Abstract
The growth of a plant's root system depends on the continued activity of the root meristem, and the generation of new meristems when lateral roots are initiated. Plants have developed intricate signalling systems that employ secreted peptides and plasma membrane-localized receptor kinases for short- and long-range communication. Studies on growth of the vascular system, the generation of lateral roots, the control of cell differentiation in the root meristem and the interaction with invading pathogens or symbionts has unravelled a network of peptides and receptor systems with occasionally shared functions. A common theme is the employment of conserved modules, consisting of a short signalling peptide, a receptor-like kinase and a target transcription factor, that control the fate and proliferation of stem cells during root development. This review intends to give an overview of the recent advances in receptor and peptide ligand-mediated signalling involved in root development.
Keywords: plant, root, meristem, signalling, receptors, peptides
1. Cell communication in root meristems
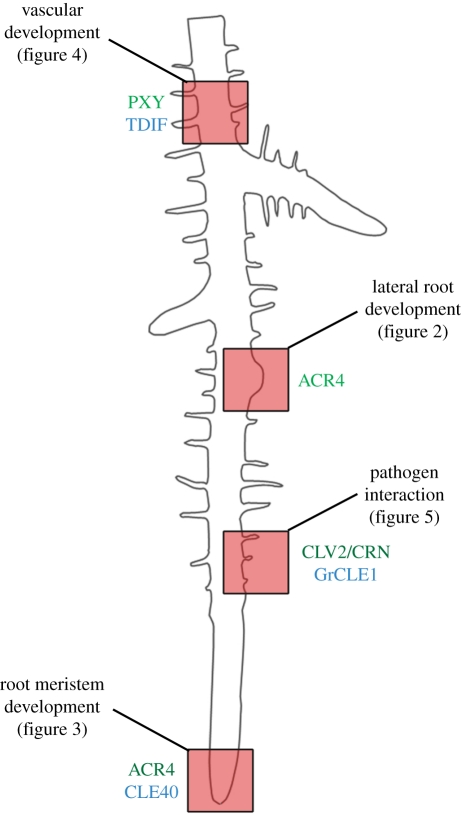
Plant meristems are the sites that generate the majority of the plant body by continuously initiating the formation of new organs. The primary meristems are formed during embryogenesis at opposite ends of the embryo. New meristems will be initiated at later stages of development, for example, when flower meristems are formed that produce all floral organs. In many grasses, the primary root is active only for a short period of time, and the majority of the expanded root system is generated from lateral roots. The highly ordered cell arrangement in these root meristems and the generation of very discrete cell files by specific stem cells, together with the observation of a quiescent centre as a cell group that regulates stem cell behaviour in its immediate vicinity, have stimulated the search for molecules that mediate intercellular communication to coordinate cell division and differentiation patterns in the root meristem (figures 1–3). Furthermore, secondary growth in width requires the activity of a cambium meristem that gives rise to xylem and phloem in a coordinated manner (figures 1 and 4). Roots are also contacted by a wide spectrum of micro-organisms that can act as pathogens or undergo close interactions for mutual benefit. Root nodule formation by Leguminosae is triggered by nitrogen-fixing bacteria, and parasitic nematodes induce the growth of root cysts or knots (figures 1 and 5). Communication with these organisms occurs via evolutionarily conserved signalling pathways that were readapted for the specific type of interaction. Research in recent years has elucidated how peptides and their corresponding receptor systems mediate the extensive communication required for an organized growth and development of the root, but also for the appropriate responses to other organisms (figure 1). Peptides belonging to the so-called CLV3/EMBRYO SURROUNDING REGION (CLE) family act as signals in several of these pathways, which are in most cases recognized by leucine-rich-repeat (LRR) receptor-like kinases. The paradigmatic prototype for such signalling cascades is the CLAVATA pathway, which, although primarily understood from analysis of the shoot system, will be discussed first. Importantly, many of the key players operating in the shoot stem cell systems were later found to control similar cell fate decisions in the root.
Figure 1.
Schematic of an Arabidopsis root. Different developmental processes and pathogen interactions are highlighted by red boxes and will be explained in more detail in the respective figures. Involved peptide ligands (blue) and receptors (green) are indicated.
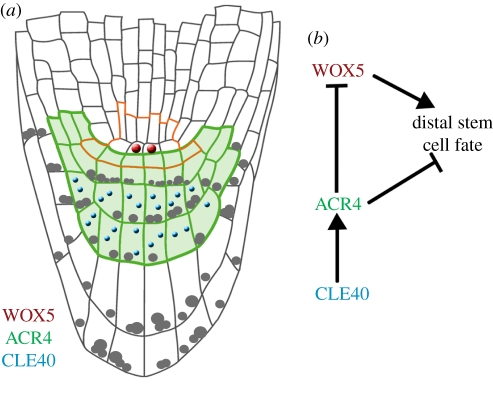
Figure 3.
Root meristem development. (a) Schematic of the Arabidopsis root meristem. Colours represent the localization of the homeodomain transcription factor WOX5 (red) in the quiescent centre, the receptor kinase ACR4 (green) in columella stem cells and columella cells and the peptide ligand CLE40 (blue) in columella cells. The stem cell niche is outlined in orange. (b) Conceptual model of distal stem cell fate regulation in the root meristem. Arrows indicate positive regulation and barred arrows indicate negative regulation.
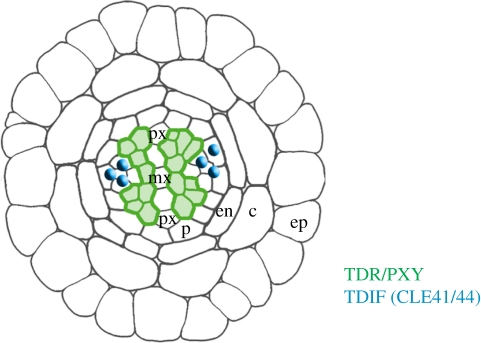
Figure 4.
