In the beginning—even before the Human Genome Project—there were chromosomes. Notwithstanding their primary role in determining the transmission of genes and alleles during mitosis and meiosis and their clear relevance to major forms of human pathology such as cancer and birth defects, chromosomes in complex eukaryotic organisms remain relatively poorly understood. The molecular basis for functional chromosomal elements such as telomeres, origins of DNA replication and the centromere are under investigation in a host of organisms ranging from the unicellular yeast Saccharomyces cerevisiae to humans. This task has been both complicated and motivated in part by the fascinating observation that the cellular, genomic, and molecular mechanisms of some of these functional elements are remarkably variable from organism to organism (1). Nowhere is this more evident than for the centromere, the chromosomal locus responsible for attachment of the chromosome to the mitotic and meiotic spindle apparatus and thus for segregation of chromosomes to daughter cells during cell division. Centromeres can be small, unitary elements (as in S. cerevisiae), localized repetitive elements stretching on for millions of base pairs (as in many plants and animals, including humans), or dispersed elements distributed along the length of the chromosome (as in the nematode Caenorhabditis elegans) (1). And most recently, as illustrated by the paper of Saffery et al. (2) in this issue of PNAS, it has become apparent that, even in organisms characterized by a localized centromere, other chromosomal sequences that are located far away from the normal centromere and that normally have nothing to do with centromere function can fulfill the role of a centromere. These so-called “neocentromeres,” once formed, appear to behave functionally in all respects as if they are bona fide centromeres. A lingering debate in the field has been whether the existence of neocentromeres, especially in humans, implies the existence of many different sequences equally capable of forming a functional centromere, or whether it reflects a strong selection for sequences—no matter how inefficient—capable of conferring mitotic stability (3, 4). Saffery et al. (2) have addressed this question by attempting to generate human artificial chromosomes with neocentromeres. Neocentromeres were first described in maize as subtelomeric heterochromatic “knobs” that could be transformed into functioning centromeres capable of mobilization on the spindle (5). However, unlike maize neocentromeres, which are comprised primarily of tandemly repeated satellite DNA with homology to maize centromere DNA (6), human neocentromeric DNA is not repetitive (7) and appears not to share obvious homology with the 171-bp repeated DNA family, alpha satellite, that characterizes normal human centromeres (3). Furthermore, several dozen neocentromeres have been documented in the human genome, from many different chromosomal regions (8–10). Together, these observations have been used to argue in favor of the model that the basis for centromere activity in human chromosomes is epigenetic, rather than determined solely by primary sequence (3, 4, 8, 11). The basis for such an epigenetic mark is not clear and may reflect a functional determinant created by secondary structure (4, 12, 13) or some other characteristic of centromere behavior such as late DNA replication (3, 14).
Neocentromeres appear to be indistinguishable from normal, alpha satellite-based centromeres in terms of the chromatin and kinetochore complex that provides functional activity.
Although neocentromeres are the most striking example of the functional plasticity of centromeres in higher organisms, they are by no means the only example. Human centromeres, for example, can adopt a wide range of apparently equally functional configurations (4). A typical centromere appears to consist of several million base pairs of alpha satellite DNA, although there is substantial variation both in the amount of alpha satellite as well as its primary sequence from chromosome to chromosome. Dicentric chromosomes—generally presumed to be unstable because of their susceptibility to chromosome breakage (15)—are in fact frequently stabilized by centromere inactivation, the epigenetic modification of one of the two centromeres to render it incapable of forming a kinetochore and binding to spindle microtubules (4, 16). Although the molecular basis of centromere inactivation is unknown, it is accompanied by absence of centromere and kinetochore proteins that are normally associated with functionally active centromeres (16). Among these is centromere protein A (CENP-A), a centromere-specific histone H3 variant that normally associates with alpha satellite DNA and marks centromeric chromatin (17). CENP-A homologues have been described at yeast, worm, and fly centromeres, indicating an evolutionarily highly conserved chromatin component at functional centromeres despite the wild variation in centromeric DNA content among eukaryotic chromosomes (1, 18). It is thus particularly notable that neocentromeres are associated with all of the known, functionally implicated centromere proteins (9, 10, 19). Furthermore, in the chromatin of the one neocentromere examined to date, CENP-A is associated over a nearly half million base pair region of genomic DNA that is, at least at the level of primary sequence, not detectably different from that of surrounding non-CENP-A-binding DNA (20). Thus, neocentromeres appear to be indistinguishable from normal, alpha satellite-based centromeres in terms of the chromatin and kinetochore complex that provides functional activity.
