Abstract
Phytohormones are important plant growth regulators that control many developmental processes, such as cell division, cell differentiation, organogenesis and morphogenesis. They regulate a multitude of apparently unrelated physiological processes, often with overlapping roles, and they mutually modulate their effects. These features imply important synergistic and antagonistic interactions between the various plant hormones. Auxin and cytokinin are central hormones involved in the regulation of plant growth and development, including processes determining root architecture, such as root pole establishment during early embryogenesis, root meristem maintenance and lateral root organogenesis. Thus, to control root development both pathways put special demands on the mechanisms that balance their activities and mediate their interactions. Here, we summarize recent knowledge on the role of auxin and cytokinin in the regulation of root architecture with special focus on lateral root organogenesis, discuss the latest findings on the molecular mechanisms of their interactions, and present forward genetic screen as a tool to identify novel molecular components of the auxin and cytokinin crosstalk.
Keywords: auxin, cytokinin, forward genetic screen, lateral roots
1. Introduction
Root is a complex organ necessary to fix the above-ground plant body to the soil and to enable uptake of water and nutrients from the soil. Although the root is established already during embryogenesis at the basal pole of the embryo, the root system starts to develop massively during the post-embryonic plant life. The root system architecture results from two parallelly occurring processes, primary root growth and recurrent branching. In this manner, the plant root occupies the surrounding soil niche and uses the available nutritional resources. Thus, the recurrent initiation of the lateral root (LR) organogenesis by plants is the key process in the dynamic formation of the functional root system.
In Arabidopsis thaliana, LRs originate in the pericycle cell layers adjacent to the xylem pole [1,2]. After certain pericycle cells have acquired founder cell properties, LR primordia are initiated by several anticlinal divisions and they develop as a consequence of coordinated cell division and differentiation. Later on, the LR primordia emerge from the parent root, mainly by cell elongation. The LR meristem, of which structure and function are similar to the primary root meristem, is then activated [3].
The positive role of auxin at all stages of the LR organogenesis, including initiation and development, is well established. Accumulation of auxin and activation of auxin responses in individual pericycle cells stimulate founder cell specification and LR initiation (LRI) [4,5]. This positive impact of auxin on LRI was evidenced by modulation of auxin levels [4,6] as well as of auxin responses [7–9]. In the later developmental phases of LR primordia, auxin distribution gradients with maxima at the primordia tips determine the proper primordia organogenesis [10]. This auxin gradient is generated by the concerted action of AUXIN RESISTANT1 (AUX1)–like AUX1 (LAX) auxin influx carriers [11], PIN-FORMED (PIN) auxin efflux carriers [12–16], and members of the multi-drug-resistant/P-glycoprotein subfamily of ATP-binding cassette proteins [17]. When polar auxin transport is disturbed genetically or by chemical inactivation, the development of the LR primordia is severely affected [10,18,19]. Besides its action as an important patterning factor, auxin also controls the interaction between the LR primordia and the neighbouring tissues and mediates the non-invasive emergence of LR primordia through adjacent tissue layers by stimulation of cell wall-remodelling genes expression [20].
Although auxin seems to be a general regulator through the different phases of the LR organogenesis, the transduction cascade and downstream response might be very specific for every developmental phase and mediated through specific pairs of auxin signalling components, such as AUX/INDOLE-3-ACETIC ACID (AUX/IAA) repressors and AUXIN RESPONSE FACTORS (ARFs). Whereas priming is under the control of IAA28, ARF5, ARF6, ARF7, ARF8 and ARF19, founder cell specification and progress towards formative division resulting in LRI require the regulatory modules IAA14–ARF7 and IAA14–ARF19, acting in parallel with IAA12–ARF5 [21,22].
As the LR organogenesis programme is activated repetitively over time in simultaneously growing primary roots, the question arises as to the nature of the mechanisms spatio-temporally controlling these recurrent initiations. Oscillations of the auxin activity in the protoxylem cells of the basal root meristem were found to correlate with the subsequent LRI in a more proximal zone of the root [23,24]. Priming has been proposed as one of the earliest events that predetermines the LRI through regular auxin activity peaks [23]. Regular root bending as a consequence of the gravity-sensitive root behaviour generates auxin maxima in the pericycle cells at the convex bent side and activates LRI. Therefore, these auxin oscillations might be triggered by mechanisms that depend on the auxin reflux controlling the root gravity response [23,25] and/or the mechanical deformation of tissues by stretching during the curve formation [6]. Conceptually different is the hypothesis considering existence of endogenous oscillatory mechanisms. Regular oscillations in a set of genes including transcriptional regulators detected in the roots might be the primary mechanism recurrently activating processes that result in LRI [24].
Besides auxin, several other plant hormones have been found to regulate the LR organogenesis [26–29], among which cytokinin exhibits one of the strongest inhibitory effects. Any increase in cytokinin activity, either by exogenous manipulation of cytokinin levels [30,31] or endogenous modulation of the activity of genes involved in cytokinin metabolism, results in changed LRI frequencies and developmental defects [31,32]. Suppression of the cytokinin signalling pathway either by interfering with the receptor ARABIDOPSIS HISTIDINE KINASE4 (AHK4)/CYTOKININ RESISTANT1 (CRE1) and its homologues AHK2 and AHK3, or the positive components, the B-type ARABIDOPSIS RESPONSE REGULATOR (ARR) genes, typically enhances LR organogenesis [33,34].