Vascular development. Schematic of Arabidopsis root vasculature. Colours indicate the localization of the receptor TDR/PXY (green) in the phloem and the peptide ligand TDIF (CLE41/44) (blue) in the procambium. Individual cell layers are labelled: mx, metaxylem; px, protoxylem; p, pericycle; en, endodermis; c, cortex; ep, epidermis.
Figure 5.
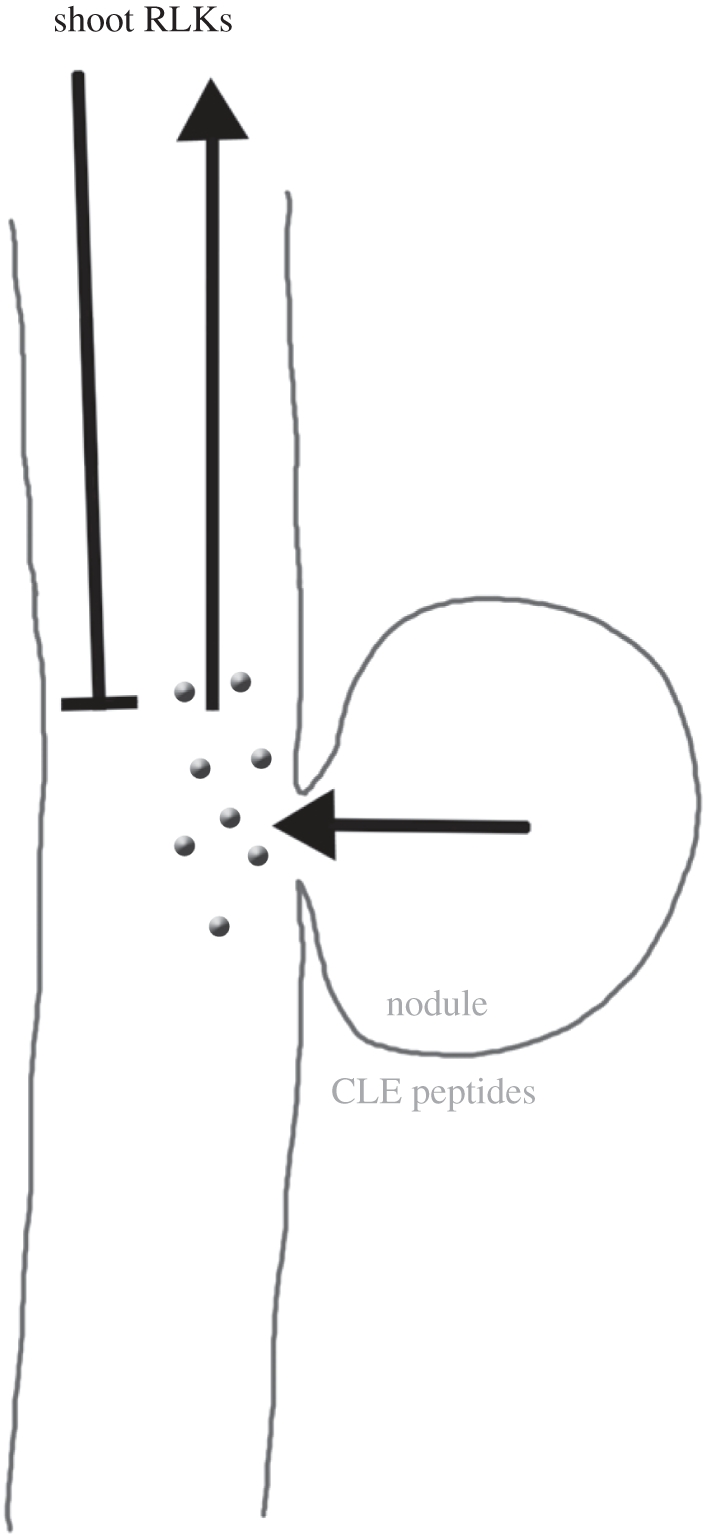
Autoregulation of nodulation. Schematic of nodule/root signalling. Nodule formation induces CLE peptide production, which act as long-range signals that are perceived by receptor-like kinases (RLKs) in the shoot. This triggers the generation of a secondary signal (unknown) that restricts nodule initiation.
