Abstract
Root system architecture is a trait that displays considerable plasticity because of its sensitivity to environmental stimuli. Nevertheless, to a significant degree it is genetically constrained as suggested by surveys of its natural genetic variation. A few regulators of root system architecture have been isolated as quantitative trait loci through the natural variation approach in the dicotyledon model, Arabidopsis. This provides proof of principle that allelic variation for root system architecture traits exists, is genetically tractable, and might be exploited for crop breeding. Beyond Arabidopsis, Brachypodium could serve as both a credible and experimentally accessible model for root system architecture variation in monocotyledons, as suggested by first glimpses of the different root morphologies of Brachypodium accessions. Whether a direct knowledge transfer gained from molecular model system studies will work in practice remains unclear however, because of a lack of comprehensive understanding of root system physiology in the native context. For instance, apart from a few notable exceptions, the adaptive value of genetic variation in root system modulators is unknown. Future studies should thus aim at comprehensive characterization of the role of genetic players in root system architecture variation by taking into account the native environmental conditions, in particular soil characteristics.
Keywords: root system architecture, natural genetic variation, quantitative trait locus, Brachypodium, Arabidopsis
1. Introduction
Plants express a particularly high degree of phenotypic response to the environment since they are continuously exposed to fluctuating environmental conditions. This plasticity manifests itself in specific developmental, physiological and reproductive adjustments that are thought to optimize plant fitness. Excellent examples for this are the exaggerated elongation growth response of some species that allows them to escape shading by neighbouring competitors [1,2], or the control of flowering time as a function of annual or perennial lifestyle as well as climatic conditions [3]. The adaptive value of variation in these and other shoot traits can be rather obvious at times. This is often less clear when it comes to root system traits, although the root system plays a fundamental role in plant growth and survival by providing support, water and nutrients for the shoot [4,5]. Moreover, roots participate in secondary functions, including hormone biosynthesis and storage of photoassimilates. Root system architecture is considered a highly plastic trait [6,7] and is determined by developmental and environmental factors that interact to optimize exploration of the soil and the capture of edaphic resources [8]. For example, nitrogen, phosphorus, iron and sulphur are among the nutrients that have been reported to alter post-embryonic root development and, therefore, root system architecture [9]. The modulation of root system architecture and its response to biotic and abiotic stresses is of particular interest in crop science, because it is considered that optimization of crop root systems through breeding has been largely neglected as a result of the fertilizer-intensive agriculture of the last decades [10].
2. Root system architecture in dicotyledons versus monocotyledons
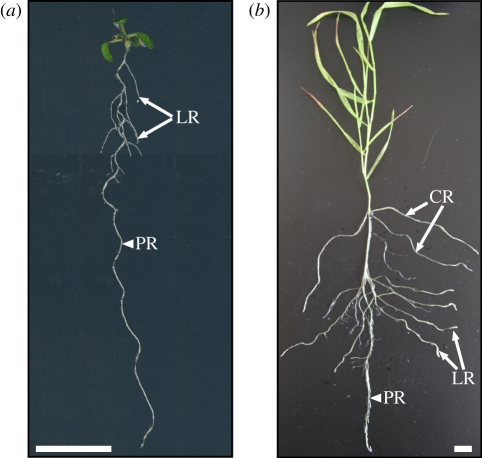
Root system architecture reflects the shape, three-dimensional distribution, branching pattern and age of the primary and post-embryonically generated roots [5,11]. Variation of root system architecture between different species is common, but the pronounced split between dicotyledons and monocotyledons is most prominent (figure 1). The typically allorhiz root system of dicotyledonous species consists of a single primary root and a network of lateral roots, which remain active during the plant's life cycle [5,12] (figure 1a). Here, root system architecture is mainly determined by cell division and elongation during primary and lateral root growth, as well as the extent of root branching. The system is occasionally completed by adventitious roots, which are much more important in the typically homorhiz root systems of monocotyledons (figure 1b). Their root systems are characterized by a complex architecture that emerges during successive stages of development [13]. During embryogenesis, a primary root and, sometimes, a variable number of seminal roots are formed and emerge shortly after germination [13]. After the seedling stage, this system is gradually replaced by a complex shoot-borne root system that is largely composed of crown roots. While mutagenesis approaches have succeeded in the identification of a large number of loci that are essential for the making and elaboration of the root system, non-essential alleles or novel genes that modulate the architecture of functional root systems are only beginning to be identified. This is mostly through the analysis of intra-specific natural genetic variation, an approach that has been successfully applied in the dicotyledon model system, Arabidopsis thaliana (Arabidopsis).