The opposing contributions of both auxin and cytokinin pathways to regulate the LR organogenesis put special demands on the mechanisms that balance their activities and mediate their interaction. Auxin control over the cytokinin signalling repressors ARR7 and ARR15 transcription appear to be critical for proper early embryo root pole establishment [35]. Similarly, auxin and cytokinin activities in the shoot apical meristem are counterbalanced through transcriptional regulation of ARR7 and ARR15 by AUXIN RESPONSE FACTOR5/MONOPTEROS (MP) [36]. Besides crosstalk between auxin and cytokinin signalling pathways, cytokinin also interferes with the auxin distribution by modulating the polar auxin transport activity. This mode of interaction is particularly important for root apical meristem maintenance and LR organogenesis [31,37–40]. In the root apical meristem, the auxin–cytokinin crosstalk circuit is mediated through the AHK3 receptor and the downstream transcription factor ARR1 that adjusts the expression of the IAA3/SHORT HYPOCOTYL2 (SHY2) auxin signalling repressor and attenuates the expression of several PIN genes as a consequence [37]. In addition to the transcriptional control, cytokinin also impacts on the PIN1 intracellular trafficking [40–42]. This regulatory mode is important in view of the controlled leaf initiation and positioning and LR organogenesis [41,42]. Although there are several hints at the molecular nature of the auxin–cytokinin crosstalk, our knowledge on the key players is still very limited. Hence, novel approaches are necessary to identify the molecular components of the auxin–cytokinin interaction network.
Here, we describe a forward genetic screen as an approach to characterize intersections of the auxin and cytokinin signalling pathways. By using LR organogenesis as a model, we designed a mutant screen that specifically targets the interactions between auxin and cytokinin. Mutants were screened that produce LRs after application of auxin simultaneously with inhibiting concentrations of cytokinin. Twenty-two novel mutant alleles, designated primordia on auxin and cytokinin (pac) were recovered and classified based on their LRI and response to auxin and cytokinin. Important candidates as crosstalk components are considered primarily mutants in which the basal LRI process was not affected and the cytokinin resistance phenotype occurred only in the presence of auxin. Interestingly, detailed characterization of the pac mutant phenotypes suggested that some mutants might represent molecular components that control the cytokinin-dependent expression of the PIN auxin efflux carriers and photomorphogenesis.
2. Material and methods
(a). Plant material and growth conditions
Ethyl methanesulphonate (EMS)-mutagenized and non-mutagenized transgenic Arabidopsis thaliana (L.) Heynh. lines harbouring PIN1::PIN1:GFP [10], etr1 [43] and cre1–12 [44] were used. Seeds were sterilized with chloral gas, sown in Petri dishes on 0.8 per cent agar with 1 per cent sucrose-containing 0.5× Murashige and Skoog (MS) medium, stored for 2 days at 4°C, and grown on vertically oriented plates in growth chambers under a 16 L : 8 D cycle photoperiod at 18°C. Seven days after germination, seedlings were harvested and processed.
(b). Ethyl methanesulphonate mutagenesis and screening of mutants
Seeds of transgenic Arabidopsis plants (ecotype Columbia-0) harbouring PIN1::PIN1:GFP were soaked in 0.2 or 0.3 per cent EMS solution for 8 h. M2 seeds were bulk-harvested from approximately 20 M1 plants and pooled. Approximately 600 M2 seedlings from each pool were used for screening. Four-day-old seedlings germinated on 0.5× MS media supplemented with 1 per cent sucrose were overlaid with 0.5 × MS liquid medium containing 1 µM IAA and 7 µM 6-benzylaminopurine (BAP) and cultivated for 48 h and 72 h, respectively. To record the efficiency of the hormonal treatments in every experiment, non-mutated PIN1::PIN1:GFP seedlings were analysed treated only with control media, supplemented with 1 µM IAA and 1 µM IAA plus 7 µM BAP. The numbers of LR primordia were scored with a fluorescence stereomicroscope MZ16F (Leica Microsystems) and mutants with more LR primordia than the control background were selected.
(c). Analyses of root growth, organogenesis of LR primordia and etiolated seedlings
Mutants and control seedlings were grown on 0.5× MS medium without or supplemented with hormones: 0.1 µM BAP, 50 nM 1-naphthaleneacetic acid (NAA), 1 µM 1-aminocyclopropane-1-carboxylic acid (ACC). Seven days after germination, the plant material was cleared as described [3]. Root lengths were measured on scanned slides. LR primordia were counted with a differential interference contrast microscope BX51 (Olympus). Hypocotyl lengths in etiolated seedlings were analysed after 6 days of cultivation in the dark. Petri dishes with etiolated seedlings were scanned and hypocotyl lengths were measured with the ImageJ software (http://rsbweb.nih.gov/ij/). At least 20 seedlings were analysed and the experiments were repeated twice independently. For the statistical evaluation, the t-test was done with the Excel statistical package. For calculation of the relative change of LRI after hormone treatment, LRI was expressed as the ratio of treated to untreated plants and the ratio of mutant to control plants was calculated. In each case, the total error was propagated [45]. One way analysis of variance combined with Holm–Sidak method was applied to evaluate a statistical significance using SigmaPlot software.
(d). Analysis of PIN1::PIN1:GFP expression
The cytokinin impact on the PIN1::PIN1:GFP expression was examined in root meristems exposed for 6 h to control medium or liquid 0.5× MS medium supplemented with 10 µM BAP. At least 10 seedlings were analysed with the confocal laser-scanning microscope TCS SP2 AOBS (Leica Microsystems). The images from the obtained micrographs were processed in Adobe Photoshop.