2. The CLAVATA pathway as a paradigm for intercellular communication
Genetic screens served to identify the first genes that contribute to cell communication pathways. Ease of access and the sometimes drastic developmental phenotypes of mutants have allowed isolation of genes encoding receptor kinases and signalling peptides that control the stem cell populations of shoot and floral meristems. Similar genes and functions were later discovered to play a role in root meristem development. However, because our knowledge on peptide signalling pathways is more detailed for the shoot system, we will first discuss the CLAVATA signalling pathway acting in the above-ground parts of the plant as a paradigm. The CLAVATA1 (CLV1) gene of Arabidopsis encodes a receptor protein composed of an extracellular domain with 21 LRR domains and a cytoplasmic kinase domain of the serine–threonine type, connected by a single-pass transmembrane domain (TMD) [1,2]. CLV1 is expressed in a central domain of shoot and floral meristems, where it serves to restrict the activity of the homeodomain transcription factor WUSCHEL (WUS) [3]. WUS functions are required to maintain an active and non-differentiating stem cell population at the tip of the meristem. Here, stem cells are located that divide rarely, but provide new transit-amplifying (TA) cells for organ initiation and further growth in the peripheral regions of the meristem. If WUS activity fails, stem cells differentiate prematurely, and the entire meristem ceases to generate new plant organs, such as leaves or floral structures [3–5]. However, loss of function mutants of CLV1 allow higher levels of WUS expression, and also expansion of the WUS expression domain towards the meristem tip [5]. This increased WUS activity inhibits the release of cells from stem cell fate, and also reactivates stem cell-specific gene expression profiles in TA cells [6]. In consequence, the stem cell zone of the meristem enlarges and extra floral organs are generated [5]. clv-mutant flowers generate only club-shaped (clavata in latin) siliques, hence the name. Initial genetic screens had identified three different CLAVATA loci (CLV1, CLV2 and CLV3) [1,7,8]. CLV3 is a founding member of the CLE gene family. The CLV3 gene is expressed exclusively in stem cells of the aboveground meristems [9,10], and acts as a secreted signalling peptide. All predicted CLE peptides share a conserved C-terminal motif, which forms the core of the active peptide [11,12]. Overexpression of CLE genes in transgenic plants grown in liquid cultures and purification of extracellular peptides from the medium allowed determination of the structure of the active peptide [13,14]. For all cases shown so far, CLE peptides are processed from larger precursors by proteolytic digestion [15,16]. One or several of the prolines residing in the core CLE-motif are hydroxylated and can carry arabinofuranose sugars [13,17–19]. Such peptide glyosylation could serve to protect from proteolysis in the extracellular space, or could change the binding interface with their corresponding receptors. The arabinosylated form of CLV3 was indeed found to bind with a higher affinity to the extracellular domain of CLV1. Furthermore, the glycosylated CLV3 and CLE2 peptides were capable of fully rescuing the shoot meristem phenotype of a clv3 mutant upon addition to the medium [13], while the non-glycosylated peptides only partially restored shoot stem cell control when employed at the same concentrations. This study strongly indicates that sugar modifications of CLE peptides can be assigned a major role in determining signalling activity.
Secretion of CLV3 from stem cells and the resulting activation of the CLV1 receptor provide a strong feedback signal for stem cell homeostasis. Any increase in stem cell number causes increased CLV3/CLV1 signalling; the resulting repression of WUS expression would immediately cause a corresponding decrease in stem cell number, allow differentiation of stem cells in the outer stem cell domain, and thereby decrease CLV3 signalling again [5,9]. Several mathematical models have meanwhile simulated this feedback system in operation, and shown that it can serve to maintain a stable and long-lasting stem cell population in meristems [20–22].
The activity of the CLV3/CLV1 system described here so far is restricted to shoot and floral meristems, and prompted the search for other peptide–receptor pairs that modulate stem cell maintenance or cellular differentiation in other parts of the plant. Additional genetic screens by analysing mutants with clv1-like phenotypes identified CLV2, encoding a receptor-like protein that, in contrast to CLV1, lacks the cytoplasmic serine–threonine kinase domain [8,23]. Because mutations in CLV2 enhance mutations in CLV1, two parallel acting pathways must exist that transmit the CLV3 signal [24]. Consistent with this, both CLV1 and CLV2 proteins were shown to bind CLV3 [13,19,25].
3. A CLV-like pathway acting in the root
A first role for CLV2 in root development was discovered when Arabidopsis seedlings were grown on medium containing micromolar concentrations of CLE peptides [26–28], or in transgenic plants overexpressing CLE genes. In both cases, root growth arrested and differentiation of root meristem cells was observed, suggesting that CLE peptides can act similarly to CLV3 in the shoot and restrict stem cell maintenance. Mutant screens based on this CLE overexpression phenotype ended with the isolation of two novel suppressor mutations, sol1 and sol2 (for SUPPRESSOR of LLP1/CLE19 overexpression). The SOL1 gene encodes a Zn2+-carboxypeptidase that is proposed to process CLE peptide precursors [28]. SOL2 was also identified as a suppressor of CLV3 overexpression in the shoot, and, for its phenotypic resemblance to clv-mutants, termed CORYNE (CRN; greek for club-shaped). Interestingly, neither clv2 nor crn/sol2 mutants displayed a root phenotype that was distinct from wild-type in the absence of increased CLE peptide availability. Cloning of CRN/SOL2 uncovered that it encodes a kinase that carries a TMD and a short extracellular sequence [24,29]. This predicted serine–threonine kinase domain of CRN may be non-functional, because kinase assays have been unsuccessful so far, and at least partial complementation of a crn mutant was achieved with an engineered mutant kinase version predicted to lack any kinase activity [30,31]. The mutant phenotypes of crn/sol2 and clv2 are indistinguishable, and double-mutant combinations revealed no additional phenotypic alterations [24,32]. Together with further genetic studies, this indicated that CLV1 and CLV2 together with CRN/SOL2 act in two parallel pathways that control stem cell maintenance in the shoot. In root tissues, a requirement for CRN/SOL2 and CLV2 was only shown when CLE peptides were added, or when CLE genes were overexpressed in transgenic plants because the mutants do not show a growth arrest after CLE peptide treatment [24,27,29].
Molecularly, CRN and CLV2 can interact via their TMDs and adjacent juxtamembrane amino acids [32,33]. Using fluorescence resonance energy transfer between receptor protein fusions, it was shown that coexpression and interaction between CRN and CLV2 are required to localize both proteins at the plasma membrane. If expressed individually, CRN or CLV2 will be retained in the endoplasmic reticulum [32]. Thus, the main role for the kinase-defective CRN protein may lie in its ability to guide CLV2 receptor to the plasma membrane. Signal transmission to the nucleus could then be mediated by additional membrane-associated kinases that interact with CLV2 through CRN.
