figure 1.
PKD induces autophagy. 293T cells were transfected with mDsRed-LC3 and PKD-GFP or GFP as control and fixed 72 h post transfection. (a) Representative images of mDsRed-LC3 in control (i) or PKD-GFP-transfected (ii) cells. Bar=20 μm. (b) Quantification of cells with punctate mDsRed-LC3 staining. Data are representative of three independent experiments. *P<0.002. (c and d) HeLa cells were transfected with the tandem RFP-GFP-LC3 construct and either empty vector or HA-PKD. (c) Representative images of vector or HA-PKD-transfected HeLa cells showing both RFP and GFP puncta staining in each cell. (d) Quantification of RFP-only LC3 puncta (representing autolysosomes) per total cell area of transfected cells (either showing puncta or not) measured by Metamorph. Data represent mean+S.D. of three individual experiments. *P≤0.02. (e) 293T cells were transfected with either vector or HA-PKD and where indicated stimulated with EBSS (4 h) as amino-acid starvation conditions. Cell extracts were immunoblotted with the indicated antibodies. The p62 protein levels (% of control) and the ratios of LC3-II/total LC3 are indicated
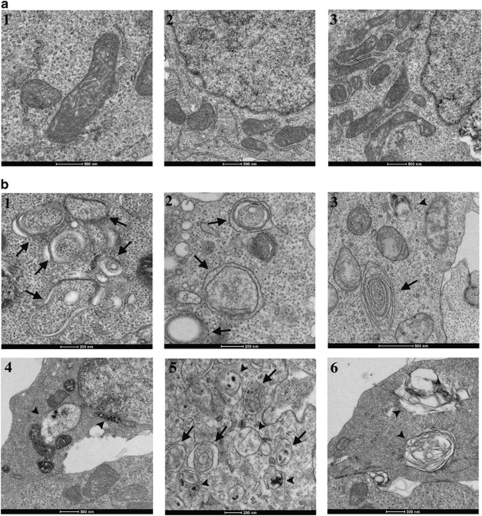
To further examine the effect of PKD on autophagy, 293T cells ectopically expressing PKD-GFP, or GFP as control, were fixed and visualized by electron microscopy (EM). Autophagosomes are identified in electron micrographs by the presence of double-membrane vesicles containing cytoplasmic constituents. Furthermore, different stages of the autophagic vesicles can be distinguished: early autophagosomes appear as a double-limiting membrane surrounding cytoplasm, late-autophagic compartments are characterized by degraded or electron-dense material, and in autolysosomes, the inner limiting membrane of the autophagic vesicle is degraded.14 Although GFP expression had no effect on cell morphology (Figure 2a), in cells expressing PKD-GFP, an extensive accumulation of autophagosomes was apparent (Figure 2b1–6). Notably, PKD expression induced the accumulation of not only early autophagosomes (Figure 2b1–3) but also late-autophagic compartments (Figure 2b3–6). These data indicate that PKD is an activator of autophagy whose ectopic expression is sufficient to induce autophagosome formation and maturation.
Figure 2.
PKD induces autophagosome formation. Transmission electron micrographs of 293T cells transfected with either GFP (a) or PKD-GFP (b). Arrows indicate autophagosomes and arrowheads indicate late-autophagic compartments
PKD and DAPk induce autophagy under oxidative stress
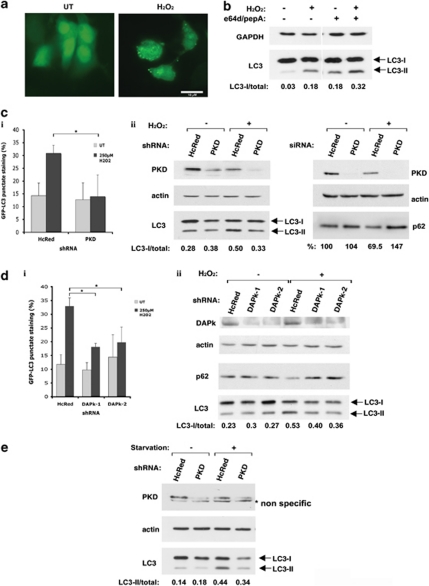
Oxidative stress has been implicated in the induction of autophagy. Treatment of 293T cells with H2O2 stimulated autophagy as observed by the accumulation of LC3 puncta and lipidation (Figures 3a and b), and the observed LC3 shift was increased when cells were treated with the lysosomal inhibitors e64d and pepstatin A during exposure to oxidative stress (Figure 3b). This indicates that in these cells, the accumulation of lipidated LC3 during oxidative stress reflects increased autophagic flux rather than a block in autophagosome maturation or fusion with the lysosome. As PKD is activated under oxidative stress,6, 7, 8 we wished to determine whether PKD is involved in oxidative stress-induced autophagy. To this end, autophagy during oxidative stress was evaluated by measuring GFP-LC3 punctate staining and LC3 lipidation in cells treated with H2O2, in which PKD expression was knocked down by shRNA. As control, cells were transfected with shRNA vectors targeting HcRed, a non-mammalian protein. Knockdown of PKD inhibited the accumulation of GFP-LC3 punctate staining under oxidative stress (Figure 3ci). The knockdown also reduced the oxidative stress-induced elevation in the lipidated LC3-II/total LC3 ratio, and the reduction in p62 protein levels (Figure 3cii). This suggests that PKD is required for the induction of autophagy under oxidative stress.