3. Results
(a). Forward genetic screen for mutants defective in auxin/cytokinin crosstalk
The antagonistic auxin–cytokinin interaction is strongly visible in the regulation of the LR organogenesis. Whereas auxin promotes both LRI and LR development, cytokinin exhibits inhibitory effects [4,30,31,46]. Thus, to identify new molecular components required for balancing the auxin/cytokinin activities, we decided to use the LR organogenesis as a suitable model system. We designed the forward genetic screen to look for mutants that produce LRs when auxin is applied simultaneously with cytokinin at inhibiting concentrations. As best crosstalk candidates, we considered mutants in which the basal LRI process was not affected and the cytokinin resistance phenotype occurred only in the presence of auxin.
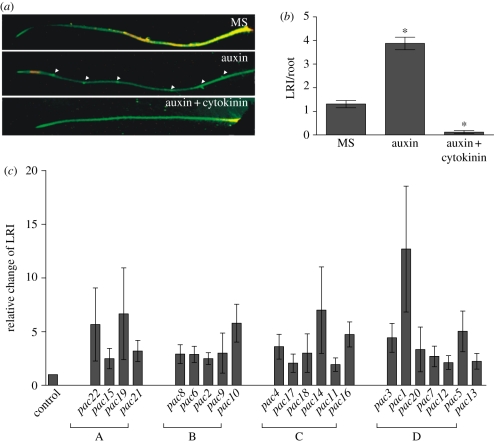
To determine the optimal screening conditions, different auxin and cytokinin concentrations were applied, separately or simultaneously, on 4 days old Arabidopsis seedlings (for details, see §2). The LRI was evaluated 48 h and 72 h after treatment (electronic supplementary material, figure S1a). Application of 1 µM IAA enhanced the LRI almost threefold when compared with control seedlings (8 ± 1.4 versus 2.9 ± 0.86 LR primordia per centimetre; electronic supplementary material, figure S1b). When applied simultaneously with cytokinin, 7 µM BAP most efficiently inhibited the auxin-stimulated LRI when compared with 1 or 5 µM BAP (electronic supplementary material, figure S1b). Thus, 1 µM IAA and 7 µM BAP applied together were used to screen for mutants initiating LRs under these restrictive conditions (figure 1a,b). M1 families (1700) of EMS-mutagenized PIN1::PIN1:GFP lines were harvested into 72 pools (approx. 20–25 individuals per pool). Approximately, 600 seedlings from each pool were examined for their sensitivity to auxin/cytokinin and mutants resistant to the hormonal treatment were propagated. From 150 lines selected in the first round of the screen, 22 mutant lines were recovered with obvious resistance to auxin and cytokinin in the next generation and designated primordia on auxin and cytokinin (pac; figure 1c).
Figure 1.
Forward genetic screen for mutants defective in auxin/cytokinin crosstalk. (a,b) Strong stimulatory effect of auxin (1 µM IAA) application on LRI observed after 48 h on four-day-old PIN1::PIN1:GFP seedlings. Simultaneous application of cytokinin (7 µM BAP) counteracted the stimulatory auxin effect. LRI was scored in PIN1::PIN1:GFP seedlings 48 h after treatment with control media (MS) and media supplemented with 1 µM IAA or 1 µM IAA and 7 µM BAP media (*p < 0.05, n = 20 seedlings). (c) pac mutants recovered in the forward genetic screen exhibiting a reduced sensitivity to the simultaneous auxin and cytokinin treatment (p < 0.05, n = 10 seedlings). Error bars mark standard errors. LRI scored as total number of LR primordia per root.
(b). Identification of pac mutants defective in auxin–cytokinin crosstalk
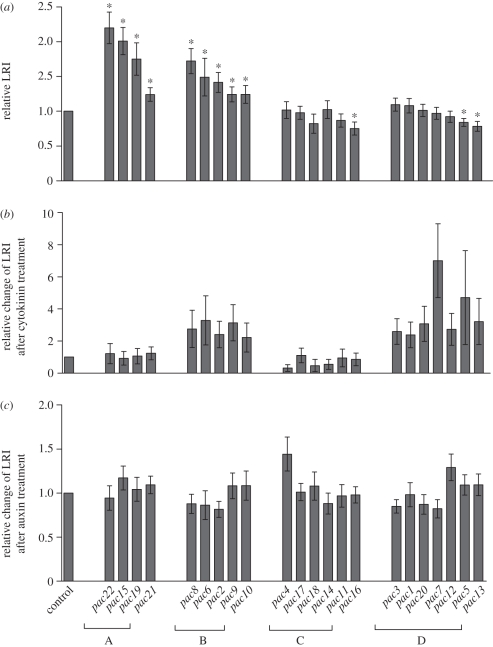
To distinguish pac mutants exhibiting an enhanced LRI exclusively under simultaneous auxin/cytokinin treatments from the mutants defective in LRI or altered auxin or cytokinin sensitivity, the LRI phenotypes were analysed thoroughly. Based on the LRI and its cytokinin sensitivity, we grouped the pac mutants into four subgroups: subgroups A (pac22, pac15, pac19 and pac21) and B (pac8, pac6, pac2, pac9 and pac10) exhibited an increased LRI. By contrast, mutants in subgroups C (pac4, pac17, pac18, pac14, pac11 and pac16) and D (pac3, pac1, pac20, pac7, pac12, pac5 and pac13) did not show enhanced LRI (figure 2a).
Figure 2.
