Abstract
The development of root systems may be strongly affected by the symbiotic interactions that plants establish with soil organisms. Legumes are able to develop symbiotic relationships with both rhizobial bacteria and arbuscular mycorrhizal fungi leading to the formation of nitrogen-fixing nodules and mycorrhizal arbuscules, respectively. Both of these symbiotic interactions involve complex cellular reprogramming and profound morphological and physiological changes in specific root cells. In addition, the repression of pathogenic defence responses seems to be required for successful symbiotic interactions. Apart from typical regulatory genes, such as transcription factors, microRNAs (miRNAs) are emerging as riboregulators that control gene networks in eukaryotic cells through interactions with specific target mRNAs. In recent years, the availability of deep-sequencing technologies and the development of in silico approaches have allowed for the identification of large sets of miRNAs and their targets in legumes. A number of conserved and legume-specific miRNAs were found to be associated with symbiotic interactions as shown by their expression patterns or actions on symbiosis-related targets. In this review, we combine data from recent literature and genomic and deep-sequencing data on miRNAs controlling nodule development or restricting defence reactions to address the diversity and specificity of miRNA-dependent regulation in legume root symbiosis. Phylogenetic analysis of miRNA isoforms and their potential targets suggests a role for miRNAs in the repression of plant defence during symbiosis and revealed the evolution of miRNA-dependent regulation in legumes to allow for the modification of root cell specification, such as the formation of mycorrhized roots and nitrogen-fixing nodules.
Keywords: microRNA, legumes, symbiosis, evolution, plant defence, transcription factor
1. Introduction
The symbiotic interactions that plants establish with soil organisms may alter the development of root systems. Legumes are able to develop symbiotic interactions with both rhizobial bacteria and arbuscular mycorrhizal (AM) fungi leading to the formation of nitrogen-fixing nodules and mycorrhizal arbuscules, respectively. Both symbiotic interactions involve complex cellular reprogramming, that results in profound morphological and physiological changes to specific root cells. In particular, nitrogen-fixing symbiosis leads to the formation of a new organ, the root nodule, in which atmospheric nitrogen is fixed by the bacteria. This symbiosis allows the legumes to assimilate atmospheric nitrogen and thus to grow in nitrogen-depleted soils, which is an important agricultural trait. Indeed, legumes (Fabaceae) are the second most important crop family after cereals and are an essential nutrient resource for humans and animals.
Nodule development depends on the establishment of a complex molecular dialogue between the bacteria and the plant [1]. Flavonoids that are exuded by the roots induce the biosynthesis of specific lipochitooligosaccharides by the rhizobia, the Nod factors, which are recognized by a family of plant LysM receptor kinases (NRF1 and NFR5) [2,3]. The genetic host–symbiont recognition relies on specific interactions between the Nod factors and the corresponding receptors [4]. Once perceived by the root, Nod factors trigger the nodule developmental process by eliciting cell dedifferentiation and division in the cortex and pericycle to produce the nodule primordium. Thus, the development of the nodule integrates plant regulatory pathways that are related to lateral root organogenesis, for which the reactivation of differentiated root cells and subsequent divisions occur in the pericycle to form a new lateral organ [5,6]. Molecular mechanisms governing nodule organogenesis implicate phytohormones and complex gene regulation through the activation of several transcription factors ([5,6], and references therein). For example, the nodulation-specific pathway mutants (nsp1 and nsp2) in the legume model Medicago truncatula show inhibitions in both the infection thread growth and the expression of nodulation markers and do not initiate the development of nodule primordia. MtNSP1 and MtNSP2 belong to the GRAS family of transcription factors that have diverse functions in plant growth and development, such as gibberellin signal transduction, axillary meristem formation, cytochrome a signal transduction and gametogenesis.
Concomitant with the activation of the nodule primordium in front of the infection site, the bacteria penetrate root hairs through an infection thread. They are then released by endocytosis into the cytoplasm of the nodule primordium to form bacteroids, in which atmospheric N2 fixation finally takes place [7]. Maillet et al. [8] showed that mutants of the Nod factor signalling pathway are also affected in response to the Myc factor [9], leading to the formation of arbuscular mycorrhiza. Rhizobia should thus have co-opted ancient widespread mycorrhizal signalling pathways to enable the establishment of endosymbiotic interactions with legumes. Furthermore, Myc and Nod factors stimulate lateral root branching [4], suggesting that in the course of evolution, signals for modifying root system architecture were used to facilitate symbiotic infections or nodule organogenesis [6,10].
Although the relationships that are established in plant–symbiotic and plant–pathogen interactions differ, it is possible that defence reactions take place in legume–rhizobia symbiosis [11,12]. After the initial nodule formation, the plant undergoes a hypersensitive-like (HR-like) response and inhibits subsequent rhizobial infections to limit the number of nodules [13]. During both the response to pathogen infection and rhizobia–legume interaction, NADPH-oxidases and plasma membrane-bound ion channels are activated in the plant, triggering an initial oxidative burst [14]. The accumulation of reactive oxygen species leads to cell death and the necrosis of the area of infection, which prevents bacterial progression in the plant tissue. These observations suggest that Nod factors may also be important in the evasion of the plant immune response and the protection of cells from reactive oxygen species [10,15]. However, other components, such as receptor-like kinase with leucine-rich repeat (LRR) domains, have been identified to be essential for nodulation signalling in addition to the perception of pathogen signals [16,17]. The modulation of defence response pathways must occur during symbiosis to permit the invasion of the plant host cell by the bacteria without triggering cell death.
