Abstract
Roots are highly responsive to environmental signals encountered in the rhizosphere, such as nutrients, mechanical resistance and gravity. As a result, root growth and development is very plastic. If this complex and vital process is to be understood, methods and tools are required to capture the dynamics of root responses. Tools are needed which are high-throughput, supporting large-scale experimental work, and provide accurate, high-resolution, quantitative data. We describe and demonstrate the efficacy of the high-throughput and high-resolution root imaging systems recently developed within the Centre for Plant Integrative Biology (CPIB). This toolset includes (i) robotic imaging hardware to generate time-lapse datasets from standard cameras under infrared illumination and (ii) automated image analysis methods and software to extract quantitative information about root growth and development both from these images and via high-resolution light microscopy. These methods are demonstrated using data gathered during an experimental study of the gravitropic response of Arabidopsis thaliana.
Keywords: root development, image analysis, automated image acquisition
1. Introduction
Understanding the development of root systems is vital to efforts towards food security. Roots provide anchorage and facilitate acquisition of water and nutrients from the soil. Growing roots explore their local environment in order to exploit those resources [1]. Quantitative data describing the structure and development of root systems are central to the study of plant growth and function. The ability to monitor the growth of plant organs provides valuable information about how those organs respond in or adapt to different situations [2,3].
One of the earliest studies of root response to an environmental stimulus, gravity, was published in the Philosophical Transactions of the Royal Society some 200 years ago. Knight [4] tied garden bean seeds to a small waterwheel, whose rotation produced a counterforce to gravity, and found that regardless of their initial orientation the emerging plants aligned themselves with the radii of the wheel. The analysis was entirely qualitative, and the experiment recorded only in notes and sketches. Supporting technology has improved immeasurably since Knight's experiment, but problems remain. Though time-based measurements are key to the detailed understanding of root growth, traditional root bioassays are based on at best a small number of measurements, and often only endpoint analyses [5]. These are informative, but have the limitation of only examining long-term effects on root growth. Transient events and subtle temporal changes can be missed.
Image acquisition and analysis provide a potential solution. Image sequences provide a rich source of data on plant growth. Implicit in each image is a detailed description of a plant's state of development at the time of acquisition, and images can be captured at high speeds. Once captured, they can be stored and re-examined to extract further information, perhaps for a different scientific purpose, at a later date. Time-lapse photography was used as early as the 1930s [6,7] to measure the heights of seedlings after application of the phytohormone ethylene, providing important information about the timing of its effects on growth regulation.
Today, a wide variety of image acquisition devices are available which can be deployed to analyse root growth. Confocal laser scanning microscopy provides high-quality digital images at the molecular and cellular scale [8]. Standard light microscopes can be used to detail the development of individual roots in high-resolution (again digital) images. Digital cameras are now of sufficient quality that even consumer devices can be used to gather data on sets of plant roots growing together on growth-room plates [9]. Modern data storage techniques allow large repositories of digital images to be constructed, browsed and examined, often remotely. As biological experiments often require large numbers of samples to be examined, a key requirement of many tools providing data on plant growth is that they be high-throughput. High-throughput systems can process large numbers of samples in short time periods with minimal user involvement. To achieve high-throughput recovery of data on root growth, automatic image acquisition methods are required.
The simplest automated image acquisition approach employs individual imaging and illumination equipment for each sample. For example, Brooks et al. [10] studied root gravitropism in Arabidopsis using a batch of seven identical image stations. However, hardware costs are high if imaging large numbers of samples and higher throughput imaging is usually achieved via automation, moving either the sample or the imaging hardware. Static sample systems image multiple samples using a single acquisition system by moving the camera(s) in front of each subject in turn using linear actuators, turntables or multi-axis positioners. This approach is adopted in the camera-positioning robot developed by the Phytomorph project, which uses a gantry arrangement to image banks of 36 Petri plates arranged in a 6 × 6 grid [11]. In contrast, static camera systems translocate each sample to an imaging station, typically by using motorized carousels, turntables or conveyor belts. Static camera methods have been constructed to support the GROWSCREEN-Root system [12] at FZJ Julich and the aeroponics-based root phenotyping platform under development at UCL Louvain [13]. This approach is advantageous in that a single imaging station is required but care must be taken to ensure that movement to the imaging station does not disturb the samples.
This rapid expansion in the range and amount of image data available begs the question: how is it to be analysed? The human analysis of images employed in early time-lapse photography work is time-consuming, subjective and prone to error. Analysis ‘by eye’ can produce measurements that are difficult to replicate and may result in subtle phenotypes, such as a delayed response, being missed. Full, or even partial, automation of the image analysis stage can address these problems, and a number of root image analysis methods and tools have been proposed. Such tools allow objective measurements to be made, and at a much higher frequency than is possible with manual approaches.
