Abstract
In Arabidopsis, lateral root initiation occurs in a subset of pericycle cells at the xylem pole that will divide asymmetrically to give rise to a new lateral root organ. While lateral roots never develop at the phloem pole, it is unclear how the interaction with xylem and phloem poles determines the distinct pericycle identities with different competences. Nevertheless, pericycle cells at these poles are marked by differences in size, by ultrastructural features and by specific proteins and gene expression. Here, we provide transcriptional evidence that pericycle cells are intimately associated with their vascular tissue instead of being a separate concentric layer. This has implications for the identification of cell- and tissue-specific promoters that are necessary to drive and/or alter gene expression locally, avoiding pleiotropic effects. We were able to identify a small set of genes that display specific expression in the phloem or xylem pole pericycle cells, and we were able to identify motifs that are likely to drive expression in either one of those tissues.
Keywords: pericycle, tissue specificity, cell identity, lateral root initiation, Arabidopsis
1. Introduction
The Arabidopsis primary root presents a radial pattern of tissue types that are arranged in concentric layers: epidermis, ground tissues (cortex and endodermis) and vascular cylinder (pericycle, xylem and phloem) [1]. This organization finds its origin in the embryonic development of the root with periclinal (radial) and anticlinal (circumferential) cell divisions giving rise to new tissue layers and expanding these tissue layers, respectively. The first rounds of periclinal and anticlinal divisions within the proembryo give rise to an inner cell mass surrounded by epidermal precursors. Further divisions of these inner cells lead to the formation of central vascular precursors encircled by a layer of ground tissues. The pericycle is first observed in between the globular and heart-shaped embryo stage, where the provascular tissue divides into outer pericycle and inner diarch cores of xylem and phloem vascular precursors. Finally, divisions of the ground tissues complete the patterning of the root, with the creation of an inner layer of endodermis and an outer layer of cortex [2–4].
Within the pericycle tissue, genes involved in various physiological processes are expressed: thioredoxin [5], biosynthetic enzymes or gene transcripts involved in the initial and final steps of tropane alkaloid biosynthesis [6,7], sulphate transport [8], cytokinin biosynthesis [9,10], morphine biosynthesis [11], ascorbate homeostasis [12], xylem loading of potassium and boron [13,14], etc. But, most importantly, a subset of pericycle cells is giving rise to a new lateral root, which is associated with spatially and temporally controlled gene expression in these cells [15,16].
(a). Distinct pericycle cell populations
Even though pericycle cells initiate from the same subset of initial cells as the rest of the stele, which present a diarch symmetry, their regular shape and radial organization around the vascular tissues made them traditionally regarded as a concentric homogeneous layer [1]. A growing number of reports, however, emphasize differences between pericycle cells according to their position adjacent to the xylem or the phloem poles, such as cell division potential [17], cell cycle progression [18], cell size [17,19], cell surface arabinogalactan–protein distribution and cell wall thickening [20–22], plasmodesmatal connections [23,24], microtubule content [25], cytoplasm content [26,27], marker gene expression [27–31] and differential gene transcription [32,33].
Marker lines have always been useful to address cell identity and, in this respect, a collection of enhancer trap lines has been extremely helpful [27,28,34]. For example, the marker line J2661 shows an expression pattern generalized to the entire pericycle [35], whereas two other marker lines, J0121 and S17, show expression in the pericycle cell populations associated with the xylem and the phloem poles, respectively [28,30]. Using another marker line, i.e. RM1007, it is already possible to observe pericycle differentiation in at least two different cell populations in the heart-stage embryo [27]. In this marker line, GFP expression starts in the pericycle cells which are in contact with the quiescent centre and which are later associated with the xylem poles, indicating that bilateral symmetry of the pericycle and genetic distinction between two different populations of cells occurs as early as the formation of the pericycle cells [27]. Consequently, these observations indicate that there is most probably no time frame in plant development where the full pericycle consists of a highly homogeneous set of cells [27].
(b). Xylem pole pericycle-associated lateral root initiation in Arabidopsis
Another particularity distinguishing pericycle cells adjacent to the xylem poles in comparison with those adjacent to the phloem poles is their competence for lateral root initiation. Even though lateral root initiation has been extensively studied and many molecular players were discovered in the past decade [34,36–39], the mechanisms controlling xylem pole pericycle (XPP) cell identity and competence for lateral root initiation are largely unknown.
The strong association between lateral root initiation and the xylem pole is supported by the lonesome highway (lhw) mutant, which displays only one xylem pole and one-sided lateral root formation [27]. Nevertheless, a differentiated protoxylem element within the xylem pole is not required as impaired vasculature development (ivad) and arabidopsis histidine phosphotransfer protein 6 (ahp6) mutants still display lateral root initiation [27]. On the other hand, the wooden leg (wol) mutant lacks phloem and all pericycle cells have xylem pole-associated identity, but it hardly produces lateral roots [27]. However, these wol pericycle cells respond to auxin with respect to their division potential [27]. At present, it is unclear how the interaction with xylem and phloem pole determines the distinct pericycle identities with differing competence. The study of mutants concomitantly impaired in vasculature, pericycle and lateral root development could, however, show that the determination of both the pericycle and the vasculature is regulated by a common genetic pathway [27].
(c). Cis-regulatory elements
The temporal and spatial transcriptional activity of genes is—to some extent—influenced by cis-regulatory elements [40,41]. Transcription factors will bind in a sequence-specific manner to cis-regulatory elements in promoters of genes (through activation or repression) or act through protein–protein interactions to regulate their gene expression [40]. In Arabidopsis, there are more than 60 families containing more than 2000 genes encoding for transcription factors [42–44]. Various methods are available to identify or confirm cis-regulatory elements and their interaction with particular transcription factors in silico, in vitro and in vivo [45], but this has only limitedly been addressed in the context of distinct expression patterns in the root [46].