In recent years, small riboregulators, such as the microRNAs (miRNAs) miR166 and miR169, have been added to the complex signalling pathways that control nodulation, mainly as repressors of key transcription factors [18,19]. In plants, several pathways are known to generate endogenous small non-coding RNAs [20,21], which share common biochemical reactions. A long double-stranded RNA (dsRNA) is formed, which is processed into small RNAs (sRNAs) that are 18–25 nucleotides in length. Then, after 3′-O-methylation, these sRNAs can be incorporated into an effector complex that mediates gene silencing by base pairing with a target RNA or DNA [20]. In addition to these common steps, each pathway presents with its own specificities. The miRNA pathway mainly produces 21 nucleotide (nt) sRNA from endogenous transcripts with internal stem-loop structures. These transcripts, called precursors or pri-miRNAs, are processed by a DICER-like 1 (DCL1) protein into a sRNA duplex containing the mature miRNA and its opposite strand, known as the miRNA*. Next, the miRNA is loaded into an AGO1-containing RISC complex that mediates mRNA target regulation in the cytoplasm [22], mainly through cleavage but also translation inhibition [23]. Trans-acting small interfering RNAs (ta-siRNAs) are produced from TAS non-protein coding genes that are targeted by specific miRNAs [24]. The TAS cleavage products are recognized by the RNA-dependent RNA polymerase 6 (RDR6), which converts them into long dsRNA. Finally, the sequential cleavage of these dsRNAs by DCL4 produces a set of phased 21 nt sRNAs that can be incorporated into RISCs to direct the cleavage of several mRNA targets [20]. Natural antisense short interfering RNAs (nat-siRNAs) originate from dsRNAs that are formed by the transcription of two partially overlapping genes in the sense and antisense orientations and repress the expression of one of them [25]. Plants also produce a large set of heterochromatic-associated DCL3-dependent siRNAs (hc-siRNAs) that are typically 24 nt in length, are associated with repetitive genomic sequences (transposons, retroelements, centromeric repeats, etc.) and direct DNA and histone methylation at the corresponding loci via an AGO4-dependent pathway. In addition, 21 and 24 nt siRNAs that are derived from transposons have also been recently implicated in transposable element silencing during gametogenesis through AGO9-dependent pathways [26]. Analyses of AGO and DCL protein families in legumes confirmed their highly conserved natures and the functional redundancy that probably exists within each family [27]. The interplay of DCLs, RDRs and AGOs determines the specific modes of action of the sRNAs on their targets, and it is clear that the most important class of riboregulatory sRNA are the miRNAs.
Animal and plant miRNAs were initially discovered because most miRNA-deficient mutants exhibit severe developmental defects [22]. However, compared with animal miRNAs, which usually have hundreds of targets, plant miRNAs tend to have fewer targets, often with regulatory functions such as transcription factors or F-box proteins [28]. This observation led to the introduction of miRNA-target nodes, miRNA nodes, which are interfaces that comprise the regulatory relationships between a specific locus from a miRNA family and its direct target [29]. It is proposed that miRNA nodes play crucial roles in governing plastic behaviour during plant development and are organized into complex networks within signalling pathways that control development [28,29]. For instance, the regulation of lateral root initiation in response to soil nitrogen availability involves at least two miRNA nodes that are associated with auxin signalling, miR167/auxin response factor 8 (ARF8) and miR393/auxin-binding factor 3 (ABF3). miRNA nodes have been embedded within signalling pathways over the course of evolution, and their modifications may underlie morphological and physiological diversities.
As mentioned above, miRNAs are processed from hairpin-containing transcripts that are transcribed from MIR genes. In contrast to animals, plants have fewer but larger families of MIR genes that have expanded recently. Indeed, for a given plant miRNA family, sequence similarities may be found not only in the conserved miRNA/miRNA* region but also throughout the whole MIR precursor. Moreover, sequence analyses of the ath-miR161 and ath-miR163 genes revealed that the foldback arms shared extensive similarities (even outside of the miRNA/miRNA* regions) with their target genes [30]. These observations suggest that miRNA genes may arise from inverted duplications of the corresponding target genes [30]. The initial duplication event would generate a siRNA through perfect or near-perfect self-complementarity, and in fact it has been shown that several young miRNAs in Arabidopsis thaliana, such as ath-miR822, ath-miR839 and ath-miR869, produce miRNA-like siRNAs that are processed by DCL4 rather than DCL1. The identification of miRNAs by high-throughput sequencing in several plant species has led to the conclusion that MIR genes are rapidly evolving. Indeed, each plant species has specific MIR genes that are not present in other, even closely related species. For example, a comparison of miRNA families from A. thaliana and Arabidopsis lyrata genomes revealed that, although the majority of miRNA families are conserved, 24–33% of the families were gained or lost, respectively, by one of the two species since they diverged 10 million years ago [31]. Using genomic and deep-sequencing data from a more divergent Brassica, Capsella rubella, the net flux rate (birth–death) of miRNAs in the Arabidopsis lineage was found to range from 1.2 to 3.3 genes per million years. Some plant miRNA families are conserved even in the moss Physcomitrella patens, indicating their very ancient origins, and play roles in fundamental processes, such as meristem function, organ polarity and cell division (e.g. miR156, miR160, miR390 and miR319 [31]). These deeply conserved miRNAs have probably co-evolved with their corresponding targets because the nucleotide divergence between orthologous miRNA genes was highest in the loop regions and lowest in the miRNA/miRNA* regions. Other miRNAs, called novel miRNAs, are not phylogenetically conserved and are considered to be evolutionarily recent. They are typically encoded by single-copy genes and expressed at moderate levels. The number of novel miRNAs largely exceeds that of conserved miRNAs [32,33], and their predicted targets include a large set of proteins. However, the vast majority of young miRNAs have few, if any, functions but could have been selected to become active under particular conditions (e.g. in response to environmental signals).
The ability of legumes to form nitrogen-fixing nodules is a process that probably evolved quite recently from ancient symbiotic interactions between roots and AM fungi. The morphogenetic processes that are associated with nodule formation suggest that it uses regulatory mechanisms that are involved in lateral root organogenesis. This example of evolutionary diversion of an ancient process calls into question the underlying molecular mechanisms that may be involved. Due to their rapid evolution, miRNA-target nodes may play roles in the acquisition of new regulatory networks for the generation of novel functions or structures. In this study, we reviewed recent literature describing certain legume miRNAs that have been functionally associated with nodulation and reanalysed genomic and deep-sequencing data to determine the diversity of miRNA-dependent processes in legume root symbiosis in an evolutionary context.