Figure 3.
PKD and DAPk are required for oxidative stress-induced autophagy. (a and b) 293T cells were treated with H2O2 (250 μM, 16 h) and where indicated also with e64d and pepstatin A (10 μg/ml each) or DMSO as control. (a) Representative GFP-LC3 punctate staining of untreated (UT) or H2O2-treated cells. (b) Cell extracts were immunoblotted with the indicated antibodies and the ratios of LC3-II/total LC3 are indicated. (c and d) 293T cells were transfected with GFP-LC3 and PKD or DAPk expression was knocked down by specific shRNA (top) or siRNA (bottom)-targeting PKD (c), two different shRNAs targeting DAPk (d), or HcRed as control (cand d). At 56 h post transfection, cells were treated with H2O2 (250 μM) and fixed 16 h later. (i) Quantification of cells with punctate GFP-LC3 staining. Data represent mean+S.D. of three individual experiments. *P<0.05. (ii) Cell extracts were immunoblotted with the indicated antibodies. The ratios of LC3-II/total LC3 are indicated. Data are representative of three experiments. (e) 293T cells were transfected with shRNA targeting either PKD or HcRed as control and stimulated with EBSS (4 h) as starvation conditions. Cell extracts were immunoblotted with the indicated antibodies. The ratios of LC3-II/total LC3 are indicated. Data are representative of three experiments
We have previously shown that PKD is a direct substrate of DAPk and that DAPk induces PKD activation under oxidative stress.8 We therefore asked whether DAPk is involved in the regulation of autophagy during oxidative stress. Knockdown of DAPk also inhibited oxidative stress-associated accumulation of GFP-LC3 punctate staining (Figure 3di), elevation in the lipidated LC3-II/total LC3 ratio, and reduction in p62 protein levels (Figure 3dii), suggesting that like PKD, DAPk is required for the induction of autophagy under oxidative stress.
The fact that PKD mediates the induction of autophagy during exposure to oxidative stress led us to examine whether it could be required for autophagy triggered by other stimuli such as amino-acid starvation. Remarkably, knockdown of PKD also inhibited the accumulation of LC3II in response to amino-acid starvation, thus further extending the role of PKD to additional cellular settings besides oxidative stress (Figure 3e).