(d). Genome-wide cell-specific transcript data of the Arabidopsis root
Recently, gene expression data for various tissues and cell types in Arabidopsis became available through the ‘digital in situ’ [32,47,48]. Cell sorting and transcriptome analysis of various tissue-specific marker lines, including J2661, J0121 and S17, allowed the comparison of the transcriptome of these cell types with each other's but also with that of the other layers of the root [32,33].
We propose here a simple approach to use the microarray data—generated on these cell-sorted marker lines in combination with other available data—to establish transcriptional differences between the xylem- and the phloem-pole-associated pericycle and the other tissues of the root, and to reveal a subset of genes that display a differential or an enriched expression in those populations of pericycle cells. Genes with such a specific expression pattern will be crucial in the future to target gene expression to a particular group of cells. In addition, we identified motifs that likely drive XPP- or phloem pole pericycle (PPP)-specific gene expression.
2. Material and methods
(a). Microarray retrieving and normalization
The following microarray hybridization files were retrieved from the gene expression omnibus database: GEO series GSE8934 (S17, S32, COBL9, J0121, S4, SUC2, J2501, RM1000), GEO series GSE7641 (PET111, AGL42), GEO Series GSE5749 (LRC, GL2, SCR, J0571, WOL) and GEO series GSE16468 (CORTEX, APL, S18). J2661 hybridization files were kindly provided by the author [35]. Normalizations of the respective datasets concerning only the pericycle (J2661, J0121 and S17) or the entire root (J2661, J0121, S17, S32, COBL9, S4, SUC2, J2501, RM1000, PET111, AGL42, LRC, GL2, SCR, J0571, WOL, CORTEX, APL and S18) have been performed using the robust multi-array average method [49]. Affymetrix probesets to Arabidopsis Genome Initiative ID assignment was performed using the affy_ATH1_array_elements-2010-12-20.txt file downloaded from The Arabidopsis Information Resource (TAIR; www.arabidopsis.org) for 21 107 probesets corresponding to unique AGI IDs.
(b). Differential expression of genes in between pericycle cell populations
Differential analysis was performed using the moderated t-test [50] within the ‘affylmGUI’ vignette for R package. ‘Present’, ‘marginal’ and ‘absent’ calls were generated using the MAS5 algorithm in the ‘affy’ vignette for R package [51]. For the first approach, a gene was considered as being differentially expressed if it could fulfil the following conditions: at least two ‘present’ calls in at least one of the microarray triplicates (J2661, S17, J0121; figure 1), a fold change of at least 2 and a maximum p-value of 0.01 in at least one of the three pairwise comparisons (J0121 versus J2661, J0121 versus S17 and J2661 versus S17) and a similar fold change sign for all three pairwise comparisons. For the second approach, a gene was considered as being differentially expressed if it showed three ‘present’ flags in one of the two microarray experiments J0121 and S17 and three ‘marginal’ or ‘absent’ flags in the other microarray experiments. Fold changes, p-values and flag calls are available in the electronic supplementary material, table S1. Gene ontology enrichment analysis was performed using the Parametric Analysis of Gene set Enrichment (PAGE) algorithm [52] with the Agri Gene Ontology (AgriGO) analysis toolkit [53], with default settings, on the list of genes significantly upregulated in the XPP or the PPP, with an average fold change calculated from the three pairwise comparisons (see the electronic supplementary material, table S1).
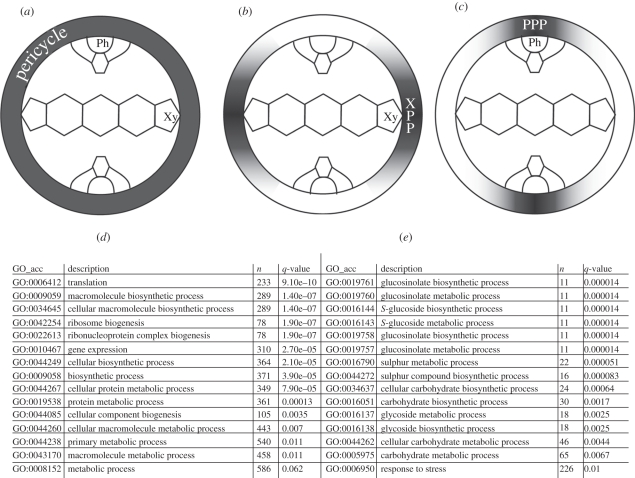
Figure 1.
Pericycle cell populations. Expression profiles of the marker lines used for the microarray experiments and the pairwise comparisons: (a) entire pericycle (XPP + PPP), (b) xylem pole pericycle (XPP) and (c) phloem pole pericycle (PPP). Gene ontology enrichment analysis for the genes significantly upregulated in the (d) XPP and (e) PPP. The symbolized vascular tissues are xylem (Xy) and phloem (Ph).
(c). Specific up- or downregulation of genes in pericycle populations in comparison with the other layers of the root
For each of the four different cell populations (pericycle and primordia), the expression value average in the selected population was individually compared with its expression value in each other selected layer (figure 2a). A p-value was evaluated from a t-test with Welch assumption of the variance, which was performed in between values of the different replicates for the selected population against all the replicate values of all other selected layers. A gene was declared significantly and specifically enriched in one population of cells if its expression level was at least two times higher or lower in comparison with each other individual selected cell layer with a maximum p-value of 0.01. For each candidate, an average enrichment was proposed by calculating the ratio between its expression value in the considered pericycle or primordia cell population against the average expression in the other layers. In case a gene could be selected to be specific of two cell populations, a best hit was finally proposed by selecting the best ratio.