2. Conserved microRNAs regulate transcription factors that are involved in nodule development
The specificity of miRNA-dependent gene regulation in a biological process may rely on the function of a specific miRNA target and/or the overlapping expression patterns between a specific miRNA gene and its targets. However, conserved miRNA-dependent gene regulation may evolve over time owing to the emergence of a new form of mature miRNA (isoform) to regulate specific targets. In M. truncatula, the enrichment of certain miRNA isoforms was reported in nodules [34], and a new isoform of miR156, which accumulates preferentially in the root apex, was observed to cleave a non-conserved target (a WD40-like protein) [35]. Based on these considerations, we have analysed the diversity of miRNA-dependent gene regulation during nodulation in legumes, although only a small number of miRNAs have been shown to be functionally involved in this process. Auxin plays a critical role in nodule initiation, and several miRNAs that have been linked to auxin signalling (miR160, miR164, miR167 and miR393, table 1) were shown to be regulated early after inoculation with rhizobia [38,44]. Because their putative involvement in nodulation has been recently reviewed elsewhere [16,17], we decided to focus on the other miRNAs that have been functionally associated with nodulation by regulating transcription factors that are involved in root or nodule development, i.e. miR156, miR169 and miR171 (table 1).
Table 1.
Overview of miRNAs associated with nodulation or symbiosis-related processes.
| miRNA | function | expression in roots | target | species (isoform) | ref. |
|---|---|---|---|---|---|
| miRNAs functionally involved in nodulation | |||||
| miR166 | nodule and lateral root formation | apical region of nodules and roots/vascular bundlea | class III HD-zip transcription factorsf | Medicago truncatula (mtr-miR 166a) | [17] |
| miR169 | nodule development | infection zone of nodulesb | CCAAT-binding transcription factor (MtHAP2.1)f | Medicago truncatula (mtr-miR 169a) | [18] |
| miR171 | Nod-factors signalling | induced during nodulationc/highly expressed in mature nodulee | GRAS family transcription factors (MtNSP2, LjNSP2)g | Medicago truncatula (mtr-miR171h), Lotus japonicaus (ltj-miR171h) | [36,37] |
| miR164 | nodule and lateral root formation | meristem and infection zone of mature nodulea, downregulated during early nodulationd | transcriptional activator (NAC1)f | Medicago truncatula (mtr-miR164c) | [38,39] |
| miRNAs linked to auxin signalling and potentially involved in nodulation | |||||
| miR160 | root cap formation, gravity sensing, adventitious rooting | downregulated during early nodulationd | auxin response factor (ARF10, ARF17, ARF16)f | Arabidopsis thaliana (ath-miR160 a/b/c) | [38,40] |
| miR167 | adventitious rooting | downregulated during early nodulationd | auxin response factor (ARF6, ARF8)e | Arabidopsis thaliana (ath-miR167 a/b/c/d) | [38,41] |
| miR393 | response to nitrate availability primary and lateral root growth, antibacterial resistance | upregulated during early nodulationd | auxin receptors (TIR1, AFB2, AFB3)f, bHLH transcription factor (bHLH77)f | Arabidopsis thaliana (ath-miR393) | [38,42,43] |
Experimental evidence for expression regulation or target validation are from:
aIn situ hybridization.
bTranscriptional reporter.
cmiRNA library sequencing.
dNorthern blot.
eqPCR.
fRACE–PCR.
gDegradome analysis.
In A. thaliana, miR166 controls xylem and pericycle cell differentiation. Produced in the endodermis, this miRNA acts non-cell autonomously and diffuses towards the stele, which produces a gradient of its target mRNAs, which encode class III HD-ZIP transcription factors [45]. In M. truncatula, Boualem et al. [18] showed that mtr-miR166 and its class III HD-ZIP targets are co-expressed in vascular bundles and in the apical regions of roots and nodules (table 1). The over-expression of a miR166 polycistronic precursor led to reductions in nodule and lateral root numbers in addition to the ectopic development of vascular bundles in transgenic roots. The miR166 family is large and deeply conserved in plants and even exists within some lycophytes and bryophytes [46]. For example, there are seven precursors in A. thaliana that code for a single mature isoform, and two of them form a cluster on chromosome 5. Interestingly, the clustered organization of miR166 genes is conserved even in bryophytes [46]. Analyses of publicly available data from the miRNA database miRBase (http://www.mirbase.org), in addition to sRNA sequencing data (http://medicago.toulouse.inra.fr [34]), showed that the M. truncatula genome contains nine miR166 precursors that code for five distinct, mature isoforms (diverging by less than 3 nt). Precursors for miR166 isoforms are distributed on six chromosomes and present with two distinct clusters, mtr-miR166c/mtr-miR166d on chromosome 3 and another cluster with two genes on chromosome 4 that has not been reported in miRBase (MtrV3Chr4_r1940, MtrV3Chr4_r1941, http://medicago.toulouse.inra.fr). In soybean (Glycine max), using comparative genomic and folding predictions, Zhang et al. [47] identified seven miR166 precursors that code for four distinct, mature isoforms. Again, two clusters were reported, gma-miR166a/gma-miR166b on chromosome 5 and gma-miR166f/gma-miR166g [47]. However, this genomic organization is not a legume-specific feature because the clustering of the miR166 genes is also found in distant species, such as poplar (Populus trichocarpa). Similar to other species that have been studied to date, the target search in soybean and M. truncatula using the psRNA target tool (http://plantgrn.noble.org/psRNATarget) predicted class III HD-ZIP transcription factors to be preferential miR166 targets in legumes. Moreover, degradome data from M. truncatula roots confirmed that the only targets of mtr-miR166 seem to be HD-ZIP III transcription factor mRNAs [48]. Hence, miR166 activity during nodule development most probably relies on the conserved functions of its targets in root cell differentiation. A number of recent studies have suggested that miRNAs* may also mediate transcript cleavage. Devers et al. [48] identified several transcripts that are cleaved by miR166* in M. truncatula roots and are distinct from those targeted by the mature miRNA. In some cases, the miRNA* may even preferentially accumulate compared with the miRNA, suggesting that the choice of the dominant strand of the miRNA may be an effective means to diversify miRNA-mediated gene regulation.