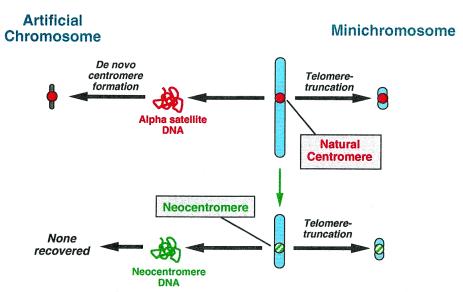
It is this premise that Saffery et al. (2) set out to explore in the context of human artificial chromosomes. The concept of artificial chromosomes was introduced nearly twenty years ago in S. cerevisiae (21), referring to construction of a fully functional chromosome from its component parts. Since then, artificial chromosomes have been used to explore many features of chromosome biology, especially in S. cerevisiae and Schizosaccharomyces pombe (22). More recently, an artificial chromosome system has been developed in human cells, based on the ability of alpha satellite DNA to provide de novo centromere function (23, 24). Indeed, the observation that alpha satellite DNA can seed formation of de novo centromeres in artificial chromosomes while other sequences cannot (25) provides the strongest argument that alpha satellite in fact comprises the functional centromere of normal human chromosomes (26). Whereas formation of mitotically stable artificial chromosomes from individual components provides an assay for the function of centromeric DNA, an alternative strategy employs engineering of already existing, natural chromosomes to provide information on the function of individual elements in their chromatin state. In this strategy, telomere-associated chromosome truncation is used to delete most or all of the chromosome arms, thus generating stable “minichromosomes” that can be as small as several million base pairs (27, 28). (By comparison, even the smallest normal human chromosome is some 50 million base pairs in length.) This approach has been used (28) to carefully engineer chromosomes for the purposes of mapping and characterizing aspects of normal centromere structure and behavior. With respect to centromere function, an important distinction between the two assays is that one tests the functionality of already competent centromeric chromatin, whereas the other tests the functionality of purified DNA, which must presumably be packaged into chromatin before becoming functional (refs. 3 and 4; Fig. 1).
Figure 1.
Formation of human artificial chromosomes and engineered minichromosomes containing natural centromeres (Top) or neocentromeres (Bottom). Minichromosomes, generated by telomere-associated truncation (27, 28), can be formed from either centromeric or neocentromeric chromatin that already possesses the epigenetic modifications necessary for functional centromere competence (18). In contrast, human artificial chromosomes can be generated de novo from isolated centromeric DNA (alpha satellite; refs. 23, 24, and 30), but not from isolated neocentromeric DNA (2). Thus, at the level of DNA, alpha satellite and neocentromeric DNA are not equivalent at centromere formation.
Saffery et al. (2) have used both approaches to examine the centromere competence of neocentromere sequences from a well characterized chromosome 10-derived marker chromosome called mardel (10). In this chromosome, the neocentromere has been previously localized to a fully sequenced ≈80-kb region originating from 10q25 (7, 29). First, Saffery et al. used the neocentromeric DNA as a candidate centromere in a human artificial chromosome assay. In over 450 clones examined, no artificial chromosomes were detected, in contrast to alpha satellite-containing input sequences that routinely generate artificial chromosomes in up to 50% of clones tested (23–25). Similarly, they were unable to generate an artificial chromosome by using a modified yeast artificial chromosome approach (30), substituting neocentromeric DNA for alpha satellite. The inability of neocentromeric sequences to form artificial chromosomes by using either approach argues strongly that these sequences—at least in the form of naked DNA—are less efficient than alpha satellite DNA at forming a de novo functional centromere.
To test the ability of chromatin at the mardel(10) neocentromere to function, they then used the minichromosome assay, truncating mardel(10) with a telomere-containing construct containing targeting DNA from the chromosome arms (2). The resulting minichromosomes were estimated to be <1–2 million base pairs in size, among the smallest human minichromosomes yet engineered. Extensive genomic characterization and functional testing provided direct evidence for the function of the mardel(10) neocentromere.
The conflicting results obtained by Saffery et al. (2) when using the artificial chromosome assay and the minichromosome assay likely reflect that the two assays measure different aspects of centromere assembly and function (Fig. 1). The biological relevance of the initial steps of centromere assembly in the de novo assay is open to some question, as naked centromeric DNA presumably does not exist in Nature. Nonetheless, remodeling of centromeric chromatin likely occurs during development and during epigenetic resolution of unusual centromere configurations, as must occur during centromere inactivation in dicentric chromosomes or in activation of neocentromeres (3, 4, 11). Formation of de novo centromeres in the artificial chromosome assay thus may provide a suitable model for exploring aspects of and requirements for centromere remodeling, which likely involves incorporation of the centromere-specific histone variant CENP-A (18). In contrast to the de novo assay, the engineering of minichromosomes does not involve generation or remodeling of centromeric chromatin; rather, it utilizes an already functional centromere (or neocentromere) as the basis for engineering smaller derivatives that are still capable of normal chromosome function. One particularly intriguing aspect of centromere organization and function raised by neocentromeres and by the aforementioned variation among centromeric DNAs from different species and organisms is the extent to which a centromere from one organism can substitute for that of another (1).