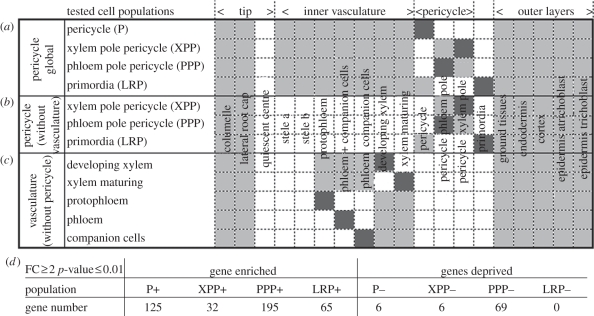
Figure 2.
Specific layer gene expression. For each tested cell population, a list of genes specifically up- or downregulated was computed by calculating the ratio between the expression value in this cell population (dark grey) with each other selected layer (light grey). (a) Global analysis involving most root tissues. (b,c) Pericycle and vasculature analysis excluding, respectively, the vasculature and the pericycle tissues. (d) Number of genes up- or downregulated in the different cell populations characterized in the global analysis. FC, fold change.
(d). Microarray clustering and enrichment analysis
The clustering was generated using the MeV [54] hierarchical clustering module, with a Pearson correlation distance metric and complete linkage clustering. We used normalized logarithmic expression values extracted from the previously described compendium of microarray data for all genes and for the marker lines depicted in the tree. It cannot be fully excluded that one tissue contaminates the neighbouring one because of incomplete cell separation before cell sorting. However, there are no high correlations between the level of expression (normalized absolute expression values) of the top 100 genes of each considered layer and their neighbouring ones, suggesting a low bias owing to such contamination on the enrichment analysis (data not shown). To perform the enrichment analysis, the number of genes being individually or commonly regulated in a given cell population of the pericycle (figure 2b) and in a layer of the vasculature (figure 2c) were calculated as described previously with a p-value threshold of 0.05 and a range of 21 different fold change thresholds varying between 1.1 and 3. The formula used to calculate the enrichment can be summarized as: enrichment (%) = (NbObsPerVasc × NbGene × 100)/(NbPer × NbVasc), NbObsPerVasc being the observed number of genes concomitantly up or downregulated in a pericycle and in a vascular cell population, NbGene being the total number of genes in the microarray, NbPer being the total number of genes up- or downregulated in the pericycle cell population and NbVasc being the total number of genes up- or downregulated in the vascular cell population. For the fold changes (from 1.1 to 3), 20 different values have been tested to assess the robustness and the consistency of the analysis. The significant enrichment values (Fisher test ≤ 0.05) were averaged to generate the radar graphic presented in this study (figure 3b).
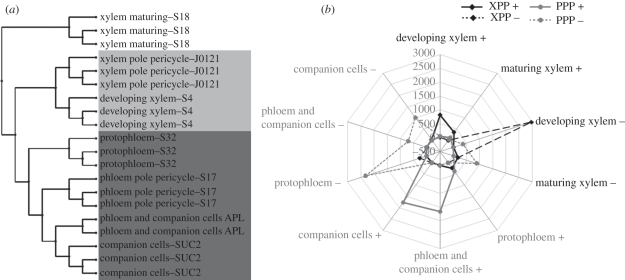
Figure 3.
Cell identity. (a) Hierarchical clustering of the microarray related with pericycle and vascular cell populations. (b) Enrichment analysis of vascular genes in the different populations of pericycle cells. Over-representation of genes concomitantly regulated is expressed as a percentage.
(e). Motif identification
To identify statistically over-represented motifs in the DNA sequences 500 bp upstream of the start codon for the xylem and phloem pericycle-specific genes, we used ymf v. 3.0 (http://wingless.cs.washington.edu/YMF/YMFWeb/YMFInput.pl) [55]. Upstream DNA sequences were retrieved from TAIR (http://www.arabidopsis.org/tools/bulk/sequences/index.jsp; electronic supplementary material, data). Motif size was set at 6, 7 and 8 with a maximum of 0 spacers in the middle and a maximum of 1 degenerate symbols [R (purine − A or G), Y (pyrimidine − C or T), W (A or T) and S (C or G)]. On the list of significant motifs generated by ymf v. 3.0, we applied FindExplanators v. 1.1.2 to extract a smaller set of ‘real’ motifs [56]. The significance of a motif was measured by the z-score of its count in the input sequences. If a motif occurs N times in the input sequences, but was expected to occur E times (with standard deviation of S) in random sequences of the same length (generated by a suitable background model), then the z-score of the motif is (N − E)/S. In addition, to confirm the 6 and 8 bp hits from ymf v. 3.0, we used Weeder v. 1.3 (http://159.149.109.9/modtools/) [57] using standard settings on the two strands. Finally, we used Pscan (http://159.149.109.9/pscan/) to identify potential transcription factors or transcription factor families that can bind to 500 bp upstream DNA sequences of xylem and phloem pericycle-specific genes.
3. Results
(a). Differential expression of genes in between pericycle cell populations
To access the differential expression of genes within pericycle cell populations, we used two different approaches based on the same dataset. Firstly, we have performed a pairwise comparison of three microarray experiments performed on the full pericycle (figure 1a), on the XPP (figure 1b) and on the PPP (figure 1c). On the basis of a minimum fold change of 2 and a maximum p-value of 0.01 (see §2), we could generate a list of genes showing higher expression in the xylem pole (1357 genes) or in the phloem pole (1763) pericycle (electronic supplementary material, table S1). These numbers already show that the two populations of pericycle cells present important differences at the transcriptomic level. An analysis of the gene ontology enrichment on these lists shows that the pericycle associated with the xylem poles presents a high enrichment of genes involved in primary metabolism processes such as translation, biosynthetic processes and ribosome biogenesis, which makes sense in view of its potential to divide (figure 1d). The population of cells constituting the pericycle associated with the phloem pole rather show an enrichment in secondary metabolites processes (figure 1e). Secondly, we have made a restrictive gene selection based on ‘present’, ‘marginal’ and ‘absent’ presence detection call flags between the XPP (figure 1b) and the PPP (figure 1c) microarray experiments (see §2). We found 1567 genes specifically present in the phloem pole and absent in the XPP population. In contrast, we found only 99 genes being present in the xylem pole and absent in the PPP populations (electronic supplementary material, table S1). These two lists, however, did not yield obvious significant gene ontology enrichment.