In addition to SOL1, CRN and CLV2, the receptor-like kinase RPK2 (RECEPTOR-LIKE KINASE 2 or TOADSTOOL2) was shown to be essential for transmitting CLE signals in the root and shoot [34,35]. Genetically, rpk2 mutants appear to be additive with clv1 and crn or clv2 mutants in shoot development, which suggests that RPK2 establishes a third independent signalling pathway for the perception of CLE peptides. Similar to CLV2 and CRN, RPK2 has multiple functions during plant development, and is required from embryogenesis to cell fate specification during pollen development [35,36]. In root development, rpk2 mutants were found to be fully epistatic to the root growth restricting activity of all CLE peptides tested. RPK2 is expressed in the root meristem, with a lower expression level in the columella domain. Interestingly, RPK2 could be dosage limited, because overexpression of RPK2 induced CLE overexpression phenotypes and corresponding meristem growth arrest in both shoot and root system [34].
4. The receptor kinase ARABIDOPSIS CRINKLY 4 controls lateral root formation and stem cell maintenance
Previously, receptor-like kinases (RLKs) were invoked in peptide signalling in the root owing to CLE peptide misexpression phenotypes in roots [26,28,37]. However, loss-of-function phenotypes were not observed for the receptors CLV2, CRN or RPK2. Studies on lateral root development and the distal root meristem revealed a role for ARABIDOPSIS CRINKLY 4 (ACR4), which was previously identified as an RLK that coordinates layer integrity in the epidermis and protoderm [38–41].
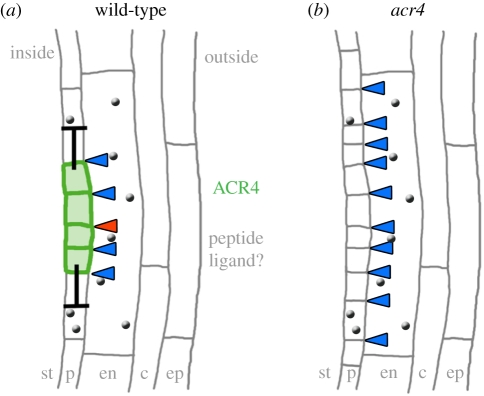
The first indication of lateral root initiation is two asymmetric divisions of pericycle cells, which occur opposite xylem poles. Four small cells are generated that undergo further periclinal divisions to generate a small mound of cells, the lateral root primordium. Detailed and high-resolution transcriptome analysis identified ACR4 as a candidate regulator that is specifically expressed in the two small daughter cells after the first asymmetric pericycle cell divisions [41]. In acr4 mutants, new lateral roots are initiated with a tighter spacing, and sometimes immediately adjacent to each other. This suggests that ACR4 is required to suppress lateral root initiation by lateral inhibition (figure 2). A peptide activating ACR4 during this process has not yet been identified. However, acr4 mutants are also affected in the maintenance of the root stem cell niche. In Arabidopsis, four cells of the quiescent centre (QC) are surrounded by stem cells that give rise to specific cell types (figure 3). Abutting the QC on the distal side is a single layer of columella stem cells (CSCs), which divide to generate several layers of starch accumulating columella cells (CCs). Differentiation of CSC daughter cells is delayed in acr4 mutants, so that roots maintain more stem CSC layers. The mutant phenotype allows linkage of ACR4 function with that of CLE40, a peptide of the CLE family that is most closely related to the shoot stem cell restricting CLV3 peptide [37,42]. CLE40 is expressed in CCs and can thus provide a feedback signal that regulates CSC activity and proliferation. ACR4 is normally expressed in CSCs and CCs, and is rapidly transcriptionally upregulated upon treatment of roots with high levels of CLE40 peptide, suggesting that ACR4 expression is itself regulated by ACR4 signalling. Although speculative, a model can be envisaged whereby upregulation of ACR4 serves to bind and sequester excessive CLE40 ligand [43]. Increased CLE40 expression results in a premature differentiation of CSCs into CCs, indicating that the CLE40 dosage must be tightly controlled for normal root meristem function. Because all described effects of CLE40 depend on ACR4 function, it is possible that CLE40 peptide binds and directly interacts with ACR4.
Figure 2.
Lateral root development. Schematic of lateral root initiation. (a) Wild-type of stage I lateral root. Colours represent the localization of the receptor kinase ACR4 (green) in the dividing pericycle cells and putative peptide ligands (grey). Barred arrows indicate an ACR4-mediated negative regulation of cell divisions. Arrowheads indicate cell divisions. (b) acr4 mutant stage I lateral root showing supernumerary cell divisions. Individual cell layers are labelled: st, stele; p, pericycle; en, endodermis; c, cortex; ep, epidermis.
The parallels between the shoot and root stem cell signalling pathways are extensive and include target genes regulated by CLE peptides. In the root meristem, a close homologue of WUS, the homeodomain transcription factor WUSCHEL-LIKE HOMEOBOX 5 (WOX5) [44], is expressed in the QC cells and serves to maintain the identity of the surrounding stem cells. In cle40 or acr4 mutants, the expression domain of WOX5 expands, while it is shifted to a more proximal location (and therefore repressed in the QC domain) when CLE40/ACR4 signalling is strongly activated [42].