Lastly, as pointed out by Saffery et al. (2), the generation of minichromosomes based on neocentromeres provides an alternative to strategies that consider the use of artificial chromosomes as potential nonviral gene therapy or gene transfer vectors (23, 26, 31). In principle, it may be possible to make neocentromere-based minichromosomes (2) that are smaller than alpha satellite-based minichromosomes (27, 28, 32). However, it will be necessary to test this premise directly, as the lower limit of neocentromeric DNA required hasn't been established. Notwithstanding the conceptual advantages of this approach, it is important to recall just how inefficient chromosome engineering in mammalian cells is compared with other, high-efficiency systems (21). Natural or unnatural, the potential of human artificial chromosomes may not be fully realized until more tractable approaches for chromosome engineering, handling, and transfer are developed and explored.
Acknowledgments
I thank Brenda Grimes, Katie Rudd, and Bala Balakumaran for helpful discussions. Work on artificial chromosomes in our laboratory is supported by a Franklin Delano Roosevelt Research Award from the March of Dimes Birth Defects Foundation.
Footnotes
See companion article on page 5705.
References
- 1.Tyler-Smith C, Floridia G. Cell. 2000;102:5–8. doi: 10.1016/s0092-8674(00)00004-0. [DOI] [PubMed] [Google Scholar]
- 2.Saffery R, Wong L H, Irvine D V, Bateman M A, Griffiths B, Cutts S M, Cancilla M R, Cendron A C, Stafford A J, Choo K H A. Proc Natl Acad Sci USA. 2001;98:5705–5710. doi: 10.1073/pnas.091468498. . (First Published May 1, 2001; 10.1073/pnas.091468498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willard H F. Curr Opin Genet Dev. 1998;8:219–225. doi: 10.1016/s0959-437x(98)80144-5. [DOI] [PubMed] [Google Scholar]
- 4.Choo K H A. Trends Cell Biol. 2000;10:182–188. doi: 10.1016/s0962-8924(00)01739-6. [DOI] [PubMed] [Google Scholar]
- 5.Rhoades M M, Vilkomerson H. Proc Natl Acad Sci USA. 1942;28:433–443. doi: 10.1073/pnas.28.10.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfenito M R, Birchler J A. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barry A E, Howman E V, Cancilla M R, Saffery R, Choo K H. Hum Mol Genet. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]
- 8.Choo K H A. Am J Hum Genet. 1997;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depinet T W, Zackowski J L, Earnshaw W C, Kaffe S, Sekhon G S, Stallard R, Sullivan B A, Vance G H, Van Dyke D L, Willard H F, et al. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 10.Warburton P E, Dolled M, Mahmood R, Alonso A, Li S, Naritomi K, Tohma T, Nagai T, Hasegawa T, Ohashi H, et al. Am J Hum Genet. 2000;66:1794–1806. doi: 10.1086/302924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karpen G H, Allshire R C. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 12.Koch J. Hum Mol Genet. 2000;9:149–154. doi: 10.1093/hmg/9.2.149. [DOI] [PubMed] [Google Scholar]
- 13.Floridia G, Zatterale A, Zuffardi O, Tyler-Smith C. EMBO Rep. 2000;1:489–493. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Csink A K, Henikoff S. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- 15.McClintock B. Proc Natl Acad Sci USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan B A, Schwartz S. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- 17.Vafa O, Sullivan K F. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan K F. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 19.Saffery R, Irvine D V, Griffiths B, Kalitsis P, Wordeman L, Choo K H. Hum Mol Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 20.Lo A W, Magliano D J, Sibson M C, Kalitsis P, Craig J M, Choo K H. Genome Res. 2001;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray A W, Szostak J W. Nature (London) 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- 22.Carbon J, Clarke L. New Biol. 1990;2:10–19. [PubMed] [Google Scholar]
- 23.Harrington J J, van Bokkelen G, Mays R W, Gustashaw K, Willard H F. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 24.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill N I, Cooke H J, Masumoto H. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 25.Ebersole T A, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B. Hum Mol Genet. 2000;9:1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- 26.Willard H F. Science. 2000;290:1308–1309. doi: 10.1126/science.290.5495.1308. [DOI] [PubMed] [Google Scholar]
- 27.Farr C J, Bayne R A, Kipling D, Mills W, Critcher R, Cooke H J. EMBO J. 1995;14:5444–5454. doi: 10.1002/j.1460-2075.1995.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heller R, Brown K E, Burgtorf C, Brown W R. Proc Natl Acad Sci USA. 1996;93:7125–7130. doi: 10.1073/pnas.93.14.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DuSart D, Cancilla M R, Earle E, Mao J I, Saffery M, Tainton K M, Kalitsis P, Martyn J, Barry A E, Choo K H A. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 30.Henning K A, Novotny E A, Compton S T, Guan X Y, Liu P P, Ashlock M A. Proc Natl Acad Sci USA. 1999;96:592–597. doi: 10.1073/pnas.96.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown W R, Mee P J, Hong Shen M. Trends Biotechnol. 2000;18:218–223. doi: 10.1016/s0167-7799(00)01438-4. [DOI] [PubMed] [Google Scholar]
- 32.Yang J W, Pendon C, Yang J, Haywood N, Chand A, Brown W R. Hum Mol Genet. 2000;9:1891–1902. doi: 10.1093/hmg/9.12.1891. [DOI] [PubMed] [Google Scholar]