Figure 1.
Comparative root architecture of Arabidopsis and Brachypodium. (a) Typical dicotyledon allorhiz root system architecture in 10-day-old Arabidopsis plants. Arabidopsis forms only one primary root during its development, which branches out through lateral roots. (b) Typical monocotyledon homorhiz root system architecture in a 30-day-old Brachypodium plant, composed of a primary root, crown roots and lateral roots. PR, primary root; LR, lateral root; CR, crown root. Scale bars = 1 cm.
3. Natural variation of root system architecture in Arabidopsis
Natural strains of Arabidopsis, so-called accessions, display rich variation in morphological and physiological features, including root system architecture [3,14]. This trait can be easily observed and scored in in vitro culture, with most studies focusing on the growth vigour of the primary root. Isolation of allelic variants that underlie observed phenotypic differences can be achieved by quantitative trait locus (QTL) mapping and subsequent map-based cloning aided by complementing, for instance, reverse genetic approaches. So far, these studies have been moderately successful in that a limited number of mostly large effect QTLs (i.e. QTLs that explain the vast majority of the genetically determined variance in a given cross) were isolated. Notable exceptions from the large effect loci include allelic variation in a vacuolar invertase gene that explains ca 20 per cent of the primary root growth variance between the Cvi and Ler accessions [15], or allelic variation in the BREVIS RADIX (BRX) gene that explains ca 10 per cent of the variance between Eil-0 and Lc-0 [16]. However, the BRX gene was originally identified as a major effect QTL for root growth based on a loss-of-function allele in the Uk-1 accession [14]. Subsequent analyses implicated BRX in the hormonal control of primary root meristem growth [17]. In brx mutants, this translates into a significantly shorter primary root, more densely spaced lateral roots and more frequent adventitious roots, thus giving rise to an overall more compact and more branched adult root system. Interestingly, the later identified allele present in Lc-0 and a few other accessions encodes a hyperactive BRX protein that confers increased root growth vigour [16]. Thus, BRX is a rare example for an experimentally verified natural allelic series ranging from loss-of-function to hyperactive alleles when compared with other isolated QTLs, which are mostly loss-of-function only [3]. A straightforward explanation for the bias towards isolation of loss-of-function QTLs would be a focus on accessions that display the opposite extremes of a phenotype of interest. Moreover, a systematic bias towards loss-of-function alleles could be introduced by the genome evolution of Arabidopsis. This is suggested by first glimpses of genetic variation from comparison of accessions at the whole genome sequence level, based on re-sequencing using short read technology [18,19]. These analyses demonstrate an expected prevalence of single nucleotide polymorphisms, but also indicate a surprising number of sizeable deletions and insertions (indels) that frequently affect coding regions. It thus appears that compared with the Col-0 reference sequence, divergent accessions carry numerous null alleles, which might be prone to detection in QTL mapping [19]. This indel history of Arabidopsis can give rise to complicated scenarios, as exemplified by diminished root growth and eventual impaired viability arising from interaction between paralogous loci of an essential gene in amino acid biosynthesis that are located on two different chromosomes [20]. Differential evolutionary trajectories of those paralogues in Col-0 and Cvi have resulted in their differential expression and deletion of one paralogue in Cvi. The functional paralogue in Col-0 is located in the position of the deleted Cvi allele, whereas the remaining functional Cvi paralogue on a different chromosome is located in the position of the hypoactive (i.e. lowly expressed) Col-0 allele. Consequently, only certain allelic combinations produce sufficient overall protein activity, whereas other combinations impair growth rate or are not even viable.