5. Root growth factors regulate auxin signalling in root meristems
Peptides such as those of the CLE family are post-translationally modified by proteolytical processing and by coupling of sugar residues to their hydroxyprolyl groups. Other peptides that pass through the secretory pathway receive a sulphate group on the hydroxyl group of tyrosine, mediated by the golgi-resident enzyme tyrosylprotein sulphotransferase (TPST). TPST is encoded by a single-copy gene in Arabidopsis that is positively regulated by auxin, and knock-out mutants (tpst-1) display pleiotropic phenotypes, including short roots, extra QC cells and differentiation of CSCs [45,46]. At least some of these developmental defects are due to reduced expression of transcription factors of the PLETHORA (PLT) family. The PLT genes respond to auxin presence in the root meristem, and are required to maintain the expression of PIN auxin efflux carriers and normal growth patterns in the root meristem [47,48]. Searches for putative short peptides that carry a secretion signal and a predicted tyrosine-sulphation motif led to the identification of the ROOT MERISTEM GROWTH FACTOR (RGF) peptide family [46]. When tpst-1 mutant roots were grown in the presence of RGF1 peptide, root meristem architecture was partially restored and root meristem length increased, together with expanded PLT expression. Thus, PLT genes appear to be regulated by an RGF-dependent signalling pathway. However, PLT transcript levels were largely unaltered in tpst-1 mutants or upon RGF addition, suggesting that a RGF pathway could control PLT translation or stability. Obviously, RGF peptides are not the only peptides requiring sulphation for activity, and RGF1 showed synergistic effects upon addition of the 5-amino acid peptide hormone phytosulphokine (PSK) and PSY1 (PLANT PEPTIDE CONTAINING SULFATED TYROSINE1), a tyrosine-sulphated 18-amino acid glycopeptide [49]. PSK and PSY1 promote cell division and cell expansion. For their biological activity, the sulphate group and the l-arabinose side chain (for PSY1) are essential. Specific LRR-RKs that bind to and are activated by PSK or PSY1, respectively, were identified previously [49,50]. But a receptor protein binding to and activated by RGFs still awaits identification. Another class of small, secreted, 15 amino acid-long hydroxyprolinated peptides, the CEP (C-TERMINALLY ENCODED PEPTIDE) peptides, has also been suggested to play a role in root development. Overexpression of CEP1 and treatment with synthetic CEP1 results in root meristem arrest. Most of the CEP family members are expressed in the root (CEP1 is expressed in lateral root primordia), but so far no functional genetic analyses have been described [14].
6. CLE peptides control cell fates in root vascular meristems
The stem cells of the vascular system are found in the procambium, which are highly polarized strands that generate phloem and xylem cells. TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF), a CLE peptide first isolated from a Zinnia cell culture system that allows in vitro tracheary element differentiation, was identified owing to its name-giving characteristics. The Arabidopsis orthologues are CLE41/CLE44, which are normally expressed in the phloem [18,51] (figure 4). Addition of CLE41/44 to liquid-cultured seedlings caused an overproliferation of phloem cells [51]. Procambium cells express PHLOEM INTERCALATED WITH XYLEM (PXY), also known as TDR (TDIF receptor), encoding a CLV1-related receptor-like kinase [52], which was shown to bind CLE41 via its extracellular LRR domains [51]. pxy mutants fail to maintain the typical order of phloem cells (outside of the procambium), separated from xylem cells (inside). A cle41 mutant formed a thinner stele than wild-type plants [53], whereas increased expression of CLE41 from the phloem caused procambium proliferation, albeit with normal vasculature organization [54]. Misexpression of CLE41 from other cell types disrupted normal tissue organization, indicating that not the absolute concentration of the CLE peptide, but rather its concentration gradient controls tissue organization and cellular identities. A target gene regulated by the TDIF/PXY module is WOX4, another member of the WOX gene family [53]. Expression of WOX4 in the procambium serves to maintain this stem cell system, a function that appears to be shared among many WOX genes studied so far. However, in contrast to WUS and WOX5 regulation, procambial WOX4 expression is under positive control by the TDIF/PXY system. Detailed transcriptome analysis of interfascicular cambium that was isolated via laser capture microdissection uncovered two additional LRR-RLKs, REDUCED IN LATERAL GROWTH (RUL1) and MORE LATERAL GROWTH (MOL1), that antagonistically control cambial growth [55]. MOL1 functions as a repressor of both WOX4 and RUL1, but phenotypes of mol1 rul1 double-mutants resemble wild-type, indicating that vascular development is subject to multiple, and possibly parallel acting ligand-triggered RLK pathways.
The two pathways controlled by TDIF in the vasculature and CLV3 in the shoot display extensive similarities. In both cases, CLE peptides act via LRR-RLKs to control the expression levels and domains of closely related WOX-family homeodomain transcription factors. However, no functional overlaps or receptor cross-activation by these peptides has been observed. A series of alanine-substituted CLV3 and TDIF peptides allowed mapping of the specificity determinants for binding to CLV1 and root growth restriction to N- and C-terminal amino acids [56]. In addition, the proline residue at position 7, which is glycosylated in CLV3 and drastically increases affinity to CLV1, is not required for root growth inhibition, suggesting mechanistic differences between the interaction of CLE peptides with CLV1 or the unidentified root receptor.
7. Communication with root pathogens and symbionts via peptides
A plant's root system represents the primary entry point for a number of pathogens, including parasitic nematodes. These nematodes fall into two major classes, which induce the formation of either root knots or cysts by the plant tissue that act as feeding sites, providing both nutrients and a housing for the proliferating nematodes. Their generation is induced by signalling molecules that the invading nematodes inject into the plant tissues. CLE peptides isolated from the oesophageal glands of the soybean cyst nematode Heterodera glycines could functionally rescue a clv3 mutant, if expressed from the CLV3 promoter sequences, suggesting that the parasite communicates with the plant cells by activation of receptor kinase signalling pathways [57]. In line with this, Arabidopsis mutants for CLV2 or CRN were found to be less susceptible to parasitic nematodes [58,59]. Genes encoding CLE peptides were previously thought to be plant-specific, but parasitic nematodes provide the notable exception. Their predicted CLE peptides lack an N-terminal secretion signal, and are therefore expected to be found in the cytoplasm of the cell upon injection. Interaction with the extracellular domain of receptor proteins such as CLV2 is only possible if the peptides become secreted. Interestingly, the nematode CLE peptides carry a variable domain N-terminal to the CLE-motif, which directs the peptide towards the apoplast via a so far unknown secretion pathway [60,61]. GrCLE1, a CLE peptide from the nematode Globodera rostochiensis, is processed from a larger precursor in the plant cell, and binds to the extracellular domains of CLV2 and the CLV1-related RLKs BAM1 and BAM2 [58].