Impact of pac mutations on LRI. (a) Increased LRI in pac mutants of subgroups A and B, but not C and D. (b) Reduced LRI cytokinin sensitivity in pac mutants of subgroups B and D, but not A and C. (c) Increased auxin sensitivity in pac15 (subgroup A), pac4 (subgroup C) and pac12, (subgroup D) mutants and moderate auxin resistance in pac2 (subgroup B), and pac7 (subgroup D). Seven-day-old seedlings were analysed germinated on control media or media supplemented with cytokinin (0.1 µM BAP) or auxin (50 nM NAA; *p < 0.05, n = 20 seedlings). Error bars mark standard errors. LRI scored as a number of LR primordia per centimetre of root length.
Based on the cytokinin response, mutants of subgroups B and D were resistant to cytokinin. Whereas in wild-type seedlings germinated on 0.1 µM BAP, the LRI was approximately 80 per cent lower than that of the untreated control, mutants of these two subgroups were able to initiate LRs (figure 2b).
Interestingly, the effect of the pac mutations on the auxin sensitivity was not very pronounced and only a few of the pac mutations modulated the auxin sensitivity. When compared with control seedlings with a 2.5–3-fold increased LRI after auxin (50 nM NAA) treatment, the pac15 (subgroup A), pac4 (subgroup C) and pac12 (subgroup D) mutants showed an enhanced auxin sensitivity, whereas the mutants pac2 (subgroup B) and pac7 (subgroup D) were moderately resistant to auxin (figure 2c).
Visual observation of cleared roots did not reveal any severe defects in LR primordia patterning in pac mutants. However, more detailed analyses using tissue-specific markers are needed for final conclusion on the role of PAC genes in LR primordia patterning.
Based on the detailed LRI phenotype analysis, we identified the subgroup of pac mutants corresponding to our primary requirements. The pac mutants of subgroup C were not affected in LRI and exhibited neither increased cytokinin resistance nor a dramatically changed auxin sensitivity. Thus, they represent the best candidates as crosstalk components that might be involved in the fine-tuning of auxin/cytokinin activities to ensure a relevant developmental output.
(c). Pac mutations modulate root sensitivity to cytokinin
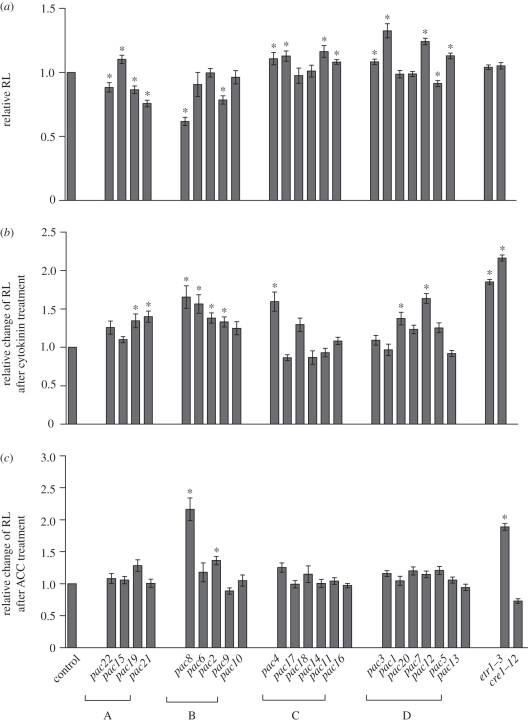
To get insight into the general effect of pac mutations on root growth and cytokinin sensitivity, we analysed root growth on control and cytokinin-supplemented media. Overall, root growth of pac mutants was variably affected. In a few mutants of subgroups A (pac22, pac19 and pac21) and B (pac8 and pac9), root growth was reduced significantly, whereas root length increased moderately in mutants of subgroup C (pac4, pac17, pac11 and pac16) and subgroup D (pac1, pac3, pac12 and pac13) (figure 3a).
Figure 3.
Modulation of root sensitivity to cytokinin but not to ethylene in pac mutants. (a) Root length analysis of pac mutants. Root growth decreased significantly in pac22, pac19 and pac21 (subgroup A) and pac8 and pac9 (subgroup B), but roots elongated in pac15 (subgroup A), pac4, pac17, pac11, pac16 (subgroup C) and pac1, pac3, pac12, pac13 (subgroup D) mutants. (b) Reduced root cytokinin sensitivity in pac mutants. (c) Root sensitivity to ethylene moderately affected in pac mutants. Seven-day-old seedlings were analysed germinated on control media or media supplemented with cytokinin (0.1 µM BAP) or 1 µM ACC (*p < 0.05, n = 20 seedlings). Error bars mark standard errors.
The cytokinin sensitivity of pac mutant roots was significantly altered. However, differently from LRI, cytokinin sensitivity was changed not only in mutants of subgroups B and D, but also of subgroup A (pac19 and pac21). Mutants of subgroup C, apart from pac4, did not show any dramatic change in root growth response to cytokinin (figure 3b).
Cytokinin is known to enhance ethylene production. Therefore, part of the cytokinin effects on the root phenotype might be mimicked by ethylene. As a consequence, mutants defective in the ethylene transduction pathway exhibit root growth insensitive not only to ethylene but also to cytokinin [47,48]. To dissect whether some of the pac mutants might be defective in the ethylene-related pathway, we analysed root growth on ACC [49], the precursor of the ethylene biosynthesis. As expected, the ethylene receptor mutant etr1 [43] was resistant to ethylene as well as to cytokinin but the cytokinin receptor mutant cre1 [44] was resistant to cytokinin, but not to ethylene (figure 3b,c). Interestingly, most pac mutants, except pac8, were not affected in the response to ethylene or exhibited mild ethylene insensitivity (pac2) (figure 3c).