4. Brachypodium as a tractable monocotyledon model system
Compared with dicotyledons in general and Arabidopsis in particular, little is known at the molecular level regarding the genetic basis of natural root system architecture variation in grasses. In part, this might reflect the fact that genetic and molecular studies in monocotyledons typically focus on crops, mainly rice and maize [12,13,21]. Arguably, the size and generation time of these species and the growth conditions required for their rearing are limiting the throughput of genetic analyses and render these systems impractical for research groups lacking appropriate and sizeable growth facilities. The calls for an easily tractable monocotyledon model for basic research therefore appear justified and have led to the promotion of Brachypodium distachyon (Brachypodium) in this capacity, culminating in the recent publication of its draft genome [22]. Brachypodium belongs to the family Poaceae (the grasses), which includes the economically most important crops, notably rice, maize, wheat and barley. Diverse phylogenetic analyses have revealed that Brachypodium is closer to wheat and barley than rice, corn or sorghum [23,24]. Together with its many favourable characteristics as an experimental system [25], this makes Brachypodium a credible model species for temperate cereals and grasses, which could aid in understanding the molecular basis of important agronomic traits that are exclusively present in grass crops. Like Arabidopsis, Brachypodium possesses numerous attributes, including a small (ca 300 Mb) genome with a low chromosome number, diploidy in most (but not all) accessions, developed genomic resources, Agrobacterium-mediated transformation, small size, self-fertility and a rapid life cycle [25–28]. Moreover, its size and ease of cultivation in large numbers under simple controlled conditions make it an accessible system for smaller laboratories, which can grow Brachypodium in the same facilities as Arabidopsis, one species next to the other. So can Brachypodium also serve as a model system for monocotyledon root system architecture and its natural variation? The answer is yes.
5. Brachypodium root system morphology
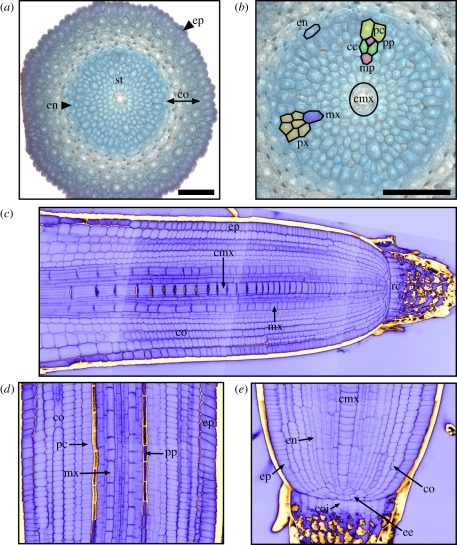
Similar to rice and maize, Brachypodium develops only one primary axillary root that emerges from the base of the embryo and breaks the coleorhiza upon germination. Moreover, compared with other cereals within the Pooideae subfamily, Brachypodium displays a much simpler seedling root system as exemplified by the reference accession, Bd21 [29] (figure 1b). For example, whereas wheat typically forms three to five primary roots, Brachypodium has only one and is in this sense more similar to rice or maize [27–29]. However, at later, adult stages of development Brachypodium and wheat display very similar root systems, which are dominated by branch roots [29]. At the tissue level, cellular organization is more complex in Brachypodium than in Arabidopsis with respect to cell types and numbers. For instance, Brachypodium has several layers with a variable number of cortical cells, whereas Arabidopsis has only one layer with a fixed number of cells. However, the cellular organization of Brachypodium roots seems to be less complex than that of other cereals in terms of the number of cells and layers in each tissue, which facilitates histological analysis in this grass species. Cross sections of Brachypodium primary roots show a single layer of epidermal cells, ground tissue consisting of five cortical and one endodermal cell layer, and the vasculature (figure 2a). The central vascular cylinder consists of one or sometimes two large central metaxylem tracheary element(s) and six peripheral xylem tracheary elements. The latter are alternating with phloem, which is typical of monocotyledon species. The phloem is composed of a protophloem sieve tube associated with two companion cells and the metaphloem sieve element and is organized in a symmetrical pattern. Finally, the external cell layer of the central cylinder, the pericycle, inter-crosses with protoxylem vessels at the sites of peripheral xylem bundles (figure 2b).
Figure 2.