In M. truncatula, miR169 regulates the expression of a transcription factor from the CCAAT-binding family (or NFYA), called MtHAP2-1 [19] (table 1). The over-expression of the mtr-miR169a precursor significantly affects nodule development [19]. The nodulation process is delayed, and nodule growth is arrested for 8–10 days following inoculation with a concomitant defect in nitrogen-fixing ability. Furthermore, the downregulation of MtHAP2-1 by RNAi led to a similar arrest in nodule development. The complementary expression patterns of MtHAP2-1 and mtr-miR169 strongly suggest that the miR169-mediated restriction of HAP2-1 to the meristematic zone is essential for the differentiation of root nodule cells into bacteroid-containing nitrogen-fixing cells. According to miRBase, the miR169 family is a large family that is conserved in various plants, including monocots, eudicots and some gymnosperms and ferns [46]. The miR169-dependent regulation of CCAAT-binding transcription factors is also highly conserved and has been shown to be involved in several biological processes, such as floral organ development [49] and responses to environmental stresses [50,51]. In A. thaliana, there are 14 miR169 genes that are distributed over four chromosomes and develop into four distinct, mature isoforms [52]. In miRBase, 17 miR169 genes that develop into 10 distinct, mature isoforms are listed for M. truncatula, while only six miR169 genes corresponding to five distinct isoforms are available for soybean. Phylogenetic analyses of the miR169 precursors in legume and non-legume species (i.e. A. thaliana, rice, soybean and M. truncatula) did not reveal any legume-specific miR169 precursor or mature isoform (not shown). However, the function of miR169 in the regulation of nodulation may be associated with the specific spatio-temporal expression of one or several members of the family. Indeed, in A. thaliana, miR169 genes show distinct or overlapping expression patterns in all stages of plant development [52]. Hence, the miR169 family diversification in legumes may have led to the generation of miR169 genes that become responsive to nodulation, such as those reported in Combier et al. [19]. Interestingly, a deep-sequencing analysis of miRNAs during soybean nodulation revealed a downregulation of miR169 accumulation from 3 h after inoculation of the symbiotic bacteria [38]. Moreover, according to the available microarray data from soybean (eFP browser, http://bar.utoronto.ca), one target (Glyma19g38800) of gma-miR169, which is nearly identical to MtHAP2-1, is strongly upregulated in root hairs during early nodulation (not shown), suggesting that the miR169-dependent regulation of this transcription factor may also play a role in the early stages of the establishment of symbiosis. Altogether, these data suggest that the recruitment of the conserved miR166 and miR169 miRNAs in nodule development should more certainly involve the coordination of miRNA/target expression compared with the appearance of new specific miRNA/target nodes.
In contrast, novel miRNA isoforms that target specific mRNAs have been identified in other conserved miRNA families, such as miR156 [35] and miR171 [48]. In A. thaliana and several other plants, miR171 is known to target members of the scarecrow-like (GRAS domain) family of transcription factors that are predominantly expressed in inflorescence and floral tissues [46]. A recent study on miRNA-mediated regulation during AM symbiosis in M. truncatula showed that MtNSP2, which is a transcription factor that is necessary for nodulation signalling [53], is a specific target of an atypical isoform of miR171 (mtr-miR171h; figure 1b; table 1) in roots [36,48]. The authors also demonstrated that mtr-miR171h accumulation was higher in roots at 3 weeks after inoculation with the symbiotic bacteria Sinorhizobium meliloti. Interestingly, the same phenomenon was previously reported in another legume model, Lotus japonicus, where a non-canonical isoform of miR171 (ltj-miR171c) was predicted to target LjNSP2, the homologue of MtNSP2, which is highly expressed in mature nodules [37]. According to miRBase, this isoform is also present in soybean (gma-miR171b; figure 1a), which supports its conserved role in nodulation. However, further analyses of the miR171 family using both miRBase and comparative genomics (figure 1a) revealed that this specific isoform is present in several non-legume species, such as P. trichocarpa (ptc-miR171) and Citrus sinensis (csi-miR171b), while it is absent in other species, such as A. thaliana, Brassica napus and Pinus taeda, that are not able to form root endomycorrhiza. Interestingly, target prediction in P. trichocarpa revealed that the poplar homologue of MtNSP2 is a preferential target of this isoform, suggesting the conservation of the miR171/NSP2 node. However, in rice, only a close homologue of this isoform was deposited in miRBase (osa-miR171h), and target prediction did not suggest rice NSP2 as a putative target. Recently, Maillet et al. [8] reported that NSP2 should be considered to be a shared component of the signalling pathways of nodulation and AM symbiosis and that it is not linked solely to nodulation. Hence, we speculate that this diversification event in the miR171 family may have been selected during evolution in plants that are able to interact with soil micro-organisms using a NSP2-dependent signalling pathway. Despite the fact that a similar miRNA node exists in different plants, the evolution of specific miRNA isoforms may incorporate related transcription factors (such as the NSP2 genes) into novel regulatory networks (e.g. nodulation). Similar to mutations in promoter cis-elements, these point mutations may then contribute to the evolution of gene regulation across species.
Figure 1.
An atypical miR171 isoform targets the nodulation and mycorrhizal-associated NSP2 transcription factor gene. (a) Nucleotide alignment of distinct miR171 mature isoforms that are deposited in miRBase. Arabidopsis thaliana (ath), Brassisca napus (bra), Citrus sinensis (csi), Glycine max (gma), Medicago truncatula (mtr), Oryza sativa (osa), Populus tricocarpa (ptc) and Pinus taeda (pta). Highlighted nucleotides represent differences compared with consensus sequences. (b) Alignment of MtNSP2 mRNA with the canonical and novel isoforms of miR171, ath-miR171a, mtr-miR171b and mtr-miR171h. In each alignment, two dots represent a perfect match, and one represents a mismatch. The arrow indicates the cleavage site that was located by Devers et al [1].