These detailed analyses of root cytokinin and ethylene sensitivity indicate that the mutant screen as designed targeted primarily genes involved in the control of the cytokinin activity and the pac mutations do not seem to interfere significantly with the ethylene pathway.
(d). A subgroup of pac mutants exhibits defects in photomorphogenesis
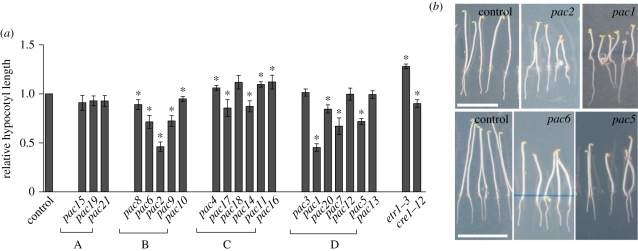
Next, we examined whether PAC genes play a role exclusively in the auxin/cytokinin-controlled LR organogenesis or are involved also in other developmental processes requiring the activity of both hormonal pathways. One such process is seedling development in response to light. In the dark, seedlings undergo skotomorphogenesis and develop long hypocotyls, an apical hook and closed cotyledons. Under light, they adopt photomorphogenesis, resulting in short hypocotyls, open cotyledons and chlorophyll accumulation [50]. Exogenous cytokinin promotes light-grown phenotypes in dark-grown seedlings [51], but auxin enhances hypocotyl elongation and suppression of auxin response seems to be critical in the regulation of photomorphogenic responses [50,52]. When grown in the dark, pac mutants from subgroups A and C exhibited no or mild changes in their development, respectively, when compared with control seedlings (figure 4a). In contrast, pac mutants in subgroups B (pac6, pac2 and pac9) and D (pac1, pac7 and pac5) exhibited strong defects in skotomorphogenesis. These pac mutations promoted the light phenotype, such as shorter hypocotyls, defective apical hook formation and open cotyledons, which might result from the lack of suppression of a photomorphogenic programme (figure 4b). Interestingly, the most affected pac mutants were those belonging to subgroups B and D that were also defective in cytokinin repression of LRI (compare figures 2b and 4a). Dark phenotypes of these pac mutants hint at a link between cytokinin-regulated LRI and photomorphogenesis. An intriguing aspect of this finding is that the lack/malfunction of one molecular factor at the same time decreases the LRI cytokinin sensitivity and stimulates photomorphogenesis in the dark, the phenotype promoted by enhanced cytokinin activity.
Figure 4.
Defects in photomorphogenesis in a subgroup of pac mutants. (a) Hypocotyl length of etiolated seedlings. pac mutants of subgroups B (pac6, pac2 and pac9) and D (pac1, pac7 and pac5) exhibited a strongly reduced hypocotyl length (*p < 0.05, n = 10 seedlings). (b) Promoted light phenotype, such as short hypocotyl, defective apical hook formation and open cotyledons in dark-grown pac1, pac2, pac5 and pac6 mutants. Error bars mark standard errors. Scale bars = 1 cm.
(e). A subgroup of pac mutations interferes with cytokinin-controlled PIN1 expression
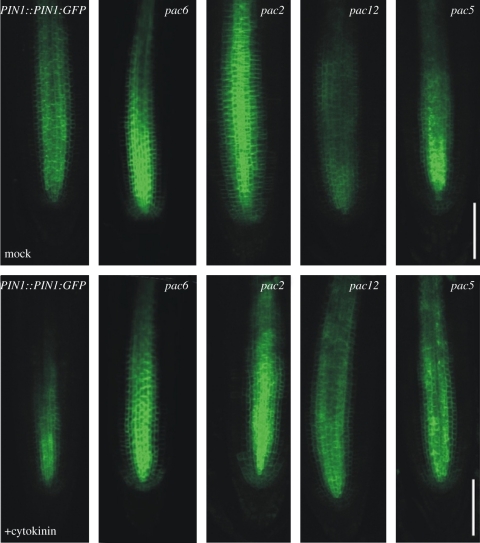
One of the recently revealed important modes of interaction between auxin and cytokinin is the cytokinin-mediated modulation of the polar auxin transport [37–42]. By modifying the expression of PIN auxin efflux carriers, cytokinin might influence the cell-to-cell auxin transport and, thus, the auxin distribution required for regulation of different developmental processes, such as LR organogenesis or root meristem activity and size [31,37,38,40]. To examine whether some of the pac genes might be involved in these regulatory pathways, the cytokinin-mediated repression of the PIN1 expression was analysed. The PIN1-GFP expression was monitored in control and pac mutant roots after cytokinin treatment and compared with untreated roots. As expected, treatment with 10 µM BAP for 6 h dramatically reduced the PIN1-GFP signal in roots of control seedlings. Several pac mutations (pac8, pac6, pac2, pac10, pac12 and pac5) interfered with the cytokinin-mediated repression of the PIN1 expression (figure 5). Interestingly, these pac mutants belonged to subgroups B and D that exhibited a cytokinin-resistant LRI and promoted photomorphogenesis under dark treatment.
Figure 5.
Interference with cytokinin-controlled PIN1 expression in pac mutants. PIN1::PIN1:GFP expression in root meristem of pac mutants exposed for 6 h to control medium or liquid 0.5× MS medium supplemented with cytokinin (10 µM BAP). The PIN1 expressions are representative of at least 10 analysed root tips of particular pac mutants. PIN1-GFP downregulation on the control media was consistently observed in pac12. Scale bars = 150 µm.
We hypothesize that these PAC genes might be the components of the pathway that regulates the polar auxin transport and, thus, underlie the control of two distant developmental processes, such as LRI and photomorphogenesis.