Cellular structure of the Brachypodium primary root. (a) Transverse section across a Brachypodium primary root at meristem level (ca 300 μm from the root tip) stained with toluidine blue in light microscopy. (b) Magnified view of the stele from the same transverse section. Vascular tissues are organized in a polyarch structure containing numerous xylem vessels alternating with phloem sieve tubes. One central metaxylem element is found in the centre of the stele, surrounded by several smaller metaxylem vessels. The phloem presents a characteristic symmetrical pattern in which protophloem is associated with two companion cells and one metaphloem cell. Scale bars in (a,b) = 50 µm. (c) Longitudinal view of a primary root stained by the mPS-PI procedure and visualized by confocal microscopy. (d) Detailed view of the vascular tissues in the stele of the same mPS-PI-stained root. Note the characteristic staining of the protophloem cell files owing to the cell wall thickening that occurs during its differentiation. (e) Detailed view of the organization of initials at the root apical meristem. The tip of the apical meristem is occupied by a quiescent centre. Columella initials are generated through an anticlinal division. The epidermis–endodermis initials undergo a first anticlinal division near the quiescent centre. The endodermis and cortex cells are produced by successive periclinal divisions. ep, epidermis; co, cortex; en, endodermis; st, stele; pc, pericycle; pp, protophloem; cc, companion cell; px, protoxylem; mx, metaxylem; cmx, central metaxylem; coi, columella initial; ee, epidermis–endodermis initial; qc, quiescent centre.
The principles of tissue layer development of Brachypodium primary roots appear to be similar to those previously described for rice [19,29] (figure 2b–e). First, a group of initials divides and differentiates into columella cells as well as peripheral root cap cells. Second, the epidermis, the endodermis and all the cortical cell layers develop from another group of initials, termed epidermis–endodermis initials, through successive asymmetric periclinal divisions. In rice, it is well documented that the number of such periclinal divisions varies between different root types, explaining the differences in the layers of cortical cells [30]. The cortical origin of epidermis is a shared feature among monocotyledons and differs from dicotyledons, where epidermal and lateral root cap cells originate from a common initial [31]. Finally, the stele initials, which are larger than the surrounding meristematic cells, undergo several divisions to produce precursor cells that will differentiate into the vascular tissues.
6. Brachypodium as a model for natural variation of root system architecture in monocotyledons
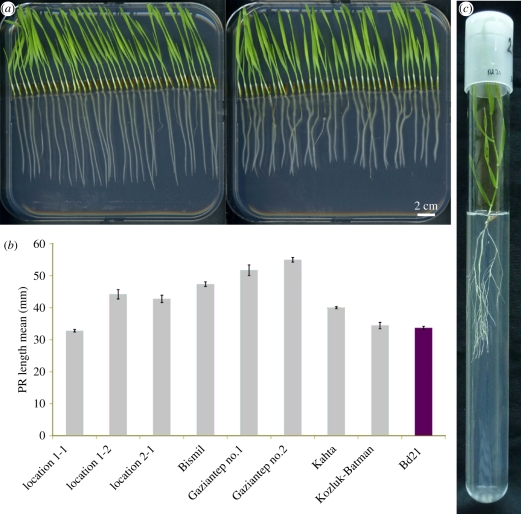
Similar to Arabidopsis, a collection of natural accessions collected from a wide range of geographical locations and derived inbred lines are currently being established for Brachypodium [32,33]. Phenotypic variation has already been found for diverse traits in a collection of Turkish accessions, indicating that this germplasm is suitable for natural variation analysis [32,34]. For example, substantial variation in flowering and vernalization time among Brachypodium accessions has been reported in one study [35], whereas broad diversity in drought tolerance was found in another [36]. Natural variation of Brachypodium root system architecture has not been investigated yet. However, a preliminary assessment of phenotypic variation in root growth in our laboratory, which monitored 178 diploid accessions from Turkey [32,34] and the community standard Bd21, looks promising (figure 3a,b). To observe Brachypodium root growth, seeds can be germinated in vertically oriented agar plates on standard media (half strength Murashige & Skoog (MS) nutrients, 1% sucrose), because they remain attached to the media surface very easily owing to their shape and small size. Depending on the growth conditions, up to 32 seedlings can be maintained on 10 × 10 cm square plates up to 10 days after germination (dag). Moreover, lateral root growth can be followed by using large square plates (24 × 24 cm), which can accommodate Brachypodium plants up to 25 dag, or by using tubes that can be adapted for new phenotype imaging methods (figure 3c) [37,38]. Finally, whole mount examination of cellular structure is possible in Brachypodium by using the modified pseudo-Schiff propidium iodide (mPS-PI) staining method combined with confocal laser scanning microscopy (figure 2c–e) [39]. With this set-up, differences in quantitative cellular parameters were found among accessions collected from eight different principal locations, indicating phenotypic diversity and the potential of this germplasm collection to reveal QTLs involved in primary root development (figure 3b). Thus, Brachypodium accessions could provide a valuable resource, enabling us to understand monocotyledon root system architecture plasticity and thereby increase our knowledge to breed grasses that can exploit soil resources more efficiently.