Nodules are formed by the root systems of Leguminosae to associate with symbiotic nitrogen-fixing bacteria. The number of nodules is controlled by the host legume in response to external cues, such as soil nitrate levels, but also through negative feedback regulation, termed autoregulation of nodulation (AON; figure 5). AON appears to be controlled by a signal generated in nodules that is transmitted to the shoot. There, a second signal is generated that suppresses the formation of further nodules. The molecular similarities between peptide-dependent stem cell signalling in the shoot system and AON were first described for soybean (Glycine max), where mutations in the CLV1-homologue GmNARK, which is preferentially expressed in leaves, were shown to induce supernodulation and the production of more lateral roots [62]. In Medicago sativa, the CLE peptides MtCLE12 and 13 [63] are expressed in root nodules and serve to inhibit nodulation. In Lotus japonicus, the CLE peptides LjCLE-Root Signal1 (LjCLE-RS1) and RS2 are upregulated by nitrate addition, and are perceived in the shoot by the receptor-like kinase HYPERNODULATION ABERRANT ROOT FORMATION 1 (HAR1) [64,65], a homologue to Arabidopsis CLV1. KLAVIER, a Lotus homologue of the Arabidopsis RPK2 receptor kinase that physically interacts with HAR1, is also required for AON [66]. In Pisum sativum, the CLV2 homologue PsCLV2 is inactivated in sym28 mutants, resulting in a typical clv2-like shoot phenotype, with altered phyllotaxis and stem fasciation, but also in failure of AON and hypernodulation [67]. The common theme here is that CLE-type peptides serve as the primary root-to-shoot signal, which is perceived by receptor complex(es) in the shoot. The nature of the secondary, nodule-repressing signal that communicates information from the shoot to the root is not yet known.
8. Conclusions
Several pathways for intercellular communication via secreted peptides have been unravelled in recent years. In many cases, the discovery of these pathways was driven by bioinformatic approaches, followed by detailed studies on predicted peptides, and the pairing through biochemical and genetic approaches with their corresponding receptors. Many of the peptides investigated so far are processed from large precursor molecules and further modified by addition of sugar residues or sulphate groups. These post-translational processing steps and modifications often change the specificity of the peptide or its overall activity, and need to be considered when studies on peptide functions are based on the addition of synthetic peptides. Determining the genome sequences of several plant species has certainly facilitated the prediction of putative peptide-encoding genes, but we have not yet gained a complete picture on the spectrum of small peptides that plant cells use for their communication.
References
- 1.Clark S. E., Running M. P., Meyerowitz E. M. 1993. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418 [DOI] [PubMed] [Google Scholar]
- 2.Clark S. E., Williams R. W., Meyerowitz E. M. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585 10.1016/S0092-8674(00)80239-1 (doi:10.1016/S0092-8674(00)80239-1) [DOI] [PubMed] [Google Scholar]
- 3.Laux T., Mayer K. F., Berger J., Jürgens G. 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96 [DOI] [PubMed] [Google Scholar]
- 4.Mayer K. F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 10.1016/S0092-8674(00)81703-1 (doi:10.1016/S0092-8674(00)81703-1) [DOI] [PubMed] [Google Scholar]
- 5.Schoof H., Lenhard M., Haecker A., Mayer K. F., Jürgens G., Laux T. 2000. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 10.1016/S0092-8674(00)80700-X (doi:10.1016/S0092-8674(00)80700-X) [DOI] [PubMed] [Google Scholar]
- 6.Reddy G. V., Meyerowitz E. M. 2005. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310, 663–667 10.1126/science.1116261 (doi:10.1126/science.1116261) [DOI] [PubMed] [Google Scholar]
- 7.Clark S. E., Running M. P., Meyerowitz E. M. 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067 [Google Scholar]
- 8.Kayes J. M., Clark S. E. 1998. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851 [DOI] [PubMed] [Google Scholar]
- 9.Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M., Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 10.1126/science.289.5479.617 (doi:10.1126/science.289.5479.617) [DOI] [PubMed] [Google Scholar]
- 10.Fletcher J. C., Brand U., Running M. P., Simon R., Meyerowitz E. M. 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914 10.1126/science.283.5409.1911 (doi:10.1126/science.283.5409.1911) [DOI] [PubMed] [Google Scholar]
- 11.Cock J. M., McCormick S. 2001. A large family of genes that share homology with CLAVATA3. Plant Physiol. 126, 939–942 10.1104/pp.126.3.939 (doi:10.1104/pp.126.3.939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oelkers K., Goffard N., Weiller G. F., Gresshoff P. M., Mathesius U., Frickey T. 2008. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8, 1. 10.1186/1471-2229-8-1 (doi:10.1186/1471-2229-8-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578–580 10.1038/nchembio.182 (doi:10.1038/nchembio.182) [DOI] [PubMed] [Google Scholar]
- 14.Ohyama K., Ogawa M., Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55, 152–160 10.1111/j.1365-313X.2008.03464.x (doi:10.1111/j.1365-313X.2008.03464.x) [DOI] [PubMed] [Google Scholar]
- 15.Ni J., Clark S. E. 2006. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140, 726–733 10.1104/pp.105.072678 (doi:10.1104/pp.105.072678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni J., Guo Y., Jin H., Hartsell J., Clark S. E. 2011. Characterization of a CLE processing activity. Plant Mol. Biol. 75, 67–75 10.1007/s11103-010-9708-2 (doi:10.1007/s11103-010-9708-2) [DOI] [PubMed] [Google Scholar]