Though automatic image analysis is required for the full potential of automated acquisition platforms to be realized, it should be stressed that the benefits are not one-directional; automating data acquisition can also ease the analysis stage. By providing a repeatable and consistent lighting, and plant- and camera-positioning systems, automated acquisition platforms produce more stable imagery. This is desirable in general, but ideal for capturing images that are to be analysed by software. Image acquisition and analysis tools should be designed as integrated experimental environments; at present, however, perhaps because of the high number of images already available, root image analysis tools are more common than automated image acquisition platforms.
Following a review of the value of related methods in computer vision [14], Roberts et al. [15] proposed an optic flow approach to the analysis of confocal microscope images which combines a robust multi-frame-likelihood model and a technique for estimating uncertainty. Quelhas et al. [16] use optic flow data extracted from confocal image sequences to identify cell division events; the key cue being the difference in velocity of pixels arising from growing and splitting cells. Sethuraman et al. [17], in contrast, analyse confocal image sequences at the tissue level, focusing on groups of cells which are tracked using a modified network snake approach.
A number of software packages exist that aim to automate aspects of the kinematic analysis of growing roots, given high-resolution microscope images. RootFlow [18] and relative elemental growth rate analysis [3] have made use of optic flow-based techniques [19,20] to recover the motion of texture features through a sequence of root images. Intensity features are identified and matched between frames, and corresponding velocity flow fields are calculated. From these vector fields, estimates of growth can be made across any part of the image, provided that reliable image features are available in the areas of interest. Multi-ADAPT [2] can be used to measure elongation and curvature of both sides of a root while KineRoot [21] tracks the tip of a growing root using a correlation-based approach. The user is, however, required to provide the initial image and mark-up the tip in some detail.
The analysis of rhizotron images attracts considerable attention from those interested in assessing root system development. Rhizotrons occur in different forms [22], all of which involve plants growing against a transparent wall through which roots are imaged. As plant roots grow against the casing wall, images of their structure and development may be captured. A good review of rhizotron-oriented image analysis methods is provided by Le Bot et al. [23]. Analysis of rhizotron images is complicated by the opaque nature of the media, hence the motivation for transparent media approaches [24].
In what follows, we describe methods and tools developed at the Centre for Plant Integrative Biology (CPIB), University of Nottingham, for the recovery of quantitative measurements of plant root growth at the organ scale. These are in two parts. We first present a high-throughput automated image acquisition platform capturing sequences of images of plated roots under natural and infrared illumination, and image analysis software capable of extracting accurate growth data from those images with minimal user interaction. We then describe TipTracker, a novel, fully automatic tool for tracking the orientation of a growing root tip. The efficacy of these methods is demonstrated using data gathered during an experimental study of the gravitropic response of Arabidopsis thaliana.
2. Material and methods
(a). Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) seeds were surface-sterilized and sown on vertical 125 × 125 mm square Petri plates as detailed previously [25]. Each plate contained 60 ml one-half strength Murashige and Skoog media (Sigma) solidified with 1 per cent (w/v) agar. After 2 days at 4°C, plates were transferred to controlled environment chambers at 23°C with a 12 h photoperiod and a photon flux density of 150 μmol m−2 s−1.
(b). Gravitropism experiments
(i). Whole plate measurements
Plants were grown for 6 days in the controlled environment chamber housing the measuring equipment. Plates were rotated by 90° and imaged every 30 min for the duration of the photoperiod (12 h) using the automated image acquisition system described in figure 3. Tip angles were measured in each image using RootTrace software [9]. Twenty-one roots across three plates were analysed in the dark condition, and 25 across three plates for the light condition. Occasional failed traces were removed from the analysis.
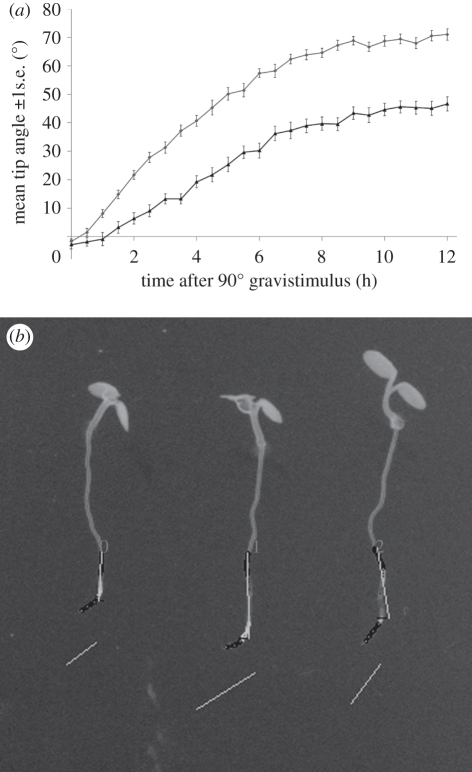
Figure 3.