Figure 3.
Brachypodium as a model for natural variation analysis of root system growth and architecture. (a) Two different Brachypodium accessions from Turkey. Note the difference in primary root length. Seeds were germinated in half strength MS-agar plates and incubated in vertical position in continuous light for 6 days. (b) Primary root length measurement of 6-day-old Brachypodium accessions from eight different locations in Turkey. The primary root length of the standard community accession Bd21 is also shown. Note the differences in root length among the locations. Vertical lines indicate standard error. (c) Bd21 plant after 30 days of growth in a tube containing half strength MS-phytagel under continuous light. The seed was germinated directly in the tube.
7. Natural variation of root system architecture—pinning down the adaptive value
In summary, abundant natural variation in root system architecture exists in both monocotyledons and dicotyledons, and its genetic basis is beginning to be unravelled. A major question relating to this variation has however hardly been addressed: what is the adaptive value of different root system morphologies? Against a background of efforts aiming towards a more sustainable agriculture, there is a general notion that the impact of the root system on plant performance has been largely neglected in crop breeding. The hopes for a positive impact of root system architecture on shoot performance in agriculture are high and has led to calls for a ‘Second Green Revolution’, which should focus on root system architecture and should be made ‘a priority for plant biology in the twenty-first century’ [40]. Indeed, root system architecture critically influences nutrient and water uptake efficiency [41,42]. For example, rooting depth impacts the efficient acquisition of soil nitrogen (and water) since nitrate leeches down the soil profile, while manipulating the root growth angle could lead to more efficient use of phosphate, which typically accumulates in top soil [43]. Although it appears that root system size generally does not limit the acquisition of mobile macronutrients [44], such as nitrate, exploration of a larger soil volume by the root system could be advantageous when immobile macronutrients, such as phosphate, or water availability are limiting growth, in particular in competitive situations [42].
However, beyond these general notions, surprisingly few studies address the topic of the adaptive value of root system architecture directly and in a genetically tractable manner. An example is a study that has conclusively demonstrated the effect of root morphology on nutrient uptake by comparing an Arabidopsis mutant with significantly decreased levels of root branching to its wild-type background under nutrient stress [45]. Other studies have investigated the importance of the root system for shoot performance in a competitive situation. For instance, for variation at the BRX locus, a conditional fitness effect of the root system on shoot performance depending on underground competition was demonstrated [46]. Such studies could be extended to involve more genotypes, clarifying, for instance, whether a mix of different genetically determined root system architectures within a population improves the collective soil exploration efficiency and could thus be maintained by frequency-dependent selection. However, to get to the root (sic!) of the adaptive value of root system architecture variation, future studies should focus on investigating genotypes of interest in the light of their ecological background. A prime example is the isolation of allelic variation in the expression levels of a sodium transporter in the root and thus salt accumulation in the shoot, which could be associated with beach versus inland habitats and suggests a clear adaptation to increased salt levels in a coastal cline [47,48]. Taking into account the soil environment is maybe the most important factor and is becoming feasible as numerous recently collected accessions (both of Arabidopsis and Brachypodium) from precisely mapped collection sites are becoming available. Finally, interactions with micro-organisms of the rhizosphere are likely to impact the in situ relevance of root system architecture for plant performance. Genetically tractable analysis of these environment × genotype interactions, and thereby evaluation of the adaptive value of root system architecture and the corresponding role of individual alleles, can probably only be resolved in field trial settings that involve transgenic plants.
Acknowledgements
We would like to thank the Swiss National Science Foundation and the University of Lausanne for supporting the research of our laboratory, Hikmet Budak for generously sharing his collection of Brachypodium accessions, and Ted Farmer for comments on the manuscript.