PKD functions downstream of DAPk in the induction of autophagy
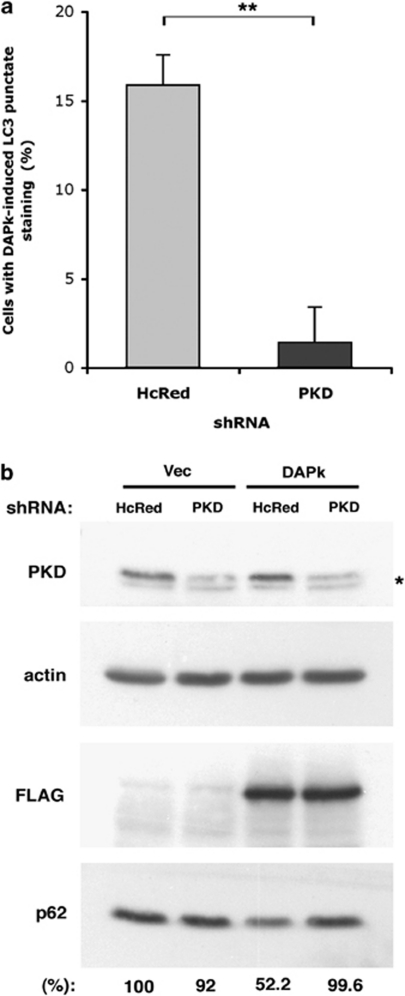
Given our findings that both PKD and DAPk are required for oxidative stress-induced autophagy, we next wished to determine the epistatic relationship of these two proteins in the regulation of autophagy. Overexpression of DAPk has been shown to induce the accumulation of autophagosomes.15 To examine whether PKD functions downstream of DAPk in the induction of autophagy, 293T cells were transfected with DAPk and either PKD-targeting or control (targeting HcRed) shRNA vectors. Although in cells transfected with control shRNA, ectopic expression of DAPk induced the accumulation of GFP-LC3 punctate staining, in cells in which PKD expression was knocked down, the stimulatory effect of DAPk on autophagy was strongly attenuated (Figure 4a). Autophagy was also evaluated by studying p62 protein levels.16 Ectopic expression of DAPk led to a reduction in p62 levels in control cells. However, knockdown of PKD inhibited the DAPk-induced p62 degradation (Figure 4b), indicating that PKD is necessary for the autophagic function of DAPk. Thus PKD, a known DAPk substrate, is a downstream mediator of DAPk-induced autophagy.
Figure 4.
PKD functions downstream of DAPk in the induction of autophagy. 293T cells were transfected with GFP-LC3, either empty vector or DAPk, and either PKD-targeting or control (HcRed-targeting) shRNA. (a) Quantification of cells with punctate GFP-LC3 staining; DAPk-induced GFP-LC3 punctate staining represents values measured in DAPk-transfected cells minus values measured in vector-transfected cells. Data represent mean+S.D. of three individual experiments. **P<0.001. (b) Immunoblot analysis of cell extracts showing reduction in p62 levels. *Nonspecific band. Data are representative of three experiments
PKD interacts with, phosphorylates, and activates Vps34
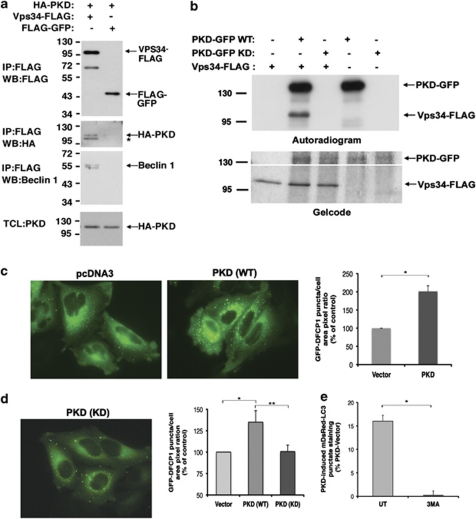
We found that the human Vps34 protein contains a sequence with high similarity to the PKD phosphorylation motif17 (Supplementary Figure 1), suggesting that PKD might act as a kinase of Vps34. To investigate the link between PKD and Vps34, we first asked whether PKD and Vps34 are physically associated in cells. To this end, co-immunoprecipitation experiments with the human proteins were performed. Immunoprecipitation of Vps34-FLAG pulled down endogenous Beclin 1 (Figure 5a), suggesting that overexpression of Vps34 is sufficient to induce the formation of the autophagy-regulating complex. Interestingly, we found that HA-PKD specifically co-immunoprecipitated with Vps34-FLAG but not with the FLAG-GFP control (Figure 5a), suggesting that PKD and Vps34 may be found in the same protein complex in cells. Next, we determined whether Vps34 is a substrate of PKD by performing in vitro kinase assays using immunopurified human Vps34 and PKD. When Vps34 was incubated with PKD in a kinase reaction containing 33P-labeled γ-ATP, Vps34 was specifically phosphorylated by PKD (Figure 5b), indicating that Vps34 is a substrate of PKD. When the kinase reaction was performed with a kinase-dead mutant of PKD (K612W/D727A), no phosphorylation of Vps34 was observed (Figure 5b). Thr677 in Vps34 corresponds to the predicted phosphorylation site. We mutated Thr677 to alanine and performed in vitro kinase assays on the immunoprecipitated mutant protein. Surprisingly, mutation of Thr677 did not inhibit Vps34 phosphorylation by PKD (Supplementary Figure 1), suggesting that other and/or additional phosphorylation sites exist. To identify such possible additional phosphorylation sites we performed mass spectrometry analysis. Notably, in this analysis, Thr677 was identified as a phosphorylation site for PKD, along with several additional sites (data not shown). Therefore, PKD phosphorylates Vps34 on multiple sites, including Thr677 within the catalytic domain.