(b). Gene expression enrichment in the pericycle cell populations
To unravel the specificity of the pericycle cell populations in comparison with the other cell layers of the root, we generated different lists of genes specifically up- or downregulated in these layers. All the radial data of the Arabidopsis high-resolution root spatio-temporal map [32] were gathered together to form a compendium representative of the main layers of the primary root. We created four different lists of genes specifically up- or downregulated in the overall pericycle (P), in one of the two xylem or phloem pole-associated populations (respectively, XPP and PPP), or in the lateral root primordia. Genes were declared specifically regulated in one of these cell populations if their expression level was at least two times higher or lower in comparison with every other individual selected cell layer with a maximum p-value of 0.01 (figure 2a; see §2). A first result appearing out of generating these four different gene lists is that a higher number of genes is specifically upregulated in comparison with genes specifically downregulated in one cell layer (figure 2d and the electronic supplementary material, table S2). Also, we could isolate a higher number of genes specifically upregulated in the PPP cell population in comparison with the XPP and the whole pericycle cell populations (this assertion remains true with different parameters of fold change and p-value tested; data not shown). To generate a final and restricted gene list, with high confidence in their XPP- or PPP layer-specific enrichment, we intersected this last analysis with the previous detection call flags analysis. A smaller list of genes could be generated, out of which 63 genes are specifically enriched in the PPP and not expressed in the XPP and seven genes are in contrast specifically enriched in the xylem pole and not expressed in the phloem pole (electronic supplementary material, table S2). Our first approach identified, for instance, GATA23 as a XPP enriched gene, which is supported by its reported expression pattern [39]. Owing to our stringent criteria, GATA23 was not retained in the final list, which nevertheless increases confidence in this list.
(c). Pericycle population transcriptomes reflect their bilateral association with neighbouring vascular tissues
The intimate association of the pericycle cell populations with their neighbouring vascular tissues also raises the question of their genetic connection. To compare their transcriptomic regulations, we performed a hierarchical clustering of the related microarray datasets. Interestingly, clustering the microarrays of the different layers constituting the stele (xylem maturing, developing xylem, protophloem, phloem and companion cells, companion cells) with the microarrays of the XPP- and PPP strikingly shows an intimate association of each pericycle cell population with its neighbouring vascular tissue (figure 3a). To reinforce this observation, we conducted an enrichment analysis by analysing the number of genes that are specifically and commonly enriched in a cell population of the pericycle (figure 2b) together with a layer of the vasculature (figure 2c), and in comparison with the other layers of the root (see §2). The enrichment was calculated as a percentage of the observed number of genes concomitantly enriched in a pericycle population together with a vascular tissue in comparison with the expected number of such genes based on the individual percentage of genes enriched in one or the other of these tissues (figure 3b). We observed a significant over-representation of genes commonly regulated in groups composed of a pericycle cell population and its neighbouring vascular tissues. Specifically, there is an over-representation of genes concomitantly regulated in the XPP and in the developing xylem (figure 3b). Similarly, there is an over-representation of genes concomitantly regulated in the PPP and a phloem tissue (mainly phloem together with the companion cells and companion cells; figure 3b).
(d). Motifs for pericycle population-specific gene expression
The identification of common motifs driving expression of XPP- or PPP-specific genes is useful to gain insight into the regulatory network of these cell populations and as a tool for targeted gene expression. While motifs can also be over-represented in intragenomic conserved non-coding sequences [58,59], we focused here on the 500 bp upstream regulatory sequence (see §2). We restricted our analyses to 500 bp as this length is often sufficient to confer expression patterns similar to longer upstream fragments [60] and is routinely used for the identification of regulatory elements enriched in promoters of target genes [61,62].
To verify whether specific motifs could be responsible for XPP- and PPP-specific expression, we searched within the final and restricted list of XPP- and PPP-specific genes for particular motifs that might be involved in regulating the tissue specific expression. We used ymf v. 3.0 on the DNA sequences 500 bp upstream of the start codon of the putative XPP- and PPP specifically enriched genes (see §2), and this resulted in the identification of several putative motifs with a 6-, 7- and 8-nucleotide length (electronic supplementary material, table S3). Several of these could be confirmed using Weeder v. 1.3 on the same set of upstream sequences (electronic supplementary material, table S3).
To determine whether particular transcription factors or transcription factor families are likely to bind these promoters we used Pscan, which revealed some interesting candidates for future analyses (electronic supplementary material, table S3). However, combining this analysis with the list of XPP- or PPP-enriched genes did not yield any obvious common candidates.
Interestingly, At5g01740, which lies close to the position of the enhancer element in J0121, is present in the restricted list of genes specifically expressed in the XPP and not expressed in the PPP. A more detailed analysis of the region showed that some identified motifs are present in this region, supporting our motif analyses (figure 4).
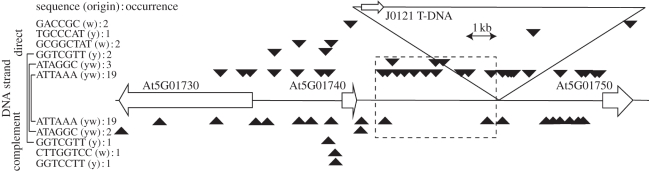
Figure 4.