References
- 1.Kami C., Lorrain S., Hornitschek P., Fankhauser C. 2010. Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66 10.1016/S0070-2153(10)91002-8 (doi:10.1016/S0070-2153(10)91002-8) [DOI] [PubMed] [Google Scholar]
- 2.Kebrom T. H., Brutnell T. P. 2007. The molecular analysis of the shade avoidance syndrome in the grasses has begun. J. Exp. Bot. 58, 3079–3089 10.1093/jxb/erm205 (doi:10.1093/jxb/erm205) [DOI] [PubMed] [Google Scholar]
- 3.Alonso-Blanco C., Aarts M. G., Bentsink L., Keurentjes J. J., Reymond M., Vreugdenhil D., Koornneef M. 2009. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21, 1877–1896 10.1105/tpc.109.068114 (doi:10.1105/tpc.109.068114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiken R. M., Smucker A. J. 1996. Root system regulation of whole plant growth. Annu. Rev. Phytopathol. 34, 325–346 10.1146/annurev.phyto.34.1.325 (doi:10.1146/annurev.phyto.34.1.325) [DOI] [PubMed] [Google Scholar]
- 5.Osmont K. S., Sibout R., Hardtke C. S. 2007. Hidden branches: developments in root system architecture. Annu. Rev. Plant Biol. 58, 93–113 10.1146/annurev.arplant.58.032806.104006 (doi:10.1146/annurev.arplant.58.032806.104006) [DOI] [PubMed] [Google Scholar]
- 6.Potters G., Pasternak T. P., Guisez Y., Jansen M. A. 2009. Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ. 32, 158–169 10.1111/j.1365-3040.2008.01908.x (doi:10.1111/j.1365-3040.2008.01908.x) [DOI] [PubMed] [Google Scholar]
- 7.Potters G., Pasternak T. P., Guisez Y., Palme K. J., Jansen M. A. 2007. Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci. 12, 98–105 10.1016/j.tplants.2007.01.004 (doi:10.1016/j.tplants.2007.01.004) [DOI] [PubMed] [Google Scholar]
- 8.Malamy J. E. 2005. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28, 67–77 10.1111/j.1365-3040.2005.01306.x (doi:10.1111/j.1365-3040.2005.01306.x) [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Bucio J., Cruz-Ramirez A., Herrera-Estrella L. 2003. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287 10.1016/S1369-5266(03)00035-9 (doi:10.1016/S1369-5266(03)00035-9) [DOI] [PubMed] [Google Scholar]
- 10.Bevan M. W., Garvin D. F., Vogel J. P. 2010. Brachypodium distachyon genomics for sustainable food and fuel production. Curr. Opin. Biotechnol. 21, 211–217 10.1016/j.copbio.2010.03.006 (doi:10.1016/j.copbio.2010.03.006) [DOI] [PubMed] [Google Scholar]
- 11.Lynch J. 1995. Root architecture and plant productivity. Plant Physiol. 109, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochholdinger F., Zimmermann R. 2008. Conserved and diverse mechanisms in root development. Curr. Opin. Plant Biol. 11, 70–74 10.1016/j.pbi.2007.10.002 (doi:10.1016/j.pbi.2007.10.002) [DOI] [PubMed] [Google Scholar]
- 13.Hochholdinger F., Tuberosa R. 2009. Genetic and genomic dissection of maize root development and architecture. Curr. Opin. Plant Biol. 12, 172–177 10.1016/j.pbi.2008.12.002 (doi:10.1016/j.pbi.2008.12.002) [DOI] [PubMed] [Google Scholar]
- 14.Mouchel C. F., Briggs G. C., Hardtke C. S. 2004. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 18, 700–714 10.1101/gad.1187704 (doi:10.1101/gad.1187704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sergeeva L. I., Keurentjes J. J., Bentsink L., Vonk J., van der Plas L. H., Koornneef M., Vreugdenhil D. 2006. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc. Natl Acad. Sci. USA 103, 2994–2999 10.1073/pnas.0511015103 (doi:10.1073/pnas.0511015103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beuchat J., Li S., Ragni L., Shindo C., Kohn M. H., Hardtke C. S. 2010. A hyperactive quantitative trait locus allele of Arabidopsis BRX contributes to natural variation in root growth vigor. Proc. Natl Acad. Sci. USA 107, 8475–8480 10.1073/pnas.0913207107 (doi:10.1073/pnas.0913207107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scacchi E., Salinas P., Gujas B., Santuari L., Krogan N., Ragni L., Berleth T., Hardtke C. S. 2010. Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc. Natl Acad. Sci. USA 107, 22 734–22 739 10.1073/pnas.1014716108 (doi:10.1073/pnas.1014716108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ossowski S., Schneeberger K., Clark R. M., Lanz C., Warthmann N., Weigel D. 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18, 2024–2033 10.1101/gr.080200.108 (doi:10.1101/gr.080200.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santuari L., Pradervand S., Amiguet-Vercher A. M., Thomas J., Dorcey E., Harshman K., Xenarios I., Juenger T. E., Hardtke C. S. 2010. Substantial deletion overlap among divergent Arabidopsis genomes revealed by intersection of short reads and tiling arrays. Genome Biol. 11, R4. 10.1186/gb-2010-11-1-r4 (doi:10.1186/gb-2010-11-1-r4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bikard D., Patel D., Le Mette C., Giorgi V., Camilleri C., Bennett M. J., Loudet O. 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323, 623–626 10.1126/science.1165917 (doi:10.1126/science.1165917) [DOI] [PubMed] [Google Scholar]
- 21.Coudert Y., Perin C., Courtois B., Khong N. G., Gantet P. 2010. Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15, 219–226 10.1016/j.tplants.2010.01.008 (doi:10.1016/j.tplants.2010.01.008) [DOI] [PubMed] [Google Scholar]
- 22.The International Brachypodium Initiative 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 10.1038/nature08747 (doi:10.1038/nature08747) [DOI] [PubMed] [Google Scholar]
- 23.Kellogg E. A. 2001. Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205 10.1104/pp.125.3.1198 (doi:10.1104/pp.125.3.1198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel J. P., et al. 2006. EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor. Appl. Genet. 113, 186–195 10.1007/s00122-006-0285-3 (doi:10.1007/s00122-006-0285-3) [DOI] [PubMed] [Google Scholar]
- 25.Draper J., et al. 2001. Brachypodium distachyon: a new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555 10.1104/pp.010196 (doi:10.1104/pp.010196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brkljacic J., et al. 2011. Brachypodium as a model for the grasses: today and the future. Plant Physiol. 157, 3–13 10.1104/pp.111.179531 (doi:10.1104/pp.111.179531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel J., Bragg J. 2009. Brachypodium distachyon, a new model for the Triticeae. In Genetics and genomics of the Triticeae, plant genetics and genomics: crops and models (eds Feuillet C., Muehlbauer G. J.), pp. 427–449 New York, NY: Springer [Google Scholar]
- 28.Garvin D. F., et al. 2008. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci. 48, S69–S84 10.2135/cropsci2007.04.0208 (doi:10.2135/cropsci2007.04.0208) [DOI] [Google Scholar]
- 29.Watt M., Schneebeli K., Dong P., Wilson W. I. 2009. The shoot and root growth of Brachypodium and its potential as a model for wheat and other cereal crops. Funct. Plant Biol. 36, 960–969 10.1071/FP09214 (doi:10.1071/FP09214) [DOI] [PubMed] [Google Scholar]
- 30.Rebouillat J., et al. 2009. Molecular genetics of rice root development. Rice 2, 15–34 10.1007/s12284-008-9016-5 (doi:10.1007/s12284-008-9016-5) [DOI] [Google Scholar]
- 31.Clowes F. A. L. 1994. Origin of the epidermis in root meristems. New Phytol. 127, 335–347 10.1111/j.1469-8137.1994.tb04284.x (doi:10.1111/j.1469-8137.1994.tb04284.x) [DOI] [PubMed] [Google Scholar]
- 32.Vogel J. P., Tuna M., Budak H., Huo N., Gu Y. Q., Steinwand M. A. 2009. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biol. 9, 88. 10.1186/1471-2229-9-88 (doi:10.1186/1471-2229-9-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mur L. A., Allainguillaume J., Catalan P., Hasterok R., Jenkins G., Lesniewska K., Thomas I., Vogel J. 2011. Exploiting the Brachypodium Tool Box in cereal and grass research. New Phytol. 191, 334–347 10.1111/j.1469-8137.2011.03748.x (doi:10.1111/j.1469-8137.2011.03748.x) [DOI] [PubMed] [Google Scholar]