- 17.Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848 10.1126/science.1128439 (doi:10.1126/science.1128439) [DOI] [PubMed] [Google Scholar]
- 18.Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. 2006. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845 10.1126/science.1128436 (doi:10.1126/science.1128436) [DOI] [PubMed] [Google Scholar]
- 19.Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294. 10.1126/science.1150083 (doi:10.1126/science.1150083) [DOI] [PubMed] [Google Scholar]
- 20.Geier F., Lohmann J. U., Gerstung M., Maier A. T., Timmer J., Fleck C. 2008. A quantitative and dynamic model for plant stem cell regulation. PLoS ONE 3, e3553. 10.1371/journal.pone.0003553 (doi:10.1371/journal.pone.0003553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohm T., Zitzler E., Simon R. 2010. A dynamic model for stem cell homeostasis and patterning in Arabidopsis meristems. PLoS ONE 5, e9189. 10.1371/journal.pone.0009189 (doi:10.1371/journal.pone.0009189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonsson H., Heisler M., Reddy G. V., Agrawal V., Gor V., Shapiro B. E., Mjolsness E., Meyerowitz E. M. 2005. Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics. 21(Suppl 1), i232–240 10.1093/bioinformatics/bti1036 (doi:10.1093/bioinformatics/bti1036) [DOI] [PubMed] [Google Scholar]
- 23.Jeong S., Trotochaud A. E., Clark S. E. 1999. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934 10.1105/tpc.11.10.1925 (doi:10.1105/tpc.11.10.1925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller R., Bleckmann A., Simon R. 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946 10.1105/tpc.107.057547 (doi:10.1105/tpc.107.057547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y., Han L., Hymes M., Denver R., Clark S. E. 2010. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63, 889–900 10.1111/j.1365-313X.2010.04295.x (doi:10.1111/j.1365-313X.2010.04295.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiers M., Hause G., Boutilier K., Casamitjana-Martinez E., Weijers D., Offringa R., van der Geest L., van Lookeren Campagne M., Liu C. M. 2004. Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327, 37–49 10.1016/j.gene.2003.11.014 (doi:10.1016/j.gene.2003.11.014) [DOI] [PubMed] [Google Scholar]
- 27.Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., Liu C. M. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17, 2542–2553 10.1105/tpc.105.034009 (doi:10.1105/tpc.105.034009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casamitjana-Martinez E., Hofhuis H. F., Xu J., Liu C. M., Heidstra R., Scheres B. 2003. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13, 1435–1441 10.1016/S0960-9822(03)00533-5 (doi:10.1016/S0960-9822(03)00533-5) [DOI] [PubMed] [Google Scholar]
- 29.Miwa H., Betsuyaku S., Iwamoto K., Kinoshita A., Fukuda H., Sawa S. 2008. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 49, 1752–1757 10.1093/pcp/pcn148 (doi:10.1093/pcp/pcn148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nimchuk Z. L., Tarr P. T., Ohno C., Qu X., Meyerowitz E. M. 2011. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell 23, 851–854 10.1105/tpc.110.075622 (doi:10.1105/tpc.110.075622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betsuyaku S., Takahashi F., Kinoshita A., Miwa H., Shinozaki K., Fukuda H., Sawa S. 2011. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 52, 14–29 10.1093/pcp/pcq157 (doi:10.1093/pcp/pcq157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bleckmann A., Weidtkamp-Peters S., Seidel C. A. M., Simon R. 2010. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176 10.1104/pp.109.149930 (doi:10.1104/pp.109.149930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y., Wang Y., Li R., Song X., Wang Q., Huang S., Jin J. B., Liu C. M., Lin J. 2010. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 61, 223–233 10.1111/j.1365-313X.2009.04049.x (doi:10.1111/j.1365-313X.2009.04049.x) [DOI] [PubMed] [Google Scholar]
- 34.Kinoshita A., et al. 2010. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920 10.1242/dev.048199 (doi:10.1242/dev.048199) [DOI] [PubMed] [Google Scholar]
- 35.Nodine M. D., Yadegari R., Tax F. E. 2007. RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev. Cell 12, 943–956 10.1016/j.devcel.2007.04.003 (doi:10.1016/j.devcel.2007.04.003) [DOI] [PubMed] [Google Scholar]
- 36.Mizuno S., Osakabe Y., Maruyama K., Ito T., Osakabe K., Sato T., Shinozaki K., Yamaguchi-Shinozaki K. 2007. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 50, 751–766 10.1111/j.1365-313X.2007.03083.x (doi:10.1111/j.1365-313X.2007.03083.x) [DOI] [PubMed] [Google Scholar]
- 37.Hobe M., Müller R., Grünewald M., Brand U., Simon R. 2003. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 213, 371–381 10.1007/s00427-003-0329-5 (doi:10.1007/s00427-003-0329-5) [DOI] [PubMed] [Google Scholar]
- 38.Gifford M. L., Dean S., Ingram G. C. 2003. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 130, 4249–4258 10.1242/dev.00634 (doi:10.1242/dev.00634) [DOI] [PubMed] [Google Scholar]
- 39.Tanaka H., Watanabe M., Watanabe D., Tanaka T., Machida C., Machida Y. 2002. ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 43, 419–428 10.1093/pcp/pcf052 (doi:10.1093/pcp/pcf052) [DOI] [PubMed] [Google Scholar]
- 40.Watanabe M., Tanaka H., Watanabe D., Machida C., Machida Y. 2004. The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 39, 298–308 10.1111/j.1365-313X.2004.02132.x (doi:10.1111/j.1365-313X.2004.02132.x) [DOI] [PubMed] [Google Scholar]