3. Micrornas associated with plant defence are also involved in symbiotic interactions
Several miRNAs are regulated during plant–microbe interactions. Their roles in plant basal defence (reviewed in [42]) were first supported by the findings that the Arabidopsis miRNA-deficient mutants dcl1 and hen1 displayed enhanced growth of pathogenic and non-pathogenic bacteria. Unfortunately, such mutants are not yet available in legumes. However, in soybean, sRNA sequencing data revealed a set of novel miRNAs that are regulated as early as 3 h post-inoculation (hpi) with the symbiotic bacteria Bradyrhizobium japonicum [38]. Intriguingly, the authors observed an overlap between the set of early nodulation-responsive miRNAs and those involved in the control of immunity that is triggered by pathogen-associated molecular patterns (PAMPs). For example, miR393, which plays a role in PAMP-triggered immunity (PTI) [43], was shown to be transiently induced during a symbiotic interaction from 3 hpi with maximum levels reached at 6 hpi. The same study also showed that two other miRNAs, miR160 and miR167, which have already been reported to be pathogen-responsive, were downregulated from 3 hpi. These three miRNAs regulate auxin signalling by targeting either auxin receptor or auxin response factor genes but show differential expression patterns during pathogenic bacterial interactions. For instance, miR160 and miR167 are highly induced upon infection with the avirulent strain of Pseudomonas syringae pv. tomato [42], while they are downregulated during symbiotic interactions in soybean. This regulation of pathogen-associated miRNAs during symbiotic interactions may reflect an adaptation of legume roots to ensure symbiosis through the downregulation of PTI-associated genes. Future studies analysing the expression of these miRNAs during the interactions of legume roots with non-nodulating rhizobial strains or during the interactions of rhizobia with non-legume species may be of interest. Nonetheless, the regulation of these types of miRNAs again highlights the importance of auxin signalling/homeostasis in the early steps of the nodulation process.
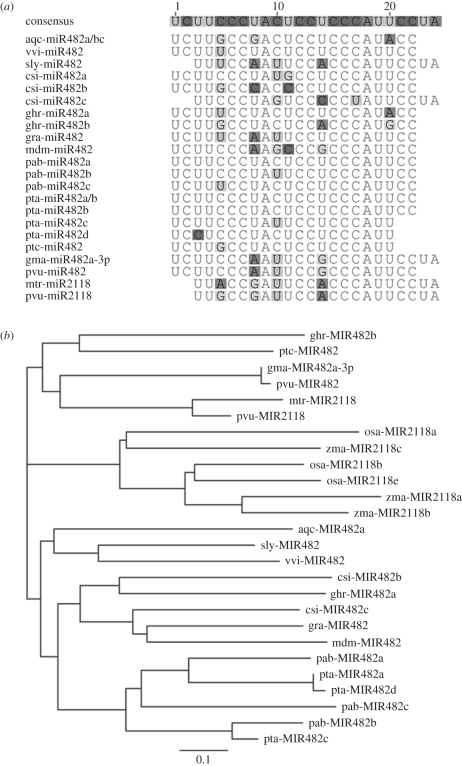
During recent years, the numbers of plant miRNAs that have been predicted to target the disease resistance genes of the NBS–LRR family have increased exponentially (review in [39]). In legumes, these miRNAs were identified by the sRNA deep sequencing of various organs (reviewed in [17]) and during pathogen and symbiotic interactions [38,54]. Lelandais-Brière et al. [34] and Kulcheski et al. [54] identified nine and five novel miRNAs that were predicted to target NBS–LRR disease resistance genes in M. truncatula and G. max, respectively. Li et al. [44] more precisely examined the expression patterns of six miRNAs during soybean nodulation, and among them, miR482 was induced early during nodulation at 6 hpi, reached maximum levels at 6 days after inoculation (dpi) and returned to basal levels at 14 dpi. Transgenic roots over-expressing the gma-miR482 precursor under the control of the plant early nodulin promoter pENOD40 produced twice as many nodules compared with the control roots without concurrent changes in their length or lateral root density, suggesting a nodule-specific role. The cleavage of two predicted targets of miR482 that encode positive regulators of plant immunity, a GSK3-like protein kinase and a putative TIR–NBS–LRR disease resistance protein, was observed. In A. thaliana, the closest homologue of the GSK3-like target is the shaggy-like protein kinase AtHIR, which is known to be strongly induced during hypersensitive responses [55]. In miRBase, miR482s genes have actually been deposited for two legumes, the common bean (Phaseolus vulgaris) and soybean, several non-legume dicots, including poplar and C. sinensis, and the conifer P. teada. However, the legume miR482s (Pvu-miR482 and Gma-miR482-3p) diverge from other dicots both at the precursor and mature sequence levels (figure 2). Although we were not able to find miR482 sequences in the M. truncatula and L. japonicus libraries, our comparative genomics and Blastn algorithm against miRBase revealed that miR2118, which is another miRNA that was first identified in M. truncatula seedlings by Jagadeeswaran et al. [56], is a close homologue to gma-miR482-3p and pvu-miR482 (figure 2b). The cleavage of three NBS–LRR protein-encoding transcripts by this miRNA was confirmed by RACE–PCR and degradome analyses [48,56] in M. truncatula roots and seedlings. Again, in addition to the P. vulgaris and M. truncatula genes, two monocots, rice and maize, show miR2118 genes in miRBase. The similarities between the mature miR482 and miR2118 and their conserved functions suggest a common origin for these two miRNA families, similar to miR159 and miR319 in flowering plants [57]. However, a more thorough analysis of their pri-miRNAs and phylogenetic tree constructions revealed a clear divergence between the miR482 and miR2118 genes in plants (figure 2b). These data suggest the existence of either a very old divergence of the two families from a common ancestor or their independent evolution to account for common functions. Moreover, the miR2118 genes in legumes apparently belong to a clade that is clearly distinct from that of monocots, suggesting a putative specialization during the nodulation process.
Figure 2.
miR482 and miR2118 are closely related miRNAs that repress disease resistance genes during nodulation. (a) Nucleotide alignment of distinct mature isoforms of miR482 and miR2118 that are deposited in miRBase. Highlighted nucleotides represent differences compared with consensus sequences. (b) Phylogenetic relationship between pri-miRNAs from the miR482 and miR2118 families according to the miRBase data. Aquilegia caerulea (aqc), Citrus sinensis (csi), Glycine max (gma), Gossypium hirsutum (ghr), Gossypium raimondi (gra), Malus domestica (mdm), Medicago truncatula (mtr), Oryza sativa (osa), Phaseolus vulgaris (pvu), Picea abies (pab), Pinus taeda (pta), Populus trichocarpa (ptc), Solanum lycopersicon (sly) and Vitis vinifera (vvi). For clarity purposes, pri-miRNAs from the same species that showed little nucleotide divergence were not represented. A phylogenetic tree was constructed with Geneious software using the neighbour joining method, a Juke-Cantor genetic distance model and a bootstrap with 100 replication events.