Xylem pole pericycle consensus sequences in the region of J0121 T-DNA insertion. Indicated are the three main open reading frames in a region of 19 kb (At5G01730, At5G01740 and At5G01750), the redundant 6-nucleotide, and all the 7- and 8-nucleotide consensus sequences that were found and the place of the T-DNA insertion in the genome of the marker line J0121. The dashed line rectangle indicates a region containing the two most represented sequences in a region 4 kb upstream of the T-DNA insertion. The arrow on the T-DNA indicates the position of the enhancer trap region. The identifier y and w indicates the origin of the sequences using ymf and Weeder algorithms, respectively.
4. Discussion
(a). Pericycle cells are intimately associated with their vascular tissue in Arabidopsis
Only a limited percentage of genes were shown to be specifically enriched in either PPP (1.5%) or XPP (4.5%) in comparison with the 21 107 genes taken into consideration on the ATH1 microarray. Notwithstanding only 23–40% of plant genes are predicted to have a strict tissue-specific expression [63], this is still surprisingly low. Still, these results are in agreement with the spatio-temporal expression map of the root ([32]; the electronic supplementary material, table S2).
Our analyses show that the pericycle and associated vasculature share a high overlap in gene expression, explaining why only very few genes are strictly enriched in the pericycle cell type. Both the clustering of the microarray and the enrichment analysis show a link between the XPP and the developing xylem. This connection is also apparent from, for example, the expression pattern of ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6), namely at the protoxylem position and in the two associated pericycle cells [29]. Interestingly, there is a less obvious link with the maturing xylem, which is a tissue undergoing deep differentiation processes leading to programmed cell death [64]. The situation is different for the PPP, which shares much more common upregulation with the cells constituting the mature phloem and its companion cells or the companion cells themselves. Considering upregulations, the PPP shares more genes with its neighbouring tissues compared with the number of genes shared by the XPP with the early associated developing xylem tissue. This observation would indicate that if both PPP and XPP populations are related with their neighbouring vascular tissues, then this link is higher for the PPP. These results are in agreement and complementary with a recent analysis characterising a stele-enriched gene regulatory network in the Arabidopsis root [32,65]. Among 11 distinct dominant transcription patterns in the stele tissues, the authors could find a pattern with PPP gene expression associated with the phloem poles but not for the XPP with the xylem. Our analysis is not restricted to the most dominant patterns and could show such a link even though a lot less strong than for the PPP. Interestingly, the authors also described two dominant patterns of genes expressed exclusively in the pericycle, which are, respectively, in front of one or the other vascular pole, but not in the overall layer [65], pointing to a similar conclusion that the pericycle presents a more heterogeneous fate than expected for a concentric tissue layer. Furthermore, the observation that xylem shares more identity with a precursor of its physically neighbouring tissue fits with the vision of the XPP as an extended meristem [18,27]. In this case, the identity of the XPP should be closer to a young tissue rather than a highly differentiated one.
(b). Cis-regulatory elements for xylem and phloem pole-associated pericycle
Over the years, there has been a necessity to use a XPP-specific marker, but hardly any expression patterns with specificity for XPP have been described. The most popular one available is the GAL4 enhancer trap line J0121 and this marker has been used as a XPP cell identity marker [27,66,67], for XPP-specific transactivation [28,39,68,69], and for cell sorting [34]. But, up to date, it is still unclear which gene causes this specific expression pattern in J0121. With respect to the PPP, the promoter from the gene At2g22850 has been shown to give PPP-specific expression (referred to as S17) [30]. This gene is present in the list of genes we describe as being differentially expressed in between the xylem and the phloem pole-associated pericycle and fit the corresponding expression pattern (flags indicate an absence from the XPP and a presence in the PPP). However, this gene was not described as being enriched in the PPP ([32] and this study). It remains to be elucidated if the S17 transcriptional fusion [30] does not reflect the entire expression pattern of the gene in planta, or if the microarray probesets of this gene present specificity problems.
Obviously, one gene expected within our dataset is the putative gene responsible for the specific expression in J0121. Recently, the position of the enhancer element in J0121 was determined to be inserted between two genes At5g01740 and At5g01750, but the relation—if any—to lateral root initiation remains to be determined [28]. Interestingly, At5g01740 is present in the restricted list of genes specifically enriched in the xylem pole and not expressed in the PPP, indicating a potential role of this gene in XPP cell fate. Its annotation as a member of the nuclear transport family makes it an interesting candidate for further functional analysis. At5g01750, on the other hand, was found to be differentially expressed in the PPP in comparison with the XPP, but also in other layers of the root. This observation indicates high chances for the promoter region of At5g01740 to enhance the J0121 marker line expression pattern.
For uniform and strong control of expression in different ecotypes and for possible translation of this knowledge to crops, the identification of cell-type-specific promoters or motifs that drive expression locally is essential. Here, we identified a number of cis-regulatory elements for XPP- and PPP-specific gene expression, which could be used for various applications, and we identified putative regulatory transcription factors in silico. Obviously, this might be oversimplified as multiple cis-regulatory elements might be required to integrate signals from multiple transcription factors resulting in combinatorial control and highly specific patterns of gene expression [40]. In future, experimentally validating if these single elements are sufficient to drive specific in planta expression in distinct pericycle populations will provide an answer.
These elements can be used to create synthetic promoters to drive expression of a (trans)gene under temporal and spatial conditions, avoiding the pleiotropic effects owing to broader expression. In a systems biology approach, these cis-regulatory elements (and their binding transcription factors) can be used to identify genes belonging to the same regulatory pathways and to understand how transcriptional regulation mechanisms work and how spatial and temporal expression networks are established during organogenesis and maintained throughout the life cycle. In addition, these elements can be used as baits—for instance, using a yeast one-hybrid approach—to discover their interacting transcription factors [65,70].