Using RootTrace to examine variation in the tip angle of Arabidopsis roots during light (upper plot) and dark (lower plot) photoperiods following a 90° gravistimulus. (a) Tip angles as a function of time. (b) Sample output image from the RootTrace image analysis tool.
(ii). Single root measurements
For higher resolution imaging of individual roots, plates were transferred to the stage of a brightfield microscope (Zeiss AxioStar Plus, Carl Zeiss Ltd.) adapted to image plates vertically, and positioned on a shelf in the controlled environment chamber. The growth plate was rotated by 90° and a single root selected for imaging. Images were collected at half hourly intervals throughout the graviresponse using a machine vision camera connected to the microscope (Oscar F-810C, Allied Vision Technologies GmbH). Image sequences were subsequently analysed using the TipTracker software presented here.
3. Results and discussion
(a). Automated image acquisition
Automatic image acquisition at the CPIB is now in its second generation. The first generation imaging platform developed within the Centre used inexpensive compact cameras to image plate-grown Arabidopsis seedlings in controlled environment rooms [26]. This bespoke system used a belt-driven linear actuator to position two cameras with a capacity of twenty 125 × 125 mm plates at a temporal resolution of approximately 2 min between image acquisition.
Subsequent designs employed commercially available actuators, again used to position cameras in front of samples growing in controlled environment chambers. Combining standard components to create systems with the same basic layout eases the construction of additional machines. It also allows the design to be easily modified to suit new, and potentially physically different, growth rooms. The design of the system currently in use at CPIB is shown in figure 1.
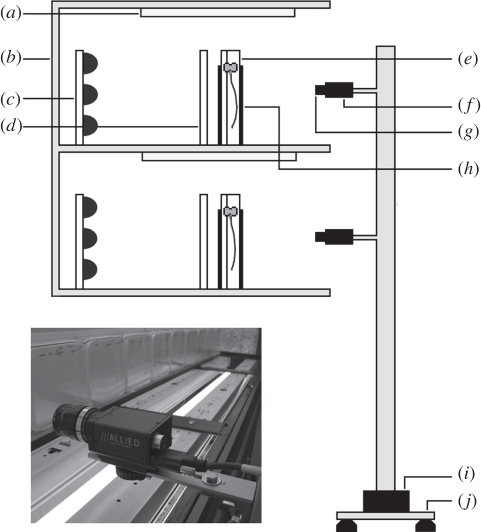
Figure 1.
The CPIB imaging platform. (a) Growth lighting, (b) controlled environment shelving, (c) NIR illuminators, (d) diffuser, (e) sample, (f) camera, (g) IR long-pass filter, (h) IR pass plastic cover, (i) linear actuator and (j) anti-vibration mounting. Inset: close-up view of one of the cameras.
The actuator is a precision leadscrew-driven linear stage with 1.5 m travel, an accuracy of ±187 µm and a repeatability of less than 2 µm (Model T-LST1500B, Zaber Technologies Inc.). The actuator translocates an imaging tower housing multiple cameras along the long axis of a growth chamber shelving unit. Machine vision cameras (Stingray F-504B, Allied Vision Technologies GmbH) are used to image vertically orientated plates, rhizotrons or growth pouches at a minimum temporal interval of approximately 5 min. Control of the actuator and cameras is via software written using the LabView graphical programming environment (National Instruments Corporation), allowing user configuration of motion, time-lapse and image acquisition parameters via a familiar graphical user interface. The assembly is positioned in front of plates or pouches growing in controlled environment chambers with minimal disruption to the growth conditions.
The single-axis CPIB robot performs a similar function to the three-axis Phytomorph machine [11]. Though the reduction in degrees of freedom must be compensated for with additional cameras, the extra camera cost is offset by the cheaper robotics, and simplicity of its design and control makes the CPIB system easy to construct, install, modify and maintain.
(b). Near-infrared illumination
The original imaging platform [26] was designed for use in controlled environment chambers in which Arabidopsis seedlings were grown under continuous light and used no additional lighting for image acquisition. If plants are to be grown under a more natural diurnal light/dark cycle, the imaging system must be capable of imaging during periods of darkness. Using suitable cameras and illuminating with near-infrared light (NIR, λ = 750–1400 nm) allow imaging throughout day/night photoperiods without impacting plant growth [27,28]. The current lighting system consists of three rows of LED flexible tape (tri-chip SMD5050, Huake Optoelectronics (Asia) Co. Ltd.) fixed to an aluminium backing plate to backlight the samples with NIR light at 850 nm. The LEDs are arranged in modules of three diodes spaced at 30 modules per metre, providing approximately 60 LEDs for each 125 × 125 mm growth plate. Two layers of diffuser film (Cinegel no. 3001, Rosco Laboratories Inc.) are attached to the rear of the sample holder to ensure even illumination of the samples. The cameras are fitted with IR long-pass filters (Type 093, Schneider Optische Werke GmbH) to exclude light below 750 nm to ensure consistency in images whether captured with growth room lights on or off. NIR imaging is possible with the Stingray cameras as they are not fitted with the visible light-only filters commonly attached to most camera sensors.