Figure 5.
PKD binds, phosphorylates, and activates Vps34. (a) 293T cells were transfected with HA-PKD and either Vps34-FLAG or FLAG-GFP as control. FLAG-tagged proteins were immunoprecipitated and eluted from the beads. The immunoprecipitated proteins and samples from the total cell lysate (TCL) were separated on SDS-PAGE, transferred to nitrocellulose, and blotted with the indicated antibodies. *Nonspecific band (verified as in control experiments). (b) Immunopurified Vps34-FLAG and either wild-type (WT) or a kinase-dead mutant (KD) of PKD-GFP were quantified against BSA standards and equal protein amounts were incubated in a kinase reaction containing 33P-labeled γ-ATP. (Top) Exposure to X-ray film (autorad); (bottom) gelcode staining (top and bottom parts are different contrast images of the same gel). (c and d) HeLa cells stably expressing GFP-DFCP1 were transfected with pcDNA3 as control and either WT (c) or KD (d) HA-PKD. At 72 h post transfection, the cells were fixed and the ratio of GFP-DFCP1 puncta per total cell area was calculated. Data represent mean+S.D. of three individual experiments. *,**P≤0.05. (e) 293T cells were transfected with mDsRed-LC3 and PKD-GFP and treated with 3MA (5 mM, 16 h) or left untreated (UT). Quantification of cells with punctate GFP-LC3 staining. Data are representative of three individual experiments. *P<0.02
To investigate whether PKD stimulates the activation of Vps34, we assessed the formation of PI(3)P, the product of Vps34 activity, in cells using a GFP-tagged version of DFCP1, a double-FYVE domain-containing PI(3)P-binding protein that has been shown to bind omegasomes.18 DFCP1 puncta specifically reflect the state of Vps34 activity and the autophagic pool of PI(3)P that it produces. Although in control cells GFP-DFCP1 appeared mostly in a diffused pattern, transfection with PKD induced the accumulation of GFP-DFCP1 punctate structures, indicating that in those cells PI(3)P formation was markedly increased (Figure 5c). Expression of a kinase-dead mutant of PKD, at levels comparable to its wild-type counterpart, had no effect on DFCP1 puncta formation (Figure 5d), suggesting that the catalytic activity of PKD is required for its ability to activate Vps34 and induce PI(3)P formation. Taken together, these data demonstrate a cross-talk between PKD and Vps34 that includes coexistence in the same protein complex in cells and phosphorylation that leads to increased Vps34 activity. If PKD induces autophagy in a canonical, Vps34-dependent manner, then blocking Vps34 should abrogate the PKD-induced autophagy. We thus measured the accumulation of autophagosomes in PKD-transfected cells in which Vps34 activity was inhibited by 3 methyladeninde (3MA). Although transfection with PKD increased the level of GFP-LC3 punctate staining in untreated cells, inhibition of Vps34 by 3MA significantly inhibited the PKD-induced GFP-LC3 punctate staining (Figure 5e). This suggests that PKD acts upstream to Vps34 in the induction of autophagy.