Acknowledgements
Ive De Smet is supported by a BBSRC David Phillips Fellowship (BB_BB/H022457/1), a Marie Curie European Reintegration grant (PERG06-GA-2009-256354) and the Research Foundation Flanders (FWO09/PDO/064 A 4/5 SDS). Boris Parizot and Ianto Roberts are supported by the Research Foundation Flanders (FWO, grant no. 3G002911). Ianto Roberts is indebted to the Agency for Innovation by Science and Technology in Flanders (IWT). This work was supported by a grant from the Interuniversity Attraction Poles Programme (IUAP VI/33), initiated by the Belgian State, Science Policy Office.
References
- 1.Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84 [DOI] [PubMed] [Google Scholar]
- 2.Scheres B., Wolkenfelt H., Willemsen V., Terlouw M., Lawson E., Dean C., Weisbeek P. 1994. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120, 2475–2487 [Google Scholar]
- 3.Jurgens G., Mayer U., Busch M., Lukowitz W., Laux T. 1995. Pattern formation in the Arabidopsis embryo: a genetic perspective. Phil. Trans. R. Soc. Lond. B 350, 19–25 10.1098/rstb.1995.0132 (doi:10.1098/rstb.1995.0132) [DOI] [PubMed] [Google Scholar]
- 4.Leyser O., Day S. 2003. Mechanisms in plant development. Oxford, UK: Blackwell Science [Google Scholar]
- 5.Lee M. Y., et al. 2005. Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol. 139, 1881–1889 10.1104/pp.105.067884 (doi:10.1104/pp.105.067884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashimoto T., Hayashi A., Amano Y., Kohno J., Iwanari H., Usuda S., Yamada Y. 1991. Hyoscyamine 6 beta-hydroxylase, an enzyme involved in tropane alkaloid biosynthesis, is localized at the pericycle of the root. J. Biol. Chem. 266, 4648–4653 [PubMed] [Google Scholar]
- 7.Suzuki K.-I., Yun D.-J., Chen X.-Y., Yamada Y., Hashimoto T. 1999. An Atropa belladonna hyoscyamine 6β-hydroxylase gene is differentially expressed in the root pericycle and anthers. Plant Mol. Biol. 40, 141–152 10.1023/a:1026465518112 (doi:10.1023/a:1026465518112) [DOI] [PubMed] [Google Scholar]
- 8.Kataoka T., Hayashi N., Yamaya T., Takahashi H. 2004. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 136, 4198–4204 10.1104/pp.104.045625 (doi:10.1104/pp.104.045625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takei K., Ueda N., Aoki K., Kuromori T., Hirayama T., Shinozaki K., Yamaya T., Sakakibara H. 2004. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45, 1053–1062 10.1093/pcp/pch119 (doi:10.1093/pcp/pch119) [DOI] [PubMed] [Google Scholar]
- 10.Bishopp A., Help H., El-Showk S., Weijers D., Scheres B., Friml J., Benkova E., Mahonen A. P., Helariutta Y. 2011. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21, 917–926 10.1016/j.cub.2011.04.017 (doi:10.1016/j.cub.2011.04.017) [DOI] [PubMed] [Google Scholar]
- 11.Weid M., Ziegler J., Kutchan T. M. 2004. The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, Papaver somniferum. Proc. Natl Acad. Sci. USA 101, 13 957–13 962 10.1073/pnas.0405704101 (doi:10.1073/pnas.0405704101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verelst W., Kapila J., De Almeida Engler J., Stone J. M., Caubergs R., Asard H. 2004. Tissue-specific expression and developmental regulation of cytochrome b561 genes in Arabidopsis thaliana and Raphanus sativus. Physiol. Plant 120, 312–318 10.1111/j.0031-9317.2004.0235.x (doi:10.1111/j.0031-9317.2004.0235.x) [DOI] [PubMed] [Google Scholar]
- 13.Dannel F., Pfeffer H., Romheld V. 1998. Compartmentation of boron in roots and leaves of sunflower as affected by boron supply. J. Plant Physiol. 153, 615–622 10.1016/S0176-1617(98)80212-5 (doi:10.1016/S0176-1617(98)80212-5) [DOI] [Google Scholar]
- 14.Takano J., Noguchi K., Yasumori M., Kobayashi M., Gajdos Z., Miwa K., Hayashi H., Yoneyama T., Fujiwara T. 2002. Arabidopsis boron transporter for xylem loading. Nature 420, 337–340 10.1038/nature01139 (doi:10.1038/nature01139) [DOI] [PubMed] [Google Scholar]
- 15.De Smet I., Vanneste S., Inze D., Beeckman T. 2006. Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60, 871–887 10.1007/s11103-005-4547-2 (doi:10.1007/s11103-005-4547-2) [DOI] [PubMed] [Google Scholar]
- 16.Peret B., De Rybel B., Casimiro I., Benkova E., Swarup R., Laplaze L., Beeckman T., Bennett M. J. 2009. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408 10.1016/j.tplants.2009.05.002 (doi:10.1016/j.tplants.2009.05.002) [DOI] [PubMed] [Google Scholar]
- 17.Dubrovsky J. G., Doerner P. W., Colon-Carmona A., Rost T. L. 2000. Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 124, 1648–1657 10.1104/pp.124.4.1648 (doi:10.1104/pp.124.4.1648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beeckman T., Burssens S., Inze D. 2001. The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 52, 403–411 10.1093/jexbot/52.suppl_1.403 (doi:10.1093/jexbot/52.suppl_1.403) [DOI] [PubMed] [Google Scholar]
- 19.Casero P. J., García-Sánchez C., Lloret P. G., Navascués J. 1989. Changes in cell length and mitotic index in vascular pattern-related pericycle cell types along the apical meristem and elongation zone of the onion root. Protoplasma 153, 85–90 10.1007/BF01322468 (doi:10.1007/BF01322468) [DOI] [Google Scholar]
- 20.Dolan L., Roberts K. 1995. Secondary thickening in roots of Arabidopsis thaliana: anatomy and cell surface changes. New Phytol. 131, 121–128 10.1111/j.1469-8137.1995.tb03061.x (doi:10.1111/j.1469-8137.1995.tb03061.x) [DOI] [PubMed] [Google Scholar]