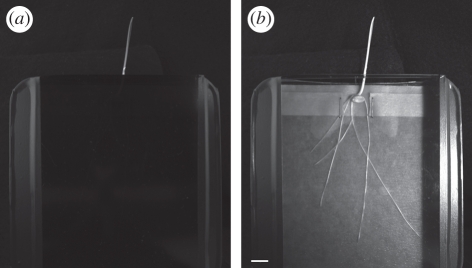
A further advantage of NIR imaging is that it allows isolation of the root system from the lighting to the aerial parts of the plants, mimicking the situation in soil. Figure 2a shows a sample plate on which a seedling of winter wheat (cv. Rialto) has been germinated. The sides of the plate are covered with IR pass plastic (Optolite IR, Instrument Plastics Ltd.) which excludes all wavelengths of light below 740 nm. The shoot is free to grow outside the plate through a hole in the lid (figure 2a). On illuminating with an NIR light source and using an NIR-sensitive camera, the root system can be imaged through the pass plastic (figure 2b). This system allows roots to be maintained in darkness throughout the imaging process and is also applicable to solid media- and soil-filled rhizotrons.
Figure 2.
Use of IR filters to maintain roots in darkness during imaging experiments. (a) Image taken using visible light illumination. Note the IR pass/visible cut filter prevents visible light from penetrating to the root. (b) Image of the same arrangement taken using NIR illumination and an NIR-sensitive camera. Scale bar, 1 cm.
(c). RootTrace: plated root analysis software
Increasing the rate of data capture shifts the bottleneck in processing to the analysis stage, motivating the development of complementary automated image analysis techniques. RootTrace [9,29] was developed to help automate the analysis of the images gathered by the CPIB image acquisition platform.
Laboratory studies of Arabidopsis root growth typically involve growing multiple plants on agar plates. Traditional measures of growth are made by physically marking the position of the root tip on the plates at regular (but infrequent) intervals. RootTrace is able to measure Arabidopsis roots growing on agar plates across image time-series, allowing growth parameters to be captured automatically. The user loads an image sequence and marks one location on each root in the first frame. No further interaction is required. The software processes the image sequence, measuring each root in each image. From this information, growth rates can be calculated for each root [9]. Local curvature estimates are made and the combined growth and curvature data presented in simple but effective heatmaps. In addition, the software is able to estimate the number of emerged lateral roots [29].
Underpinning RootTrace is a model-based, top-down approach to image analysis. A target-tracking method is adopted, which traces down each root from the user-specified start point to the root tip. A probabilistic tracking model is used to explore the area around the root, before a graph-based procedure estimates its centre line; for details, see French et al. [9]. The length of the main root can then be determined at each time point, providing growth information over time. Local curvature can also be estimated, supporting studies of gravitropism. Lateral roots are detected by noting protrusions from the main root [29]. The tool's user interface is designed to hide advanced parameters and features from the user until they are ready to investigate them. These parameters and features are designed to provide an intuitive way for the user to improve the results of tracing on ‘difficult’ images (perhaps noisy or with distracting reflections). Additionally, advanced image processing features are available, for performing background removal for example, as well as pre-processing steps for rotating and cropping input images. Output is in the form of comma-separated text files of root length, tip angle, curvature information, etc., for each root, as well as annotated output images detailing the measurements made.
Figure 3a shows data obtained from RootTrace and shows the tip angle of Arabidopsis roots during light and dark photoperiods following a 90° gravistimulus. Roots are positively gravitropic (i.e. grow towards the gravity vector) and negatively phototropic (grow away from directional light). The response curve in the light reveals the additive effect of these tropisms as illustrated by the steeper gradient in the initial phases of the response and the magnitude of the tip angle at the end of the recording period (figure 3a, upper plot). In darkness, where the only stimulus is gravity, both the gradient of the response and magnitude of final angle are reduced (figure 3a, lower plot). Figure 3b shows sample output from the RootTrace tool. Tip angle is overlaid on the input IR image (greyscale is inverted for display).