In their functional analysis, Li et al. [44] reported that miR1507 accumulates upon rhizobial infection in a soybean supernodulating mutant (NTS382) that is affected in the nodule autoregulation pathway. This legume-specific miRNA was also predicted to target transcripts for NBS–LRR proteins, and recently Devers et al. [48] discovered six cleavage products of mtr-miR1507 in their degradome analysis of M. truncatula roots that all corresponded to NBS–LRR transcripts. Interestingly, an over-representation of disease-resistant genes in the targets of moderately conserved miRNAs was observed together with a significant decrease in their mRNA levels in the mycorrhized roots. These observations further suggest that the repression of proteins that are involved in plant immunity, such as the NBS–LRR proteins, by specific miRNAs may have been selected during evolution to avoid plant defence-like responses to symbiotic partners during symbiotic interactions. Based on the hypothesis of the de novo generation of new miRNA genes and the sequence similarities between some novel miRNA genes and their NBS–LRR targets, it has been speculated that NBS–LRR genes may represent an active source of new miRNAs [58,59]. This could also explain the frequently observed overrepresentation of the NBS–LRR-targeting miRNA that was observed in the deep-sequencing analysis.
4. Conclusions and outlook
Genome-sequencing data from several species in addition to the direct sequencing of sRNA populations have revealed the complexity of miRNA-dependent gene regulation in plants. In the Fabacea family, the emergence of three distinct model species allows for analyses of diversities in miRNA gene regulation in the context of symbiotic interactions between roots and soil rhizobia. Numerous conserved and novel miRNAs that are potentially involved in this biological process in all model legumes were shown to control highly conserved signalling pathways. These pathways have been recruited for new functions in nodulation during evolution, whereas other miRNAs appear to regulate signalling pathways that are specifically associated with symbiotic interactions. Moreover, miRNAs may be implicated in the restriction of plant defence responses during symbiotic interactions (AM symbiosis and nodulation), although functional evidence is still lacking. In addition to the evolution of miRNA-target nodes owing to mutations leading to novel isoforms, the acquisition of specific spatio-temporal expression patterns by miRNA genes may also contribute to diversifying the regulatory networks within existing genes similar to coding genes. The simultaneous evolution of both non-coding RNA gene promoters and miRNAs (at miRNA or miR* levels) further highlight the diverse gene expression patterns that may occur in large eukaryotic genomes. Because many common genes seem to participate in symbiotic and pathogenic interactions, the evolution of gene regulation may have been a key step both to control defence responses and to allow for the modification of root cell specification for new purposes, such as the formation of mycorrhized roots and nitrogen-fixing nodules. Further studies analysing the repression of miRNA genes are necessary to determine the precise functions of specific isoforms and the corresponding miR* within a miRNA family during symbiotic interactions. We believe that strategies for the development of collections of miRNA mutants, inhibition of miRNA function using mimicry constructs or the repression of new targets using artificial miRNAs (amiR) may enable novel insights to be obtained regarding the evolution of legume symbiosis.
Acknowledgements
This work was supported by grants from the MIRMED project (Genoscope, CNRS) and the ANR DIAGNOGENE and the program Saclay Plant Sciences (SPS, ANR-10-LABX-40). P.B. was supported by the Marie Curie Postdoctoral IE Fellowship (European Commission, MEDEPIMIR, no. PIEF-GA-2010-273743).
References
- 1.Truchet G., Barker D. G., Camut S., Debilly F., Vasse J., Huguet T. 1989. Alfalfa nodulation in the absence of Rhizobium. Mol. Gen. Genet. 219, 65–68 10.1007/BF00261158 (doi:10.1007/BF00261158) [DOI] [Google Scholar]
- 2.Madsen E. B., et al. 2011. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 65, 404–417 10.1111/j.1365-313X.2010.04431.x (doi:10.1111/j.1365-313X.2010.04431.x) [DOI] [PubMed] [Google Scholar]
- 3.Madsen E. B., et al. 2003. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640 10.1038/nature02045 (doi:10.1038/nature02045) [DOI] [PubMed] [Google Scholar]
- 4.Geurts R., Fedorova E., Bisseling T. 2005. Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8, 346–352 10.1016/j.pbi.2005.05.013 (doi:10.1016/j.pbi.2005.05.013) [DOI] [PubMed] [Google Scholar]
- 5.Crespi M., Frugier F. 2008. De novo organ formation from differentiated cells: root nodule organogenesis. Sci. Signal. 1, re11. 10.1126/scisignal.149re11 (doi:10.1126/scisignal.149re11) [DOI] [PubMed] [Google Scholar]
- 6.Desbrosses G. J., Stougaard J. 2011. Root nodulation: a paradigm for how plant–microbe symbiosis influences host developmental pathways. Cell Host Microbe 10, 348–358 10.1016/j.chom.2011.09.005 (doi:1931-3128/j.chom.2011.09.005) [DOI] [PubMed] [Google Scholar]
- 7.Murray J. D. 2011. Invasion by invitation: rhizobial infection in legumes. Mol. Plant–Microbe Interact. 24, 631–639 10.1094/mpmi-08-10-0181 (doi:10.1094/mpmi-08-10-0181) [DOI] [PubMed] [Google Scholar]
- 8.Maillet F., et al. 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63 10.1038/nature09622 (doi:10.1038/nature09622) [DOI] [PubMed] [Google Scholar]
- 9.Seddas P. M. A., et al. 2009. Symbiosis-related plant genes modulate molecular responses in an arbuscular mycorrhizal fungus during early root interactions. Mol. Plant–Microbe Interact. 22, 341–351 10.1094/mpmi-22-3-0341 (doi:10.1094/mpmi-22-3-0341) [DOI] [PubMed] [Google Scholar]