- 41.De Smet I., et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597 10.1126/science.1160158 (doi:10.1126/science.1160158) [DOI] [PubMed] [Google Scholar]
- 42.Stahl Y., Wink R. H., Ingram G. C., Simon R. 2009. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19, 909–914 10.1016/j.cub.2009.03.060 (doi:10.1016/j.cub.2009.03.060) [DOI] [PubMed] [Google Scholar]
- 43.Stahl Y., Simon R. 2009. Is the Arabidopsis root niche protected by sequestration of the CLE40 signal by its putative receptor ACR4? Plant Signal Behav. 4, 634–635 10.4161/psb.4.7.8970 (doi:10.4161/psb.4.7.8970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814 10.1038/nature05703 (doi:10.1038/nature05703) [DOI] [PubMed] [Google Scholar]
- 45.Zhou W., et al. 2010. Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22, 3692–3709 10.1105/tpc.110.075721 (doi:10.1105/tpc.110.075721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067 10.1126/science.1191132 (doi:10.1126/science.1191132) [DOI] [PubMed] [Google Scholar]
- 47.Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 10.1038/nature06206 (doi:10.1038/nature06206) [DOI] [PubMed] [Google Scholar]
- 48.Aida M., et al. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 10.1016/j.cell.2004.09.018 (doi:10.1016/j.cell.2004.09.018) [DOI] [PubMed] [Google Scholar]
- 49.Amano Y. i., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. 2007. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl Acad. Sci. USA 104, 18 333–18 338 10.1073/pnas.0706403104 (doi:10.1073/pnas.0706403104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsubayashi Y. 2003. Ligand-receptor pairs in plant peptide signaling. J. Cell Sci. 116, 3863–3870 10.1242/jcs.00733 (doi:10.1242/jcs.00733) [DOI] [PubMed] [Google Scholar]
- 51.Hirakawa Y., et al. 2008. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl Acad. Sci. USA 105, 15 208–15 213 10.1073/pnas.0808444105 (doi:10.1073/pnas.0808444105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher K., Turner S. 2007. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 17, 1061–1066 10.1016/j.cub.2007.05.049 (doi:10.1016/j.cub.2007.05.049) [DOI] [PubMed] [Google Scholar]
- 53.Hirakawa Y., Kondo Y., Fukuda H. 2010. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22, 2618–2629 10.1105/tpc.110.076083 (doi:10.1105/tpc.110.076083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Etchells J. P., Turner S. R. 2010. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137, 767–774 10.1242/dev.044941 (doi:10.1242/dev.044941) [DOI] [PubMed] [Google Scholar]
- 55.Agusti J., Lichtenberger R., Schwarz M., Nehlin L., Greb T. 2010. Characterization of transcriptome remodeling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS Genet. 7, e1001312. 10.1371/journal.pgen.1001312 (doi:10.1371/journal.pgen.1001312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondo T., Nakamura T., Yokomine K., Sakagami Y. 2008. Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem. Biophys. Res. Commun. 377, 312–316 10.1016/j.bbrc.2008.09.139 (doi:10.1016/j.bbrc.2008.09.139) [DOI] [PubMed] [Google Scholar]
- 57.Wang X., Mitchum M. G., Gao B., Li C., Diab H., Baum T. J., Hussey R. S., Davis E. L. 2005. A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol. Plant Pathol. 6, 187–191 10.1111/j.1364-3703.2005.00270.x (doi:10.1111/j.1364-3703.2005.00270.x) [DOI] [PubMed] [Google Scholar]
- 58.Guo Y., Ni J., Denver R., Wang X., Clark S. E. 2011. Mechanisms of molecular mimicry of plant CLE peptide ligands by the parasitic nematode Globodera rostochiensis. Plant Physiol. 157, 476–484 10.1104/pp.111.180554 (doi:10.1104/pp.111.180554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Replogle A., et al. 2011. Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 65, 430–440 10.1111/j.1365-313X.2010.04433.x (doi:10.1111/j.1365-313X.2010.04433.x) [DOI] [PubMed] [Google Scholar]
- 60.Wang J., Joshi S., Korkin D., Mitchum M. G. 2010. Variable domain I of nematode CLEs directs post-translational targeting of CLE peptides to the extracellular space. Plant Signal Behav. 5, 1633–1635 10.4161/psb.5.12.13774 (doi:10.4161/psb.5.12.13774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., et al. 2010. Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 187, 1003–1017 10.1111/j.1469-8137.2010.03300.x (doi:10.1111/j.1469-8137.2010.03300.x) [DOI] [PubMed] [Google Scholar]
- 62.Searle I. R., Men A. E., Laniya T. S., Buzas D. M., Iturbe-Ormaetxe I., Carroll B. J., Gresshoff P. M. 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112 10.1126/science.1077937 (doi:10.1126/science.1077937) [DOI] [PubMed] [Google Scholar]
- 63.Mortier V., Den Herder G., Whitford R., Van de Velde W., Rombauts S., D'Haeseleer K., Holsters M., Goormachtig S. 2010. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153, 222–237 10.1104/pp.110.153718 (doi:10.1104/pp.110.153718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., Kawaguchi M. 2009. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 50, 67–77 10.1093/pcp/pcn194 (doi:10.1093/pcp/pcn194) [DOI] [PubMed] [Google Scholar]
- 65.Nishimura R., et al. 2002. HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429 10.1038/nature01231 (doi:10.1038/nature01231) [DOI] [PubMed] [Google Scholar]
- 66.Miyazawa H., et al. 2010. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137, 4317–4325 10.1242/dev.058891 (doi:10.1242/dev.058891) [DOI] [PubMed] [Google Scholar]
- 67.Krusell L., et al. 2011. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 65, 861–871 10.1111/j.1365-313X.2010.04474.x (doi:10.1111/j.1365-313X.2010.04474.x) [DOI] [PubMed] [Google Scholar]