(d). TipTracker: high-magnification root image analysis
While CPIB's image acquisition robot and RootTrace software support efficient analysis of many roots, the images involved are of comparatively low spatial resolution (approx. 40–50 µm per pixel). Experimental design often requires analysis of large sample sets to be supported by more detailed (i.e. higher spatial resolution) consideration of smaller numbers of plants. Microscopes allow the examination of growth responses in great detail (approx. 0.5–1 μm per pixel), and automatic analysis of time-series images again allows quantitative measurements to be made. Therefore, to complement RootTrace, a tool was developed to track the angle of growing Arabidopsis root tips through microscope image sequences: TipTracker. This higher magnification approach allows tracking of curvature at the extreme root apex (approx. 200 μm back from the tip) which is not possible with RootTrace, and may be important for identifying where curvature occurs [30].
To capture microscope images of a root responding to gravity, a light microscope was inverted to present the stage vertically to the objective. In this configuration, the root is allowed to grow vertically downwards, permitting a natural gravitropic response. Images can be captured as frequently as required; typically, capturing every 30 min allows the growth process to be recorded in sufficient detail to support high fidelity analysis. Clearly, this frequency can be increased or decreased dependant on the magnification used and the growth process under investigation.
Once images have been captured, they are simply imported into TipTracker and processed automatically. TipTracker then provides a measurement of the tip angle for each frame. Automatic processing to achieve this output proceeds as follows. Images are loaded and pre-processed using the OpenCV library [31]. The colour digital images are converted to greyscale, and smoothed using a 5 × 5 Gaussian kernel. The images are then thresholded to isolate a binary root silhouette, and a contour is fitted around the resulting boundary. This root boundary is stored as a set of ordered two-dimensional points. Further analysis uses the MatLab data analysis environment [32]. The location where the root enters the image is identified by scanning the four image boundaries (line AB, figure 4). Subsequently, an initial medial node is estimated by computing the mean of root boundary points near the start of the root (point X, figure 4). This is an estimated medial node and may not be accurate. The position of this medial node is corrected by passing a number of candidate lines through this medial node; each candidate line intersects the root boundary at two locations. The distance between the two intersection points is measured for each candidate line. The candidate line with the smallest distance is selected as the one which is optimally perpendicular to the root boundary (line Y, figure 4). The corrected medial node is identified as the point which lies on this line and is equidistant from the two intersection points formed by this line and the root boundary. The position of the next medial node is estimated by extrapolating a fixed distance along the straight line connecting the previous two rectified medial nodes. This is an estimated position and requires rectification (point Z, figure 4). The position of this second medial node is again rectified following the same procedure. This process is repeated for identification of subsequent medial nodes until the end of the root is encountered. The root tip angle is measured by fitting a least squares regression line to the subset of medial nodes near the tip of the root and measuring its inclination relative to horizontal (figure 5a; detail panel). For each timepoint, TipTracker processes each image independently.
Figure 4.
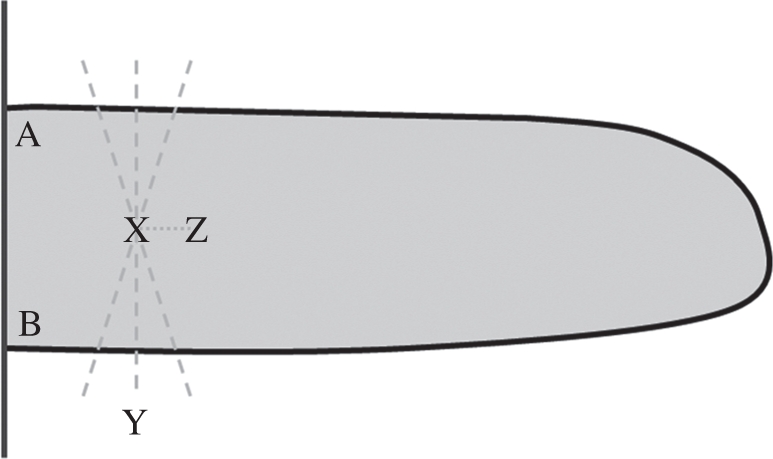
Points of interest in the processing steps of TipTracker. Vertical line AB represents the entry of the root at the edge of the image, and the thick black boundary is the root silhouette. Point X represents the initial estimated medial point; dashed lines represent candidate cross-sectional lines through the estimated medial point, the shortest of which is selected as the optimal perpendicular cross section (line Y). A corrected medial point is selected as the midpoint along this optimal cross-sectional line, and its position used to estimate the next medial point in the sequence (point Z).
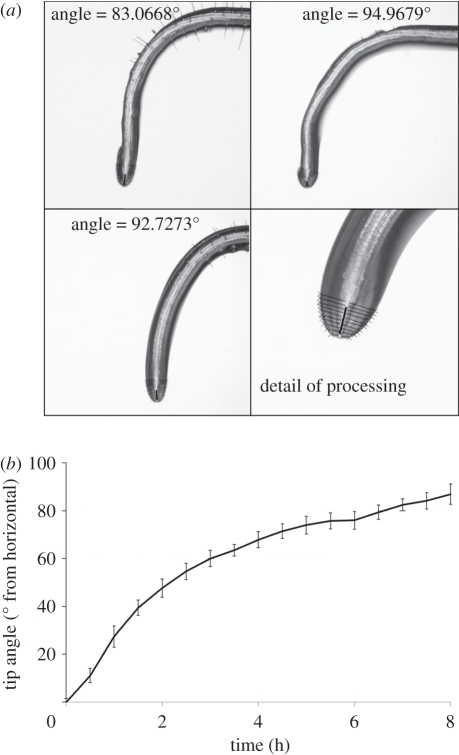
Figure 5.