4. Discussion
Forward genetic screens have proved to be very powerful tools in dissecting the molecular components and mechanisms of the different hormonal signalling pathways, including those of auxin and cytokinin [53–56]. To assess the hormonal crosstalk and to identify the molecular components that mediate the pathway interactions, genetic screens have to be designed accurately by taking into account the activities of both hormonal pathways in the regulation of common developmental processes.
The forward genetic screen that resulted in the identification of pac mutants was aimed at finding the genes that balance the auxin and cytokinin activities during LR organogenesis. LR organogenesis is a very suitable model for such screens because both auxin and cytokinin contribute to its regulation from the earliest stage on (for review, see [57]). As both auxin and cytokinin interact antagonistically, proper crosstalk is particularly important for the LR organogenesis to proceed and any deficiency in their interaction might be manifested by a defective LR organogenesis. The common feature of all pac mutants is the reduced LRI sensitivity to the simultaneous auxin/cytokinin treatment. However, among the pac mutations, several subgroups could be recognized according to additional phenotypic characteristics. The PAC genes of subgroups A and B are apparently involved in the regulation of LRI because the corresponding mutants exhibit a significantly changed LRI, while the PAC genes of the B and D subgroups might contribute to the general cytokinin signal transduction considering their cytokinin-insensitive LRI phenotype. Therefore, the PAC genes of these subgroups, although undoubtedly important factors in the regulation of LR organogenesis, might not be necessarily the components that control directly the auxin–cytokinin interaction. Importantly, the identification of the subgroup C, in which the lack of the PAC function is obvious only in the presence of both hormones, hints at the existence of genes that are specifically involved in balancing the auxin–cytokinin activities. Thus, characterization of these PAC genes might be an important start point to further investigate the regulatory pathways that mediate the auxin and cytokinin crosstalk.
In addition to the LR organogenesis, both auxin and cytokinin are involved also in the regulation of other developmental processes (for review, see [58–61]), including the developmental switch between photomorphogenesis and skotomorphogenesis (for review, see [62]), but, as for the LR organogenesis, the mechanisms underlying their communication is unknown. Recent results [41] have revealed that cytokinin might be an important integrator of the light and auxin pathways. Lack of leaf initiation in dark-grown tomato meristems can be rescued by application of cytokinin. In the dark, PIN1, the key auxin transporter that ensures the proper hormone distribution underlying phyllotaxis [63], is internalized. Cytokinin might compensate for light treatments and stabilize PIN1 on the membranes. These results imply a scenario in which light activates the cytokinin signalling that, in turn, alters the auxin distribution important for the proper phyllotaxis through the modulation of the polar auxin transport activity. Interestingly, mutants of subgroups B and D, besides the reduced sensitivity of LRI to auxin/cytokinin and cytokinin treatments, exhibit additional defects in skotomorphogenesis manifested by dark-insensitive seedling development and cytokinin-insensitive PIN1 expression. These PAC genes hint at common regulatory mechanisms that might underlie the auxin and cytokinin interactions important not only for LR organogenesis, but also, simultaneously, for other auxin/cytokinin-regulated processes, such as seedling development controlled by darkness and light. Mutant phenotypes imply that part of such a regulatory mechanism might be executed through modulation of the auxin transport. Thus, the PAC genes of subgroups B and D represent promising candidates for additional factors that integrate cytokinin, auxin and light pathways in the regulation of plant development.
Acknowledgements
We thank Marleen Vanstraelen and Candela Cuesta Moliner for helpful discussions, Jiří Friml for providing material and Martine De Cock and Annick Bleys for help in preparing the manuscript. This work was supported by the European Research Council Starting Independent Research grant (ERC-2007-Stg-207362-HCPO to E.B.).
References
- 1.Dubrovsky J. G., Doerner P. W., Colón-Carmona A., Rost T. L. 2000. Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 124, 1648–1657 10.1104/pp.124.4.1648 (doi:10.1104/pp.124.4.1648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beeckman T., Burssens S., Inzé D. 2001. The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 52, 403–411 10.1093/jexbot/52.suppl_1.403 (doi:10.1093/jexbot/52.suppl_1.403) [DOI] [PubMed] [Google Scholar]
- 3.Malamy J. E., Benfey P. N. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44 10.1234/12345678 (doi:10.1234/12345678) [DOI] [PubMed] [Google Scholar]
- 4.Laskowski M. J., Williams M. E., Nusbaum H. C., Sussex I. M. 1995. Formation of lateral root meristems is a two-stage process. Development 121, 3303–3310 [DOI] [PubMed] [Google Scholar]
- 5.Dubrovsky J. G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M. G., Friml J., Shishkova S., Celenza J., Benková E. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl Acad. Sci. USA 105, 8790–8794 10.1073/pnas.0712307105 (doi:10.1073/pnas.0712307105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laskowski M., Grieneisen V. A., Hofhuis H., ten Hove C. A., Hogeweg P., Marée A. F. M., Scheres B. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6, e307. 10.1371/journal.pbio.0060307 (doi:10.1371/journal.pbio.0060307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukaki H., Nakao Y., Okushima Y., Theologis A., Tasaka M. 2005. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44, 382–395 10.1111/j.1365-313X.2005.02537.x (doi:10.1111/j.1365-313X.2005.02537.x) [DOI] [PubMed] [Google Scholar]