PKD is recruited to LC3-positive punctate vesicles and localizes on autophagosome membranes
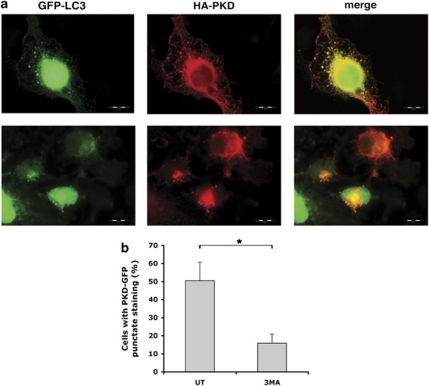
Analysis of PKD-expressing 293T cells by fluorescent microscopy demonstrated that HA-PKD is recruited to punctate structures that partially colocalized with GFP-LC3 (Figure 6a). Quantitative assessments of >100 punctate vesicles from randomly chosen cells using MetaMorph image and analysis software indicated that 50% of the PKD-positive puncta colocalize with LC3-positive puncta. The partial colocalization between PKD and LC3 could be related to the differences between the fluorescent tags of PKD and LC3 (i.e., GFP versus mDsRed). Although the GFP-fusion construct will primarily label autophagosomes, the mDsRed-fusion protein will be present on both autophagosomes and autolysosomes, and their distribution will therefore only partially overlap. Notably, treatment of the cells with 3MA, which inhibits Vps34 activity, blocked the appearance of PKD punctate staining. Together, these findings were consistent with the possibility that the recruitment of PKD to LC3-positive structures in a Vps34-dependent manner represents localization to autophagosomes (Figure 6b).
Figure 6.
PKD is recruited to LC3-positive punctate structures. (a) 293T cells were transfected with HA-PKD and GFP-LC3 and fixed 72 h post transfection. Cells expressing HA-PKD were immunostained with anti-HA antibodies. (b) 293T cells were transfected with mDsRed-LC3 and PKD-GFP and treated with 3MA (5 mM, 16 h) or left untreated (UT). Cells in which PKD-GFP was localized to punctate structures were quantified from total transfected cells. Data represent mean+S.D. of three individual experiments. *P<0.05
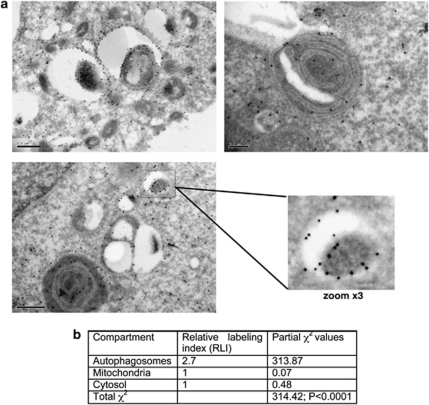
As PKD can be associated with different membranes, and Vps34 activity is also correlated with protein trafficking events, we performed electron microscopy analysis to confirm the localization of PKD on autophagosomes by immunogold labeling. To this end, 293T cells were transfected with HA-PKD and immunogold labeling was performed with anti-HA antibodies. Consistent with the observation that PKD stimulates the accumulation of autophagosomes, expression of HA-PKD induced the formation of autophagic vesicles in these cells. Remarkably, EM immunogold-labeling analysis revealed that HA-PKD is specifically localized on autophagosomal membranes (Figure 7a). The distribution of gold particles in the immunolabeled cells was statistically analyzed according to published methods,19 indicating that the observed distribution of gold particles on autophagosomes is specific (Figure 7b). In conclusion, PKD not only induces the accumulation of autophagic vesicles, but is also an integral part of the autophagic machinery, and is recruited to the autophagosome.
Figure 7.
PKD is localized on autophagosomal membranes. 293T cells were transfected with HA-PKD, fixed, and labeled with anti-HA antibodies followed by gold-conjugated secondary antibodies. Cells were visualized by electron microscopy. (a) Representative images of immunogold-labeled cells. Examples of mitochondria (M) and nucleus (N) are indicated. (b) Statistical evaluation of the gold labeling distribution was performed as follows: a lattice of test points was randomly superimposed on the EM images and three volume-based compartments (autophagosomes, mitochondria, and cytosol) were analyzed for observed and expected gold labeling distributions. Estimates of relative labeling index (RLI) and partial χ2 values indicate that of the three compartments only autophagosomes are preferentially labeled as their RLI is higher than 1 and their partial χ2 value is the significant contributor to the total χ2 value. In χ2 test analysis the null hypothesis of no difference between expected and observed gold particle distributions must be rejected at a probability level of P<0.0001
Discussion
In this study, we identify PKD as a positive mediator of autophagy and a Vps34 kinase. We show that ectopic expression of PKD induces the accumulation of early and late autophagosomes by measuring LC3 punctate staining and by electron microscopy and provide proof that PKD participates in the induction of autophagy during oxidative stress. Interestingly, Vps34 is the second lipid kinase reported as a PKD substrate; PKD also phosphorylates PI4KIIIβ.20 In addition, two other substrates of PKD, CERT and OSBP, are also involved in lipid signaling.21, 22 Furthermore, Vps34 has been shown to localize to the TGN,23 similar to these three other known PKD substrates. Thus, it is plausible that PKD has a broad role in controlling phosphatidylinositol-regulated events in the TGN.