(a) Example output frames from TipTracker, 8 h into the sequence across four roots. Bottom-right panel is a detail of the processing output, with the cross-sectional lines and fitted tip angle line clearly visible. (b) Tip angle results from TipTracker over an 8 h time lapse at 30 min intervals, with standard error bars (n = 4). Image sequences were captured using a machine vision camera on an inverted light microscope (vertical stage), and processed automatically using TipTracker. Col-0 plants were used to demonstrate the fidelity of the technique.
To demonstrate the output produced by TipTracker, time sequence images of growing Arabidopsis thaliana Col-0 roots were captured on the inverted light microscope, with a resolution of approximately 0.7 μm per pixel. The raw images were imported directly into TipTracker, and time-based angle measurements were produced (figure 5). Processing took approximately 1 min per image sequence and required no user interaction. TipTracker produces annotated images (figure 5a) as well as the angle measurements in a comma-separated text file. Figure 5b illustrates typical numerical results from the software, showing roots exhibiting a gravitropic response. TipTracker is an open source software tool, and will be available from the CPIB website (http://www.cpib.ac.uk/software/)
4. Conclusions and outlook
The technologies described here are not targeted at specific plant traits, but seek to provide lower level, more generic descriptions of plant growth from which a variety of traits can be extracted using methods appropriate to the task at hand. To date, RootTrace has been shown to be capable of identifying primary root growth at a spatial resolution of 50 µm, and reliably detecting lateral roots longer than 500 µm. Growth experiments typically operate at a temporal resolution of 30 min, while root emergence uses images captured several hours apart. Curvature and tip angle resolution is difficult to summarize, as many methods exist by which these can be extracted from RootTrace data, and each has strengths and weaknesses. A typical experiment using the image acquisition robot and RootTrace examines approximately 400 seedlings, assuming 20 seedlings per 125 × 125 mm growth plate. Higher numbers are possible if younger seedlings are grown at a higher density (up to 40 seedlings can be accommodated on a single plate). TipTracker typically operates on a single root at a temporal resolution of several minutes, providing root growth rates at a spatial resolution of 1 µm and tip angle to within 0.5°.
Typical experimental tasks which might be addressed using RootTrace include screening populations of wild-type, mutant and treated seedlings for variations in primary root growth, tip angle (both during normal growth and following a gravistimulus), lateral root number and/or root curvature. TipTracker is intended to support higher resolution determination of primary growth rate and tip angle during growth (again under normal conditions or in response to a gravity stimulus).
Interest in automatic image acquisition and analysis tools like those described here has increased significantly within the plant sciences in recent years. This is due to the emergence of the systems approach to biological research and an increasing awareness that quantitative measurement of the phenotype has fallen behind understanding of the genotype. Tools such as those presented here are becoming more widely sought and more commonly used. Core techniques from image processing, computer vision and robotics can and are being applied and combined to produce high-throughput and high-resolution data acquisition systems. These systems can form highly valuable toolkits providing biologists with quantitative data at a variety of scales. Current methods are, however, often limited, or best suited, to younger plants with comparatively simple root system architectures imaged under laboratory conditions.
The image acquisition challenge is to identify methods of visualizing the highly complex, dense root architectures produced by more mature crop plants, ideally in three-dimensions and in natural growth conditions. Methods of acquiring three-dimensional data from plant roots grown in transparent media have recently been proposed [24], and imaging modalities are being developed and deployed which allow plant roots to be assessed in a medium much more closely related to their natural environment—soil. Traditional methods of measuring the development of plants grown in soil are destructive, requiring root systems to be removed from their environment and washed. These processes disrupt the topology of the root, limiting the architectural features that can be detected and measurements that can be made. Recently, X-ray computed tomography [33], nuclear magnetic resonance imaging and positron emission tomography [34] have been used to visualize the three-dimensional structure of plant roots in a variety of media, including soil columns. The challenge for image analysis is to produce techniques and tools capable of processing the resulting images, with the minimum of user interaction, to provide the data needed to support work in systems biology and plant phenotyping.
Acknowledgements
The authors acknowledge BBSRC and EPSRC CISB programme funding to the Centre for Plant Integrative Biology (CPIB) and BBSRC Professorial Fellowship funding to M.J.B. H.H. acknowledges the support of the Al-Tajir Trust.