- 8.Vanneste S., et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17, 3035–3050 10.1105/tpc.105.035493 (doi:10.1105/tpc.105.035493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J. S., Jürgens G., Estelle M. 2005. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 10.1016/j.devcel.2005.05.014 (doi:10.1016/j.devcel.2005.05.014) [DOI] [PubMed] [Google Scholar]
- 10.Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 10.1016/S0092-8674(03)00924-3 (doi:10.1016/S0092-8674(03)00924-3) [DOI] [PubMed] [Google Scholar]
- 11.Bennett M. J., Marchant A., Green H. G., May S. T., Ward S. P., Millner P. A., Walker A. R., Schulz B., Feldmann K. A. 1996. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950 10.1126/science.273.5277.948 (doi:10.1126/science.273.5277.948) [DOI] [PubMed] [Google Scholar]
- 12.Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230 10.1126/science.282.5397.2226 (doi:10.1126/science.282.5397.2226) [DOI] [PubMed] [Google Scholar]
- 13.Luschnig C., Gaxiola R. A., Grisafi P., Fink G. R. 1998. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187 10.1126/science.282.5397.2226 (doi:10.1126/science.282.5397.2226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friml J., et al. 2002. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673 10.1016/S0092-8674(02)00656-6 (doi:10.1016/S0092-8674(02)00656-6) [DOI] [PubMed] [Google Scholar]
- 15.Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. 2002. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 10.1038/415806a (doi:10.1038/415806a) [DOI] [PubMed] [Google Scholar]
- 16.Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 10.1038/nature02085 (doi:10.1038/nature02085) [DOI] [PubMed] [Google Scholar]
- 17.Blakeslee J. J., et al. 2007. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19, 131–147 10.1105/tpc.106.040782 (doi:10.1105/tpc.106.040782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casimiro I., et al. 2001. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852 10.1105/tpc.13.4.843 (doi:10.1105/tpc.13.4.843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruegger M., Dewey E., Hobbie L., Brown D., Bernasconi P., Turner J., Muday G., Estelle M. 1997. Reduced naphthylphthalamic acid binding in the tir 3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9, 745–757 10.1105/tpc.9.5.745 (doi:10.1105/tpc.9.5.745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swarup K., et al. 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954 10.1038/ncb1754 (doi:10.1038/ncb1754) [DOI] [PubMed] [Google Scholar]
- 21.De Rybel B., et al. 2010. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706 10.1016/j.cub.2010.09.007 (doi:10.1016/j.cub.2010.09.007) [DOI] [PubMed] [Google Scholar]
- 22.De Smet I., et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl Acad. Sci. USA 107, 2705–2710 10.1073/pnas.0915001107 (doi:10.1073/pnas.0915001107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Smet I., et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134, 681–690 10.1242/dev.02753 (doi:10.1242/dev.02753) [DOI] [PubMed] [Google Scholar]
- 24.Moreno-Risueno M. A., Van Norman J. M., Moreno A., Zhang J., Ahnert S. E., Benfey P. N. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 10.1126/science.1191937 (doi:10.1126/science.1191937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucas M., Guédon Y., Jay-Allemand C., Godin C., Laplaze L. 2008. An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS One 3, e3673. 10.1371/journal.pone.0003673 (doi:10.1371/journal.pone.0003673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sponsel V. M., Schmidt F. W., Porter S. G., Nakayama M., Kohlstruk S., Estelle M. 1997. Characterization of new gibberellin-responsive semidwarf mutants of Arabidopsis. Plant Physiol. 115, 1009–1020 10.1104/pp.115.3.1009 (doi:10.1104/pp.115.3.1009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao F., Shen J., Brady S. R., Muday G. K., Asami T., Yang Z. 2004. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134, 1624–1631 10.1104/pp.103.036897 (doi:10.1104/pp.103.036897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Smet I., Zhang H., Inzé D., Beeckman T. 2006. A novel role for abscisic acid emerges from underground. Trends Plant Sci. 11, 434–439 10.1016/j.tplants.2006.07.003 (doi:10.1016/j.tplants.2006.07.003) [DOI] [PubMed] [Google Scholar]
- 29.Mouchel C. F., Osmont K. S., Hardtke C. S. 2006. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461 10.1038/nature05130 (doi:10.1038/nature05130) [DOI] [PubMed] [Google Scholar]
- 30.Li X., Mo X., Shou H., Wu P. 2006. Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47, 1112–1123 10.1093/pcp/pcj082 (doi:10.1093/pcp/pcj082) [DOI] [PubMed] [Google Scholar]
- 31.Laplaze L., et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19, 3889–3900 10.1105/tpc.107.055863 (doi:10.1105/tpc.107.055863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550 10.1105/tpc.014928 (doi:10.1105/tpc.014928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riefler M., Novak O., Strnad M., Schmülling T. 2006. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54 10.1105/tpc.105.037796 (doi:10.1105/tpc.105.037796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason M. G., Mathews D. E., Argyros D. A., Maxwell B. B., Kieber J. J., Alonso J. M., Ecker J. R., Schaller G. E. 2005. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17, 3007–3018 10.1105/tpc.105.035451 (doi:10.1105/tpc.105.035451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller B., Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097 10.1038/nature06943 (doi:10.1038/nature06943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Z., Andersen S. U., Ljung K., Dolezal K., Miotk A., Schultheiss S. J., Lohmann J. U. 2010. Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092 10.1038/nature09126 (doi:10.1038/nature09126) [DOI] [PubMed] [Google Scholar]