- 10.Soto M. J., Dominguez-Ferreras A., Perez-Mendoza D., Sanjuan J., Olivares J. 2009. Mutualism versus pathogenesis: the give-and-take in plant–bacteria interactions. Cell Microbiol. 11, 381–388 10.1111/j.1462-5822.2009.01282.x (doi:CMI1282/j.1462-5822.2008.01282.x) [DOI] [PubMed] [Google Scholar]
- 11.Vasse J., Debilly F., Truchet G. 1993. Abortion of infection during the Rhizobium meliloti–alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 4, 555–566 10.1046/j.1365-313X.1993.04030555.x (doi:10.1046/j.1365-313X.1993.04030555.x) [DOI] [Google Scholar]
- 12.Bozso Z., Maunoury N., Szatmari A., Mergaert P., Ott P. G., Zsiros L. R., Szabo E., Kondorosi E., Klement Z. 2009. Transcriptome analysis of a bacterially induced basal and hypersensitive response of Medicago truncatula. Plant Mol. Biol. 70, 627–646 10.1007/s11103-009-9496-8 (doi:10.1007/s11103-009-9496-8) [DOI] [PubMed] [Google Scholar]
- 13.Marino D., Andrio E., Danchin E. G. J., Oger E., Gucciardo S., Lambert A., Puppo A., Pauly N. 2011. A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189, 580–592 10.1111/j.1469-8137.2010.03509.x (doi:10.1111/j.1469-8137.2010.03509.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Haeze W., Holsters M. 2004. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 12, 555–561 10.1016/j.tim.2004.10.009 (doi:10.1016/j.tim.2004.10.009) [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa T., et al. 2011. From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume–Rhizobium symbiosis. Plant J. 65, 169–180 10.1111/j.1365-313X.2010.04411.x (doi:10.1111/j.1365-313X.2010.04411.x) [DOI] [PubMed] [Google Scholar]
- 16.Khan G., Declerck M., Sorin C., Hartmann C., Crespi M., Lelandais-Brière C. 2011. MicroRNAs as regulators of root development and architecture. Plant Mol. Biol. 77, 47–58 10.1007/s11103-011-9793-x (doi:10.1007/s11103-011-9793-x) [DOI] [PubMed] [Google Scholar]
- 17.Khan G. A., Hudik E., Sorin C., Hartmann C., Crespi M., Lelandais-Brière C. 2011. Non coding RNAs in plants; RNA technologies. Berlin, Germany: Springer [Google Scholar]
- 18.Boualem A., Laporte P., Jovanovic M., Laffont C., Plet J., Combier J. P., Niebel A., Crespi M., Frugier F. 2008. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 54, 876–887 10.1111/j.1365-313X.2008.03448.x (doi:10.1111/j.1365-313X.2008.03448.x) [DOI] [PubMed] [Google Scholar]
- 19.Combier J. P., et al. 2006. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20, 3084–3088 10.1101/gad.402806 (doi:10.1101/gad.402806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez F. 2006. Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 11, 460–468 10.1016/j.tplants.2006.07.006 (doi:10.1016/j.tplants.2006.07.006) [DOI] [PubMed] [Google Scholar]
- 21.Xie Z. X., Johansen L. K., Gustafson A. M., Kasschau K. D., Lellis A. D., Zilberman D., Jacobsen S. E., Carrington J. C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, 642–652 10.1371/journal.pbio.0020104 (doi:e10410.1371/journal.pbio.0020104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 10.1016/j.cell.2009.01.046 (doi:10.1016/j.cell.2009.01.046) [DOI] [PubMed] [Google Scholar]
- 23.Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y. Y., Sieburth L., Voinnet O. 2008. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190 10.1126/science.1159151 (doi:10.1126/science.1159151) [DOI] [PubMed] [Google Scholar]
- 24.Vaucheret H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771 10.1101/gad.1410506 (doi:10.1101/gad.1410506) [DOI] [PubMed] [Google Scholar]
- 25.Bardou F., Merchan F., Ariel F., Crespi M. 2011. Dual RNAs in plants. Biochimie 93, 1950–1954 10.1016/j.biochi.2011.07.028 (doi:10.1016/j.biochi.2011.07.028) [DOI] [PubMed] [Google Scholar]
- 26.Saze H., Kakutani T. 2011. Differentiation of epigenetic modifications between transposons and genes. Curr. Opin. Plant Biol. 14, 81–87 10.1016/j.pbi.2010.08.017 (doi:10.1016/j.pbi.2010.08.017) [DOI] [PubMed] [Google Scholar]
- 27.Capitao C., Paiva J. A. P., Santos D. M., Fevereiro P. 2011. In Medicago truncatula, water deficit modulates the transcript accumulation of components of small RNA pathways. BMC Plant Biol. 11, 79. 10.1186/1471-2229-11-79 (doi:10.1186/1471-2229-11-79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubio-Somoza I., Weigel D. 2011. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 16, 258–264 10.1016/j.tplants.2011.03.001 (doi:10.1016/j.tplants.2011.03.001) [DOI] [PubMed] [Google Scholar]
- 29.Rubio-Somoza I., Cuperus J. T., Weige D., Carrington J. C. 2009. Regulation and functional specialization of small RNA-target nodes during plant development. Curr. Opin. Plant Biol. 12, 622–627 10.1016/j.pbi.2009.07.003 (doi:10.1016/j.pbi.2009.07.003) [DOI] [PubMed] [Google Scholar]
- 30.Allen E., Xie Z. X., Gustafson A. M., Sung G. H., Spatafora J. W., Carrington J. C. 2004. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36, 1282–1290 10.1038/ng1478 (doi:10.1038/ng1478) [DOI] [PubMed] [Google Scholar]
- 31.Cuperus J. T., Fahlgren N., Carrington J. C. 2011. Evolution and functional diversification of MIRNA genes. Plant Cell 23, 431–442 10.1105/tpc.110.082784 (doi:10.1105/tpc.110.082784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahlgren N., et al. 2007. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2, e219. 10.1371/journal.pone.0000219 (doi:10.1371/journal.pone.0000219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B. H., Pan X. P., Cannon C. H., Cobb G. P., Anderson T. A. 2006. Conservation and divergence of plant microRNA genes. Plant J. 46, 243–259 10.1111/j.1365-313X.2006.02697.x (doi:10.1111/j.1365-313X.2006.02697.x) [DOI] [PubMed] [Google Scholar]