We have previously reported that oxidative stress induces DAPk activation, and that under these stress conditions, DAPk activates PKD through phosphorylation. Oxidative stress, resulting from the accumulation of reactive oxygen species, creates damage to cellular components that are then sequestered by autophagosomes. Thus, under such stress conditions, regulatory proteins must be activated to consequently stimulate the autophagic machinery. Here, we provide proof that the DAPk–PKD pathway plays an important role in the process of autophagy and specifically that both DAPk and PKD are required for autophagy under oxidative stress. Moreover, we establish the epistatic relationship between DAPk and PKD by showing that PKD is a downstream effecter of DAPk in the regulation of autophagy. DAPk is a tumor-suppressor protein whose expression is lost in a vast number of human malignancies. Although DAPk mediates cell death following various stress stimuli, including both apoptosis and autophagic cell death during ER stress,9 not many molecular events downstream of DAPk activation have been elucidated. In recent studies from our lab, Beclin 1, a major Vps34 regulatory protein, was identified as a direct substrate of DAPk. Phosphorylation by DAPk leads to dissociation of Beclin 1 from its inhibitors Bcl-2/Bcl-XL10, 24 and this presumably enables the binding to and activation of Vps34. We here further delineate the network of DAPk-regulated events that induce autophagy by revealing that activation of PKD under oxidative stress is essential for this process. Thus, DAPk regulates the Vps34–Beclin 1 complex by two mechanisms: the first through phosphorylation of Beclin 1, and the second indirectly through phosphorylation of PKD, which phosphorylates and activates Vps34.
Materials and Methods
Cell lines and transfections
HEK 293T cells were grown in Dulbecco's modified Eagle's medium (Biological Industries, Beit-haemek, Israel) supplemented with 2 mM glutamine (Gibco BRL), 100 U/ml penicillin and streptomycin (Gibco BRL, Carlsbad, CA, USA), and 10% fetal bovine serum (Thermo Scientific Hyclone, Logan, UT, USA). Transfections were performed by the calcium phosphate method.
DNA constructs, antibodies, and reagents
PKD1-GFP was kindly provided by K Pfizenmaier (Stuttgart, Germany). A kinase-dead mutant of PKD-GFP was generated by introducing the following point mutations: K612W and D727A. HA-PKD was kindly provided by A Toker (Boston, MA, USA) and GFP-LC3 and RFP-GFP-LC3 by T Yoshimori (Osaka, Japan); mDsRed-LC3 was generated by replacing the GFP tag with monomeric mDsRed; GFP-DFCP1 was kindly provided by N Ktistakis (Cambridge, UK); hVps34-FLAG was generated by adding FLAG sequence by PCR. The following antibodies and immunological reagents were used: anti-Flag M2 gel beads, FLAG peptide, monoclonal antibodies against actin AC-40 and DAPk, polyclonal antibodies against LC3B (Sigma-Aldrich, St. Louis, MO, USA); polyclonal antibodies against HA, PKD, and Beclin 1, protein G PLUS-Agarose (Santa Cruz Biotechnology, Santa Cruz, CA, USA); monoclonal antibodies against HA (BAbCo); monoclonal antibodies against p62 (BD Pharmingen, San Diego, CA, USA); and HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies (Jackson Immuno-Research, Suffolk, UK). Antibodies were visualized by enhanced chemiluminescence (Supersignal; Pierce, Thermo Scientific, Rockford, IL, USA).
RNA interference
DAPk expression was knocked down by the targeting sequences 5′-CATGGAGAAATTCAAGAAG-3′ or 5′-TGAGAAGCATGTAATGTTA-3′ of human DAPk (NM_004938) inserted into pSUPER. PKD1 expression was knocked down by PKD-targeting shRNA in pSUPER vector kindly provided by A Toker or by siRNA SMARTpool targeting PKD (Dharmacon, Lafayette, CO, USA). As shRNA and siRNA control, the non-mammalian protein HcRed (AF363776) was targeted by the sequence 5′-GTATGCGCATCAAGATGTA-3′.