References
- 1.Lynch J. 1995. Root architecture and plant productivity. Plant Physiol. 109, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishikawa H., Evans M. L. 1997. Novel software for analysis of root gravitropism: comparative response patterns of Arabidopsis wild-type and axr1 seedlings. Plant Cell Environ. 20, 919–928 10.1046/j.1365-3040.1997.d01-129.x (doi:10.1046/j.1365-3040.1997.d01-129.x) [DOI] [PubMed] [Google Scholar]
- 3.Walter A., Spies H., Terjung S., Kusters R., Kirchgeßner N., Schurr U. 2002. Spatio-temporal dynamics of expansion growth in roots: automatic quantification of diurnal course and temperature response by digital image sequence processing. J. Exp. Biol. 53, 689–698 [DOI] [PubMed] [Google Scholar]
- 4.Knight T. A. 1806. On the direction of the radicle and germen during the vegetation of seeds. Phil. Trans. R. Soc. 99, 108–120 [Google Scholar]
- 5.Parry G., Delbarre A., Marchant A., Swarup R., Napier R., Perrot-Rechenmann C., Bennett M. J. 2001. Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J. 25, 399–406 10.1046/j.1365-313x.2001.00970.x (doi:10.1046/j.1365-313x.2001.00970.x) [DOI] [PubMed] [Google Scholar]
- 6.Michener H. 1938. The action of ethylene on plant growth. Am. J. Bot. 25, 711–720 10.2307/2436929 (doi:10.2307/2436929) [DOI] [Google Scholar]
- 7.Van der Laan P. 1934. Der Einfluss von Aethylen auf die Wuchsstoffbildung be Aena und Vicia. Rec. Trav. Bot. Neerl. 31, 691–742 [Google Scholar]
- 8.Stephens D. J., Allan V. J. 2003. Light microscopy techniques for live cell imaging. Science 300, 82–86 10.1126/science.1082160 (doi:10.1126/science.1082160) [DOI] [PubMed] [Google Scholar]
- 9.French A. P., Úbeda-Tomás S., Holman T. J., Bennett M. J., Pridmore T. P. 2009. High throughput quantification of root growth using a novel image analysis tool. Plant Physiol. 150, 1784–1795 10.1104/pp.109.140558 (doi:10.1104/pp.109.140558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durham Brooks T. L., Miller N. D., Spalding E. P. 2010. Plasticity of Arabidopsis root gravitropism throughout a multi-dimensional condition space quantified by automated image analysis. Plant Physiol. 152, 206–216 10.1104/pp.109.145292 (doi:10.1104/pp.109.145292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phytomorph Robotic Camera. 2011. Robotic camera. See http://phytomorph.wisc.edu/hardware/robot.php. (accessed 18 September 2011)
- 12.Nagel K. A., et al. 2009. Temperature responses of roots: impact on growth, root system architecture and implications for phenotyping. Funct. Plant Biol. 36, 947–959 10.1071/FP09184 (doi:10.1071/FP09184) [DOI] [PubMed] [Google Scholar]
- 13.Lobet G., Pagès L., Draye X. 2011. A novel image analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 157, 29–39 10.1104/pp.111.179895 (doi:10.1104/pp.111.179895). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts T., McKenna S. J., Du C-J., Wuyts N., Valentine T., Bengough A. G. 2010. Estimating the motion of plant root cells from in vivo confocal laser scanning microscopy images. Mach. Vis. Appl. 21, 921–939 10.1007/s00138-009-0207-x (doi:10.1007/s00138-009-0207-x) [DOI] [Google Scholar]
- 15.Roberts T., McKenna S. J., Wuyts N., Valentine T., Bengough G. 2007. Performance of low-level motion estimation methods for confocal microscopy of plant cells in vivo. In IEEE Workshop on Motion and Video Computing (WMVC'07), Austin, TX, February 2007, pp. 13–15 Washington, DC: IEEE Computer Society 10.1109/WMVC.2007.32 (doi:10.1109/WMVC.2007.32) [DOI] [Google Scholar]
- 16.Quelhas P., Mendonça A., Campilho A. 2010. Optical flow based Arabidopsis thaliana root meristem cell division detection. Lect. Notes Comp. Sci. 6112, 217–226 10.1007/978-3-642-13775-4_22 (doi:10.1007/978-3-642-13775-4_22) [DOI] [Google Scholar]
- 17.Sethuraman V., French A., Wells D., Kenobi K., Pridmore T. In press Tissue-level segmentation and tracking of cells in growing plant roots. Mach. Vis. Appl. (doi:10.1007/s00138-011-0329-9) [Google Scholar]