- 37.Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M. T., Aoyama T., Costantino P., Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384 10.1126/science.1164147 (doi:10.1126/science.1164147) [DOI] [PubMed] [Google Scholar]
- 38.Růžička K., Šimášková M., Duclercq J., Petrášek J., Zažimalová E., Simon S., Friml J., Van Montagu M. C. E., Benková E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl Acad. Sci. USA 106, 4284–4289 10.1073/pnas.0900060106 (doi:10.1073/pnas.0900060106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pernisová M., et al. 2009. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl Acad. Sci. USA 106, 3609–3614 10.1073/pnas.0811539106 (doi:10.1073/pnas.0811539106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W., To J. P. C., Cheng C.-Y., Schaller G. E., Kieber J. J. 2011. Type-A response regulators are required for proper root apical meristem function through the post-transcriptional regulation of PIN auxin efflux carriers. Plant J. 68, 1–10 10.1111/j.1365-313X.2011.04668.x (doi:10.1111/j.1365-313X.2011.04668.x) [DOI] [PubMed] [Google Scholar]
- 41.Yoshida S., Mandel T., Kuhlemeier C. 2011. Stem cell activation by light guides plant organogenesis. Genes Dev. 25, 1439–1450 10.1101/gad.631211 (doi:10.1101/gad.631211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marhavý P., et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 21, 796–804 10.1016/j.devcel.2011.08.014 (doi:10.1016/j.devcel.2011.08.014) [DOI] [PubMed] [Google Scholar]
- 43.Roman G., Lubarsky B., Kieber J. J., Rothenberg M., Ecker J. R. 1995. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139, 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higuchi M., et al. 2004. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl Acad. Sci. USA 101, 8821–8826 10.1073/pnas.0402887101 (doi:10.1073/pnas.0402887101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bevington P., Robinson K. 2002. Data reduction and error analysis for the physical sciences, 3rd edn. New York: McGraw Hill. [Google Scholar]
- 46.Himanen K., et al. 2004. Transcript profiling of early lateral root initiation. Proc. Natl Acad. Sci. USA 101, 5146–5151 10.1073/pnas.0308702101 (doi:10.1073/pnas.0308702101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.To J. P. C., Deruère J., Maxwell B. B., Morris V. F., Hutchison C. E., Ferreira F. J., Schaller G. E., Kieber J. J. 2007. Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19, 3901–3914 10.1105/tpc.107.052662 (doi:10.1105/tpc.107.052662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cary A. J., Liu W., Howell S. H. 1995. Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107, 1075–1082 10.1104/pp.107.4.1075 (doi:10.1104/pp.107.4.1075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang S. F., Hoffman N. E. 1984. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189 10.1146/annurev.pp.35.060184.001103 (doi:10.1146/annurev.pp.35.060184.001103) [DOI] [Google Scholar]
- 50.Lau O. S., Deng X. W. 2010. Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577 10.1016/j.pbi.2010.07.001 (doi:10.1016/j.pbi.2010.07.001) [DOI] [PubMed] [Google Scholar]
- 51.Chory J., Reinecke D., Sim S., Washburn T., Brenner M. 1994. A role for cytokinins in de-etiolation in Arabidopsis—det mutants have an altered response to cytokinins. Plant Physiol. 104, 339–347 10.1104/pp.104.2.339 (doi:10.1104/pp.104.2.339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J., et al. 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 10.1105/tpc.106.047688 (doi:10.1105/tpc.106.047688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hobbie L., Estelle M. 1994. Genetic approaches to auxin action. Plant Cell Environ. 17, 525–540 10.1111/j.1365-3040.1994.tb00147.x (doi:10.1111/j.1365-3040.1994.tb00147.x) [DOI] [PubMed] [Google Scholar]
- 54.Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063 10.1038/35059117 (doi:10.1038/35059117) [DOI] [PubMed] [Google Scholar]
- 55.Leyser O. 1997. Auxin: lessons from a mutant weed. Physiol. Plantarum 100, 407–414 10.1111/j.1399-3054 (doi:10.1111/j.1399-3054) [DOI] [Google Scholar]
- 56.Kakimoto T. 1996. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274, 982–985 10.1126/science.274.5289.982 (doi:10.1126/science.274.5289.982) [DOI] [PubMed] [Google Scholar]
- 57.Fukaki H., Tasaka M. 2009. Hormone interactions during lateral root formation. Plant Mol. Biol. 69, 437–449 10.1007/s11103-008-9417-2 (doi:10.1007/s11103-008-9417-2) [DOI] [PubMed] [Google Scholar]
- 58.Müller D., Leyser O. 2011. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 107, 1203–1212 10.1093/aob/mcr069 (doi:10.1093/aob/mcr069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su Y. H., Liu Y. B., Zhang X. S. 2011. Auxin–cytokinin interaction regulates meristem development. Mol. Plant 4, 616–625 10.1093/mp/ssr007 (doi:10.1093/mp/ssr007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pernisová M., Kuderová A., Hejátko J. 2011. Cytokinin and auxin interactions in plant development: metabolism, signalling, transport and gene expression. Curr. Protein Pept. Sci. 12, 137–147 10.2174/138920311795684887 (doi:10.2174/138920311795684887) [DOI] [PubMed] [Google Scholar]
- 61.Bishopp A., Benková E., Helariutta Y. 2011. Sending mixed messages: auxin–cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 14, 10–16 10.1016/j.pbi.2010.08.014 (doi:10.1016/j.pbi.2010.08.014) [DOI] [PubMed] [Google Scholar]
- 62.Alabadi D., Blazquez M. A. 2008. Integration of light and hormone signals. Plant Signal Behav. 3, 448–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinhardt D., Pesce E. R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260 10.1038/nature02081 (doi:10.1038/nature02081) [DOI] [PubMed] [Google Scholar]