Cell extracts and immunoprecipitation
Cells were lysed in buffer A8 or immunoprecipitation lysis buffer (10 mM TRIS pH 7.4, 300 mM NaCl, 0.5% NP-40). Following preclearance in protein G PLUS-Agarose, the extracts were incubated with anti-FLAG M2 gel beads, washed extensively, and the immunocomplexes were boiled in sample buffer, separated on SDS-PAGE, and immunoblotted.
In vitro kinase assay
PKD-GFP (wild type (WT) or kinase dead (KD)), Vps34-FLAG WT, or Vps34-FLAG T677A expressed in HEK293T cells were immunopurified and/or eluted as follows: for FLAG immunoprecipitation, cells were lysed in cold B-buffer25 and immunoprecipitated as described above plus two additional washes in 54K buffer.25 The proteins were eluted with excess FLAG peptide. For GFP immunoprecipitation, cells were lysed in cold RIPA buffer and immunoprecipitated as described above. PKD-GFP (WT or KD) was incubated with either WT or T677A Vps34-FLAG in a kinase reaction buffer (30 mM TRIS pH 7.4, 30 mM MgCl2) supplemented with 10 μCi of 33P-labeled γ-ATP, 50 μM ATP, protease inhibitors, 1 mM NaF, and 40 mM β-glycerophosphate.
Immunofluorescence and punctate staining assay
Cells were fixed in 3.7% formaldehyde and viewed by fluorescent microscopy (Olympus BX41, Olympus, Center Valley, PA, USA) with 60 × (NA 1.25) or 100 × (NA 1.3) UPlan-Fl oil immersion objectives, and digital images obtained with a DP50 CCD camera using ViewfinderLite and StudioLite software (Olympus). The percentage of cells with punctate GFP-LC3 or mDsRed-LC3 fluorescence (more than 3 puncta/cell) per total GFP-LC3 or mDsRed-LC3-positive cells was quantified. Triplicates of at least 100 transfected cells were counted for each point. To calculate RFP-only puncta in cells transfected with the tandem RFP-GFP-LC3 construct, RFP and GFP dot signals were extracted using the Top Hat algorithm of MetaMorph image and analysis software (Molecular Devices, Sunnyvale, CA, USA), and the total area of the RFP-only dots was calculated and divided by the total cell area. For GFP-DFCP1, the dot signals were extracted from duplicates of at least 100 cells for each point using the Top Hat algorithm of MetaMorph, and the total area of the dots was calculated and divided by the total cell area.
Transmission electron microscopy (TEM) and immunogold staining
HEK293T cells were transfected with PKD-GFP and fixed in Karnovsky's fixative.15 Cells were embedded with agar noble (1.7%) and postfixed with 1% OsO4, 0.5% K2Cr2O7, and 0.5% C6FeK4N6+4 in 0.1 M cacodylate buffer. The pellet was stained and blocked with 2% aqueous uranyl acetate followed by ethanol dehydration, and embedded in graded Embed812 (EMS, Hatfield, PA, USA). Ultrathin sections (70–90 nm, ultramicrotome Leica UCT, Leica Microsystems GmbH, Wetzlar, Germany) were analyzed under 120 kV at Tecnai 12 TEM and imaged with Eagle 2k × 2k CCD camera FEI (Eindhoven, The Netherlands). Immunogold labeling was performed as described previously.15
Mass spectrometry analysis
Proteins on SDS-PAGE were reduced, modified with 40 mM iodoacetamide, and proteolyzed (modified trypsin or chymotrypsin (Promega, Madison, WI, USA). Peptides were resolved by reverse-phase chromatography on 0.075 × 200-mm fused silica capillaries (J&W, Hants, UK) packed with Reprosil reversed-phase material (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) and eluted with linear 90 min gradients of 5–45% and 15 min of 95% acetonitrile with 0.1% formic acid at flow rates of 0.25 μl/min. Mass spectrometry was performed by an ion-trap mass spectrometer (Orbitrap, Thermo Scientific, Waltham, MA, USA) in a positive mode using repetitively full MS scan followed by collision-induced dissociation (CID) of the seven dominant ions selected from the first MS scan. Phosphopeptides were enriched on Fe+2 columns (Sigma-Aldrich) (binding in 250 mM acetic acid, 30% acetonitrile, and elution in 400 mM ammonium hydroxide). These peptides were analyzed similarly except the usage of multistage activation in the fragmentation method. The mass spectrometry data were analyzed using the Sequest 3.31 software (J Eng and J Yates, University of Washington, Seattle, WA, USA, and Finnigan, San Jose, CA, USA) and Pep-Miner26 searching against the human part of the NCBI-NR database.