- 34.Filiz E., Ozdemir B. S., Budak F., Vogel J. P., Tuna M., Budak H. 2009. Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 52, 876–890 10.1139/G09-062 (doi:10.1139/G09-062) [DOI] [PubMed] [Google Scholar]
- 35.Schwartz C. J., Doyle M. R., Manzaneda A. J., Rey P. J., Mitchell-Olds T., Amasino R. M. 2011. Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon. Bioener. Res. 3, 38–46 10.1007/s12155-009-9069-3 (doi:10.1007/s12155-009-9069-3) [DOI] [Google Scholar]
- 36.Luo N., Liu J., Yu X., Jiang Y. 2011. Natural variation of drought response in Brachypodium distachyon. Physiol. Plant 141, 19–29 10.1111/j.1399-3054.2010.01413.x (doi:10.1111/j.1399-3054.2010.01413.x) [DOI] [PubMed] [Google Scholar]
- 37.Clark R. T., MacCurdy R. B., Jung J. K., Shaff J. E., McCouch S. R., Aneshansley D. J., Kochian L. V. 2011. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 156, 455–465 10.1104/pp.110.169102 (doi:10.1104/pp.110.169102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer-Pascuzzi A. S., Symonova O., Mileyko Y., Hao Y., Belcher H., Harer J., Weitz J. S., Benfey P. N. 2010. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 152, 1148–1157 10.1104/pp.109.150748 (doi:10.1104/pp.109.150748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthelemy J., Palauqui J. C. 2008. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20, 1494–1503 10.1105/tpc.107.056069 (doi:10.1105/tpc.107.056069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch J. P. 2007. Roots of the second green revolution. Aust. J. Bot. 55, 493–512 10.1071/BT06118 (doi:10.1071/BT06118) [DOI] [Google Scholar]
- 41.Linkohr B. I., Williamson L. C., Fitter A. H., Leyser H. M. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29, 751–760 10.1046/j.1365-313X.2002.01251.x (doi:10.1046/j.1365-313X.2002.01251.x) [DOI] [PubMed] [Google Scholar]
- 42.Schenk H. J. 2006. Root competition: beyond resource depletion. J. Ecol. 94, 725–739 10.1111/j.1365-2745.2006.01124.x (doi:10.1111/j.1365-2745.2006.01124.x) [DOI] [Google Scholar]
- 43.Ge Z., Rubio G., Lynch J. P. 2000. The importance of root gravitropism for inter-root competition and phosphorus acquisition efficiency: results from a geometric simulation model. Plant Soil 218, 159–171 10.1023/A:1014987710937 (doi:10.1023/A:1014987710937) [DOI] [PubMed] [Google Scholar]
- 44.Robinson D. 1996. Resource capture by localized root proliferation: why do plants bother? Ann. Bot. Lond. 77, 179–185 10.1006/anbo.1996.0020 (doi:10.1006/anbo.1996.0020) [DOI] [Google Scholar]
- 45.Fitter A., Williamson L., Linkohr B., Leyser O. 2002. Root system architecture determines fitness in an Arabidopsis mutant in competition for immobile phosphate ions but not for nitrate ions. Proc. R. Soc. Lond. B 269, 2017–2022 10.1098/rspb.2002.2120 (doi:10.1098/rspb.2002.2120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shindo C., Bernasconi G., Hardtke C. S. 2008. Intraspecific competition reveals conditional fitness effects of single gene polymorphism at the Arabidopsis root growth regulator BRX. New Phytol. 180, 71–80 10.1111/j.1469-8137.2008.02553.x (doi:10.1111/j.1469-8137.2008.02553.x) [DOI] [PubMed] [Google Scholar]
- 47.Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., Yakubova E., Salt D. E. 2006. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2, e210. 10.1371/journal.pgen.0020210 (doi:10.1371/journal.pgen.0020210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baxter I., et al. 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6, e1001193. 10.1371/journal.pgen.1001193 (doi:10.1371/journal.pgen.1001193) [DOI] [PMC free article] [PubMed] [Google Scholar]