- 34.Lelandais-Briere C., Naya L., Sallet E., Calenge F., Frugier F., Hartmann C., Gouzy J., Crespi M. 2009. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21, 2780–2796 10.1105/tpc.109.068130 (doi:10.1105/tpc.109.068130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naya L., Khan G. A., Sorin C., Hartmann C., Crespi M., Lelandais-Briere C. 2010. Cleavage of a non-conserved target by a specific miR156 isoform in root apexes of Medicago truncatula. Plant Signal Behav. 5, 328–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Branscheid A., Devers E. A., May P., Krajinski F. 2011. Distribution pattern of small RNA and degradome reads provides information on miRNA gene structure and regulation. Plant Signal Behav. 6, 1609–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Luis Margarit A. 2010. Etude des microRNAs impliqués dans la symbiose de Lotus japonicus. Doctoral thesis, Strasbourg University, Strasbourg, France [Google Scholar]
- 38.Subramanian S., Fu Y., Sunkar R., Barbazuk W. B., Zhu J. K., Yu O. 2008. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genom. 9, 160. 10.1186/1471-2164-9-160 (doi:10.1186/1471-2164-9-160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'haeseleer K., et al. 2011. Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol. 191, 647–661 10.1111/j.1469-8137.2011.03719.x (doi:10.1111/j.1469-8137.2011.03719.x) [DOI] [PubMed] [Google Scholar]
- 40.Wang J.-W., Wang L.-J., Mao Y.-B., Cai W.-J., Xue H.-W., Chen X.-Y. 2005. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant cell 17, 2204–2216 10.1105/tpc.105.033076 (doi:10.1105/tpc.105.033076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutierrez L., Bussell J. D., Păcurar D. I., Schwambach J., Păcurar M., Bellini C. 2009. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant cell. 21, 3119–3132 10.1105/tpc.108.064758 (doi:10.1105/tpc.108.064758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padmanabhan C., Zhang X. M., Jin H. L. 2009. Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 12, 465–472 10.1016/j.pbi.2009.06.005 (doi:10.1016/j.pbi.2009.06.005) [DOI] [PubMed] [Google Scholar]
- 43.Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J. D. G. 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439 10.1126/science.1126088 (doi:10.1126/science.1126088) [DOI] [PubMed] [Google Scholar]
- 44.Li H., Deng Y., Wu T. L., Subramanian S., Yu O. 2010. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 153, 1759–1770 10.1104/pp.110.156950 (doi:10.1104/pp.110.156950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlsbecker A., et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 10.1038/nature08977 (doi:10.1038/nature08977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Axtell M. J., Bartel D. P. 2005. Antiquity of microRNAs and their targets in land plants. Plant Cell 17, 1658–1673 10.1105/tpc.105.032185 (doi:10.1105/tpc.105.032185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang B. H., Pan X. P., Stellwag E. J. 2008. Identification of soybean microRNAs and their targets. Planta 229, 161–182 10.1007/s00425-008-0818-x (doi:10.1007/s00425-008-0818-x) [DOI] [PubMed] [Google Scholar]
- 48.Devers E. A., Branscheid A., May P., Krajinski F. 2011. Stars and symbiosis: MicroRNA- and MicroRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156, 1990–2010 10.1104/pp.111.172627 (doi:10.1104/pp.111.172627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nag A., Jack T. 2010. Sculpting the flower; the role of microRNAs in flower development. Curr. Top. Dev. Biol. 91, 349–378 (doi:10.1016/S0070-2153(10)91012-0) [DOI] [PubMed] [Google Scholar]
- 50.Zhang X., Zou Z., Gong P., Zhang J., Ziaf K., Li H., Xiao F., Ye Z. 2011. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 33, 403–409 10.1007/s10529-010-0436-0 (doi:10.1007/s10529-010-0436-0) [DOI] [PubMed] [Google Scholar]
- 51.Zhao M., Ding H., Zhu J.-K., Zhang F., Li W.-X. 2011. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190, 906–915 10.1111/j.1469-8137.2011.03647.x (doi:10.1111/j.1469-8137.2011.03647.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y. J., Fu Y. R., Ji L. S., Wu C. A., Zheng C. C. 2010. Characterization and expression analysis of the Arabidopsis mir169 family. Plant Sci. 178, 271–280 10.1016/j.plantsci.2010.01.007 (doi:10.1016/j.plantsci.2010.01.007) [DOI] [Google Scholar]
- 53.Kalo P., et al. 2005. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789 10.1126/science.1110951 (doi:10.1126/science.1110951) [DOI] [PubMed] [Google Scholar]
- 54.Kulcheski F. R., et al. 2011. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 12, 307. 10.1186/1471-2164-12-307 (doi:10.1186/1471-2164-12-307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi Y., Tsuda K., Nguyen L. V., Wang X., Lin J., Murphy A. S., Glazebrook J., Thordal-Christensen H., Katagiri F. 2011. Physical association of Arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J. Biol. Chem. 286, 31297–31307 10.1074/jbc.M110.211615 (doi:10.1074/jbc.M110.211615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jagadeeswaran G., et al. 2009. Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 184, 85–98 10.1111/j.1469-8137.2009.02915.x (doi:10.1111/j.1469-8137.2009.02915.x) [DOI] [PubMed] [Google Scholar]
- 57.Li Y., Li C. Q., Ding G. H., Jin Y. X. 2011. Evolution of MIR159/319 microRNA genes and their post-transcriptional regulatory link to siRNA pathways. BMC Evol. Biol. 11, 122. 10.1186/1471-2148-11-122 (doi:10.1186/1471-2148-11-122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klevebring D., Street N. R., Fahlgren N., Kasschau K. D., Carrington J. C., Lundeberg J., Jansson S. 2009. Genome-wide profiling of populus small RNAs. BMC Genom. 10, 620. 10.1186/1471-2164-10-620 (doi:10.1186/1471-2164-10-620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He X.-F., Fang Y.-Y., Feng L., Guo H.-S. 2008. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Letters 582, 2445–2452 10.1016/j.febslet.2008.06.011 (doi:10.1016/j.febslet.2008.06.011) [DOI] [PubMed] [Google Scholar]