Statistical analysis
Student's t-test was used to compare the differences between two groups. Statistical analysis of the immunogold labeling distribution was performed by χ2 test according to published methods.19
Acknowledgments
We thank Helena Sabanay for performing electron microscopy analysis; the Smoler Proteomic Center (Technion, Haifa, Israel) for performing the mass spectrometry analysis; and Shani Bialik for fruitful discussions. This work was supported by a grant from the Flight Attendant Medical Research Institute (FAMRI) Center of Excellence and by the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel's Ministry of Science and Technology (MOST). AK is the incumbent of Helena Rubinstein Chair of Cancer Research.
Glossary
- 3MA
3 methyladeninde
- Atg
autophagy related
- DAPk
death-associated protein kinase
- PI3K
phosphoinositide 3 kinase
- PI(3)P
phosphatydilinositol-3-phosphate
- PKD
protein kinase D
- PE
phosphatidyl-ethanolamine
- TGN
trans-Golgi network
- EM
electron microscopy
- WT
wild type
- KD
kinase dead
- TEM
transmission electron microscopy
- CID
collision-induced dissociation
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Baehrecke
Supplementary Material
References
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14:376–391. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggi M, Du C, Zhang W, Balaji KC. Protein kinase D1: a protein of emerging translational interest. Front Biosci. 2007;12:3757–3767. doi: 10.2741/2349. [DOI] [PubMed] [Google Scholar]
- Waldron RT, Rozengurt E. Oxidative stress induces protein kinase D activation in intact cells. Involvement of Src and dependence on protein kinase C. J Biol Chem. 2000;275:17114–17121. doi: 10.1074/jbc.M908959199. [DOI] [PubMed] [Google Scholar]
- Storz P, Doppler H, Toker A. Protein kinase Cdelta selectively regulates protein kinase D-dependent activation of NF-kappaB in oxidative stress signaling. Mol Cell Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Kimchi A. DAP kinase regulates JNK signaling by binding and activating protein kinase D under oxidative stress. Cell Death Differ. 2007;14:1908–1915. doi: 10.1038/sj.cdd.4402212. [DOI] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozuacik D, Bialik S, Raveh T, Mitou G, Shohat G, Sabanay H, et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008;15:1875–1886. doi: 10.1038/cdd.2008.121. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Elazar Z. Monitoring starvation-induced reactive oxygen species formation. Methods Enzymol. 2009;452:119–130. doi: 10.1016/S0076-6879(08)03608-2. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol. 2009;452:143–164. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Doppler H, Storz P, Li J, Comb MJ, Toker A. A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J Biol Chem. 2005;280:15013–15019. doi: 10.1074/jbc.C400575200. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew TM, Lucocq JM, Griffiths G. Relative labelling index: a novel stereological approach to test for non-random immunogold labelling of organelles and membranes on transmission electron microscopy thin sections. J Microsc. 2002;205 (Part 2:153–164. doi: 10.1046/j.0022-2720.2001.00977.x. [DOI] [PubMed] [Google Scholar]
- Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann T, Hausser A, Schoffler P, Schmid S, Pfizenmaier K, Olayioye MA. Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J Cell Biol. 2007;178:15–22. doi: 10.1083/jcb.200612017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhek S, Ngo M, Yang X, Ng MM, Field SJ, Asara JM, et al. Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol Biol Cell. 2010;21:2327–2337. doi: 10.1091/mbc.E10-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5:720–722. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- Shani G, Marash L, Gozuacik D, Bialik S, Teitelbaum L, Shohat G, et al. Death-associated protein kinase phosphorylates ZIP kinase, forming a unique kinase hierarchy to activate its cell death functions. Mol Cell Biol. 2004;24:8611–8626. doi: 10.1128/MCB.24.19.8611-8626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer I, Barnea E, Ziv T, Admon A. Improving large-scale proteomics by clustering of mass spectrometry data. Proteomics. 2004;4:950–960. doi: 10.1002/pmic.200300652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.