- 18.van der Weele C. M., Jiang H. S., Palaniappan K. K., Ivanov V. B., Palaniappan K., Baskin T. I. 2003. A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth: roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiol. 132, 1138–1148 10.1104/pp.103.021345 (doi:10.1104/pp.103.021345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barron J. L., Fleet D. J., Beauchemin S. S. 1994. Performance of optical flow techniques. Int. J. Computer Vision 12, 43–77 10.1007/BF01420984 (doi:10.1007/BF01420984) [DOI] [Google Scholar]
- 20.Barron J. L., Liptay A. 1994. Optical flow to measure minute increments in plant growth. Bioimaging 2, 57–61 (doi:10.1002/1361-6374(199403)2:1<57::AID-BIO5>3.3.CO;2-4) [DOI] [Google Scholar]
- 21.Basu P., Pal A., Lynch J. P., Brown K. M. 2007. A novel image-analysis technique for kinematic study of growth and curvature. Plant Physiol. 145, 305–316 10.1104/pp.107.103226 (doi:10.1104/pp.107.103226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor H. M., Upchurch D. R., McMicheal B. L. 1990. Applications and limitations of rhizotrons and minirhizotrons for root studies. Plant Soil 129, 29–35 10.1007/BF00011688 (doi:10.1007/BF00011688) [DOI] [Google Scholar]
- 23.Le Bot J., Serra V., Fabre J., Draye X., Adamowicz S., Pagè L. 2010. DART: a software to analyse root system architecture and development from captured image. Plant Soil 326, 261–273 10.1007/s11104-009-0005-2 (doi:10.1007/s11104-009-0005-2) [DOI] [Google Scholar]
- 24.Clark R. T., MacCurdy R. B., Jung J. K., Shaff J. E., McCouch S. R., Aneshansley D. J., Kochian L. V. 2011. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 156, 455–465 10.1104/pp.110.169102 (doi:10.1104/pp.110.169102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holman T. J., Wilson M. H., Kenobi K., Dryden I. L., Hodgman T. C., Wood A. T., Holdsworth M. J. 2010. Statistical evaluation of transcriptomic data generated using the Affymetrix one-cycle, two-cycle and IVT-Express RNA labelling protocols with the Arabidopsis ATH1 microarray. Plant Methods 6, 9–10 10.1186/1746-4811-6-9 (doi:10.1186/1746-4811-6-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French A., Wells D., Everitt N., Pridmore T. P. 2011. High-throughput quantification of root growth. In Measuring roots an updated approach (ed. Mancuso S.). Berlin, Germany: Springer [Google Scholar]
- 27.Wiese A., Christ M. M., Virnich O., Schurr U., Walter A. 2007. Spatio-temporal leaf growth patterns of Arabidopsis thaliana and evidence for sugar control of the diel leaf growth cycle. New Phytol. 174, 752–761 10.1111/j.1469-8137.2007.02053.x (doi:10.1111/j.1469-8137.2007.02053.x) [DOI] [PubMed] [Google Scholar]
- 28.Kiss J. Z., Mullen J. L., Correll M. J., Hangarter R. P. 2003. Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 131, 1411–1417 10.1104/pp.013847 (doi:10.1104/pp.013847) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naeem A., French A. P., Wells D. M., Pridmore T. P. 2011. High-throughput feature counting and measurement of roots. Bioinformatics 27, 1337–1338 10.1093/bioinformatics/btr126 (doi:10.1093/bioinformatics/btr126) [DOI] [PubMed] [Google Scholar]
- 30.Wolverton C., Ishikawa H., Evans M. L. 2002. The kinetics of root gravitropism: dual motors and sensors. J. Plant Growth Regul. 21, 102–112 10.1007/s003440010053 (doi:10.1007/s003440010053) [DOI] [PubMed] [Google Scholar]
- 31.Bradski G. R., Pisarevsky V. 2000. Intel's computer vision library: applications in calibration, stereo segmentation, tracking, gesture, face and object recognition. Proc. IEEE Conf. Comp. Vis. Pattern Recogn. 2, 796–797 10.1109/CVPR.2000.854964 (doi:10.1109/CVPR.2000.854964) [DOI] [Google Scholar]
- 32.MatLab 2011. Natick, MA: The MathWorks Inc [Google Scholar]
- 33.Mooney S. J., Pridmore T. P., Bennett M. J. In press Developing X-ray computed tomography to non-invasively image 3-D root system architecture in soil. Marschner Rev. Plant Soil. 10.1007/s11104-011-1039-9 (doi:10.1007/s11104-011-1039-9) [DOI] [Google Scholar]
- 34.Jahnke S., et al. 2009. Combined MRI-PET dissects dynamic changes in plant structures and functions. Plant J. 59, 634–644 10.1111/j.1365-313X.2009.03888.x (doi:10.1111/j.1365-313X.2009.03888.x) [DOI] [PubMed] [Google Scholar]