- 21.Casero P., Casimiro I., Knox J. 1998. Occurrence of cell surface arabinogalactan-protein and extensin epitopes in relation to pericycle and vascular tissue development in the root apex of four species. Planta 204, 252–259 10.1007/s004250050254 (doi:10.1007/s004250050254) [DOI] [Google Scholar]
- 22.Majewska-Sawka A., Nothnagel E. A. 2000. The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122, 3–10 10.1104/pp.122.1.3 (doi:10.1104/pp.122.1.3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright K. M., Oparka K. J. 1997. Metabolic inhibitors induce symplastic movement of solutes from the transport phloem of Arabidopsis roots. J. Exp. Bot. 48, 1807–1814 10.1093/jxb/48.10.1807 (doi:10.1093/jxb/48.10.1807) [DOI] [Google Scholar]
- 24.Complainville A., et al. 2003. Nodule initiation involves the creation of a new symplasmic field in specific root cells of medicago species. Plant Cell 15, 2778–2791 10.1105/tpc.017020 (doi:10.1105/tpc.017020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardham A. R., Gunning B. E. 1979. Interpolation of microtubules into cortical arrays during cell elongation and differentiation in roots of Azolla pinnata. J. Cell Sci. 37, 411–442 [DOI] [PubMed] [Google Scholar]
- 26.Toriyama H. 2005. On the remarkable phenomena in the root tip of leguminous plants. Root Res. 14, 35–40 10.3117/rootres.14.35 (doi:10.3117/rootres.14.35) [DOI] [Google Scholar]
- 27.Parizot B., et al. 2008. Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol. 146, 140–148 10.1104/pp.107.107870 (doi:10.1104/pp.107.107870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laplaze L., et al. 2005. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56, 2433–2442 10.1093/jxb/eri236 (doi:10.1093/jxb/eri236) [DOI] [PubMed] [Google Scholar]
- 29.Mahonen A. P., et al. 2006. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311, 94–98 10.1126/science.1118875 (doi:10.1126/science.1118875) [DOI] [PubMed] [Google Scholar]
- 30.Lee J. Y., Colinas J., Wang J. Y., Mace D., Ohler U., Benfey P. N. 2006. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl Acad. Sci. USA 103, 6055–6060 10.1073/pnas.0510607103 (doi:10.1073/pnas.0510607103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui H., Hao Y., Kovtun M., Stolc V., Deng X.-W., Sakakibara H., Kojima M. 2011. Genomewide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol. 157, 1221–1231 10.1104/pp.111.183178 (doi:10.1104/pp.111.183178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brady S. M., Orlando D. A., Lee J. Y., Wang J. Y., Koch J., Dinneny J. R., Mace D., Ohler U., Benfey P. N. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 10.1126/science.1146265 (doi:10.1126/science.1146265) [DOI] [PubMed] [Google Scholar]
- 33.Parizot B., De Rybel B., Beeckman T. 2010. VisuaLRTC: a new view on lateral root initiation by combining specific transcriptome data sets. Plant Physiol. 153, 34–40 10.1104/pp.109.148676 (doi:10.1104/pp.109.148676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Smet I., et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597 10.1126/science.1160158 (doi:10.1126/science.1160158) [DOI] [PubMed] [Google Scholar]
- 35.Levesque M. P., et al. 2006. Whole-genome analysis of the short-root developmental pathway in Arabidopsis. PLoS Biol. 4, e143. 10.1371/journal.pbio.0040143 (doi:10.1371/journal.pbio.0040143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukaki H., Tameda S., Masuda H., Tasaka M. 2002. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168 10.1046/j.0960-7412.2001.01201.x (doi:10.1046/j.0960-7412.2001.01201.x) [DOI] [PubMed] [Google Scholar]
- 37.DiDonato R. J., Arbuckle E., Buker S., Sheets J., Tobar J., Totong R., Grisafi P., Fink G. R., Celenza J. L. 2004. Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J. 37, 340–353 10.1046/j.1365-313X.2003.01964.x (doi:10.1046/j.1365-313X.2003.01964.x) [DOI] [PubMed] [Google Scholar]
- 38.Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130 10.1105/tpc.106.047761 (doi:10.1105/tpc.106.047761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Rybel B., et al. 2010. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706 10.1016/j.cub.2010.09.007 (doi:10.1016/j.cub.2010.09.007) [DOI] [PubMed] [Google Scholar]
- 40.Jeziorska D. M., Jordan K. W., Vance K. W. 2009. A systems biology approach to understanding cis-regulatory module function. Semin. Cell Dev. Biol. 20, 856–862 10.1016/j.semcdb.2009.07.007 (doi:10.1016/j.semcdb.2009.07.007) [DOI] [PubMed] [Google Scholar]
- 41.Maeda R. K., Karch F. 2011. Gene expression in time and space: additive versus hierarchical organization of cis-regulatory regions. Curr. Opin. Genet. Dev. 21, 187–193 10.1016/j.gde.2011.01.021 (doi:10.1016/j.gde.2011.01.021) [DOI] [PubMed] [Google Scholar]
- 42.Perez-Rodriguez P., Riano-Pachon D. M., Correa L. G., Rensing S. A., Kersten B., Mueller-Roeber B. 2010. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38, D822–827 10.1093/nar/gkp805 (doi:10.1093/nar/gkp805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yilmaz A., Mejia-Guerra M. K., Kurz K., Liang X., Welch L., Grotewold E. 2011. AGRIS: the Arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 39, D1118–1122 10.1093/nar/gkq1120 (doi:10.1093/nar/gkq1120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Jin J., Tang L., Zhao Y., Gu X., Gao G., Luo J. 2011. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 39, D1114–D1117 10.1093/nar/gkq1141 (doi:10.1093/nar/gkq1141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Priest H. D., Filichkin S. A., Mockler T. C. 2009. Cis-regulatory elements in plant cell signaling. Curr. Opin. Plant Biol. 12, 643–649 10.1016/j.pbi.2009.07.016 (doi:10.1016/j.pbi.2009.07.016) [DOI] [PubMed] [Google Scholar]
- 46.Chen A., Gu M., Sun S., Zhu L., Hong S., Xu G. 2011. Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol. 189, 1157–1169 10.1111/j.1469-8137.2010.03556.x (doi:10.1111/j.1469-8137.2010.03556.x) [DOI] [PubMed] [Google Scholar]
- 47.Birnbaum K., Shasha D. E., Wang J. Y., Jung J. W., Lambert G. M., Galbraith D. W., Benfey P. N. 2003. A gene expression map of the Arabidopsis root. Science 302, 1956–1960 10.1126/science.1090022 (doi:10.1126/science.1090022) [DOI] [PubMed] [Google Scholar]
- 48.Birnbaum K., Jung J. W., Wang J. Y., Lambert G. M., Hirst J. A., Galbraith D. W., Benfey P. N. 2005. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619 10.1038/nmeth0805-615 (doi:10.1038/nmeth0805-615) [DOI] [PubMed] [Google Scholar]
- 49.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 10.1093/biostatistics/4.2.249 (doi:10.1093/biostatistics/4.2.249) [DOI] [PubMed] [Google Scholar]
- 50.Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1–25 10.2202/1544-6115.1027 (doi:10.2202/1544-6115.1027) [DOI] [PubMed] [Google Scholar]
- 51.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. 10.1093/nar/gng015 (doi:10.1093/nar/gng015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S. Y., Volsky D. J. 2005. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics 6, 144. 10.1186/1471-2105-6-144 (doi:10.1186/1471-2105-6-144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du Z., Zhou X., Ling Y., Zhang Z., Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70 10.1093/nar/gkq310 (doi:10.1093/nar/gkq310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saeed A. I., et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 55.Sinha S., Tompa M. 2002. Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 30, 5549–5560 10.1093/nar/gkf669 (doi:10.1093/nar/gkf669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanchette M., Sinha S. 2001. Separating real motifs from their artifacts. Bioinformatics 17(Suppl. 1), S30–38 10.1093/bioinformatics/17.suppl_1.S30 (doi:10.1093/bioinformatics/17.suppl_1.S30) [DOI] [PubMed] [Google Scholar]
- 57.Pavesi G., Mereghetti P., Mauri G., Pesole G. 2004. Weeder web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 32, W199–203 10.1093/nar/gkh465 (doi:10.1093/nar/gkh465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freeling M., Subramaniam S. 2009. Conserved noncoding sequences (CNSs) in higher plants. Curr. Opin. Plant Biol. 12, 126–132 10.1016/j.pbi.2009.01.005 (doi:10.1016/j.pbi.2009.01.005) [DOI] [PubMed] [Google Scholar]
- 59.Freeling M., Rapaka L., Lyons E., Pedersen B., Thomas B. C. 2007. G-boxes, bigfoot genes, and environmental response: characterization of intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19, 1441–1457 10.1105/tpc.107.050419 (doi:10.1105/tpc.107.050419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berger N., Dubreucq B., Roudier F., Dubos C., Lepiniec L. 2011. Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell Online 23, 4065–4078 10.1105/tpc.111.087866 (doi:10.1105/tpc.111.087866). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koussevitzky S., Nott A., Mockler T. C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719 10.1126/science.1140516 (doi:10.1126/science.1140516) [DOI] [PubMed] [Google Scholar]
- 62.Michael T. P., et al. 2008. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4, e14. 10.1371/journal.pgen.0040014 (doi:10.1371/journal.pgen.0040014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamura J., Goldberg R. 1989. Regulation of plant gene expression: general principles. In The biochemistry of plants (eds Stumpf P. K., Conn E. E.), pp. 1–82 New York, NY: Academic Press [Google Scholar]
- 64.Zhang J., Elo A., Helariutta Y. 2011. Arabidopsis as a model for wood formation. Curr. Opin. Biotechnol. 22, 293–299 10.1016/j.copbio.2010.11.008 (doi:10.1016/j.copbio.2010.11.008) [DOI] [PubMed] [Google Scholar]
- 65.Brady S. M., et al. 2011. A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 7, 459. 10.1038/msb.2010.114 (doi:10.1038/msb.2010.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vanneste S., et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17, 3035–3050 10.1105/tpc.105.035493 (doi:10.1105/tpc.105.035493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casimiro I., et al. 2001. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852 10.1105/tpc.13.4.843 (doi:10.1105/tpc.13.4.843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laplaze L., et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19, 3889–3900 10.1105/tpc.107.055863 (doi:10.1105/tpc.107.055863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Smet I., et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl Acad. Sci. USA 107, 2705–2710 10.1073/pnas.0915001107 (doi:10.1073/pnas.0915001107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pruneda-Paz J. L., Breton G., Para A., Kay S. A. 2009. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485 10.1126/science.1167206 (doi:10.1126/science.1167206) [DOI] [PMC free article] [PubMed] [Google Scholar]