Abstract
An important missing piece in the puzzle of how plastids spread across the eukaryotic tree of life is a robust evolutionary framework for the host lineages. Four assemblages are known to harbour plastids derived from red algae and, according to the controversial chromalveolate hypothesis, these all share a common ancestry. Phylogenomic analyses have consistently shown that stramenopiles and alveolates are closely related, but haptophytes and cryptophytes remain contentious; they have been proposed to branch together with several heterotrophic groups in the newly erected Hacrobia. Here, we tested this question by producing a large expressed sequence tag dataset for the katablepharid Roombia truncata, one of the last hacrobian lineages for which genome-level data are unavailable, and combined this dataset with the recently completed genome of the cryptophyte Guillardia theta to build an alignment composed of 258 genes. Our analyses strongly support haptophytes as sister to the SAR group, possibly together with telonemids and centrohelids. We also confirmed the common origin of katablepharids and cryptophytes, but these lineages were not related to other hacrobians; instead, they branch with plants. Our study resolves the evolutionary position of haptophytes, an ecologically critical component of the oceans, and proposes a new hypothesis for the origin of cryptophytes.
Keywords: phylogenomics, plastid, haptophyte, cryptophyte, katablepharid, tree of life
1. Introduction
Eukaryotes first acquired photosynthesis through endosymbiosis, where a heterotrophic cell engulfed and retained a photosynthetic prokaryote related to modern-day Cyanobacteria, ultimately integrating it to form the highly specialized plastid organelles we see today [1–3]. This crucial event in eukaryote evolution is generally seen as unique: primary plastids probably evolved only once in the common ancestor of glaucophytes, red algae and green plants (green algae + land plants), together making the Plantae supergroup [4] (but see [5]). A much more recent case of cyanobacterium to eukaryote endosymbiosis has been reported in the rhizarian Paulinella chromatophora [6], but this event appears to have had less impact on the diversification of plastids. Photosynthesis spread further to other eukaryotic lineages by means of secondary endosymbioses, when other eukaryotes subsequently engulfed green or red algae, and, in dinoflagellates, tertiary endosymbioses [7]. On the green side, two independent cases of secondary endosymbioses are known, leading to chlorarachniophyte and euglenid algae, respectively [8]. On the red side, the situation is much more contentious.
The chromalveolate hypothesis has been regarded as a likely evolutionary framework for explaining the origin and distribution of red secondary plastids [9,10]. It posits that a single secondary endosymbiosis with a red alga gave rise to plastids in stramenopiles (or heterokonts), alveolates, haptophytes and cryptophytes, altogether forming the Chromalveolata supergroup [11]. This hypothesis is based on the fact that complex events are necessary to establish a plastid, so it is more parsimonious to limit the number of plastid origins, regardless of the number of plastid losses this implies [12]. Thus far, plastid data have generally supported the monophyly of some or all of the chromalveolate lineages where plastids are known. Molecular evidence for this includes multi-gene phylogenies [13,14], shared evolutionary history of several nucleus-encoded plastid-targeted genes [15–18], and a rare lateral gene transfer in the plastids of haptophytes and cryptophytes [19].
The chromalveolate hypothesis also predicts that the host nuclear lineages are monophyletic; so far, however, this has proven impossible to verify despite the use of substantial alignments (in the range of 30 000 amino acids). Nuclear-based phylogenomics have consistently shown that stramenopiles and alveolates are closely related, and that they form a strongly supported group with Rhizaria, altogether making the so-called SAR group [20,21]. At the same time, haptophytes and cryptophytes generally appeared together, albeit with less support and only when relatively large alignments are used [21–24]. Based on congruent plastid and nuclear data, these were proposed to be a second chromalveolate lineage, Hacrobia [25]. Other lineages that were not originally included in the chromalveolate hypothesis have since been suggested to be members of Hacrobia (namely telonemids, centrohelids, katablepharids, picobiliphytes, Palpitomonas and rappemonads), but the support for these is variable, and typically few data are available, from only a single representative of these lineages [22,25–32].
The large variations in the phylogenetic signal between plastid and nuclear data have recently been formalized in a ‘phylogenomic falsification’ of the chromalveolate hypothesis, which concluded that red algal plastids were acquired separately in different lineages [18,33]. Several alternative scenarios to the chromalveolate hypothesis have also been formulated, all attempting to explain the data by suggesting that plastids in ‘chromalveolate’ lineages originated through a single secondary endosymbiosis within a subgroup of chromalveolates, and then spread to other subgroups by multiple tertiary endosymbioses [34–36]. In these complex alternatives, haptophytes and cryptophytes emerged as key players in early plastid dissemination. Cryptophytes are also peculiar in that they are the only known lineage that still harbours the red algal endosymbiont nucleus (the nucleomorph), making them of pivotal significance to study endosymbiosis [37]. In addition, haptophytes include some of the most successful marine primary producers, which have a profound impact on global biogeochemical equilibria [38]. Despite the ongoing interest in these organisms, their phylogenetic position remains among the most uncertain of any eukaryote.
Here, we have addressed this problem by deep-sequencing a cDNA library for the last hacrobian taxon for which no genome-wide sequence data are available, the katablepharid Roombia truncata, and at the same time considerably extended the length of previously published alignments by taking advantage of newly sequenced genomes (in particular that of the cryptophyte Guillardia theta). We assembled a supermatrix to test the hacrobian question, consisting of 258 genes and 68 carefully selected operational taxonomic units (OTUs), and used it to assess the monophyly of Hacrobia and the relationships of its constituent lineages to other eukaryotes.
2. Material and methods
Roombia truncata culturing and cDNA construction, sequencing and contig assembling are described in the electronic supplementary material.
(a). Sequence alignment construction
With the presence of G. theta mandatory in each single gene, a two-step strategy was adopted to maximize the number of genes entering the final concatenated alignment (supermatrix). First, we used a dataset of 162 genes described by Burki et al. [39] and added six important newly available taxa: one katablepharid (R. truncata, this study), one cryptophyte (Rhodomonas salina, this study), one picobiliphyte (cell MS584-11) and three red algae (Calliarthron tuberculosum, Porphyridium cruentum and Eucheuma denticulatum). We also reduced the amount of missing data for G. theta as well as for the rhizarian Bigelowiella natans by using the complete nuclear genome information for these two organisms, both recently sequenced by the US Department of Energy Joint Genome Institute. Second, we used the 24 480 predicted proteins (filtered model v. 1.0) of G. theta to find additional genes suitable for phylogenomics to extend our supermatrix. In order to enrich the pool of possible new genes with those containing key taxa, we applied the constraint that both G. theta and R. truncata be present in each gene, in addition to a selection of complete genomes and expressed sequence tag (EST) datasets corresponding to the taxon sampling in figure 1. This approach led to 224 potential new genes, but only 96 remained after the quality validation (i.e. did not show any evidence for deep paralogy, lateral gene transfer or extensive lineage sorting). Overall, the dataset presented here is composed of 258 genes. See the electronic supplementary material for a detailed procedure of the alignment construction, and tables presenting the missing data information and descriptions of all genes (electronic supplementary material, table S1 and S4, respectively).
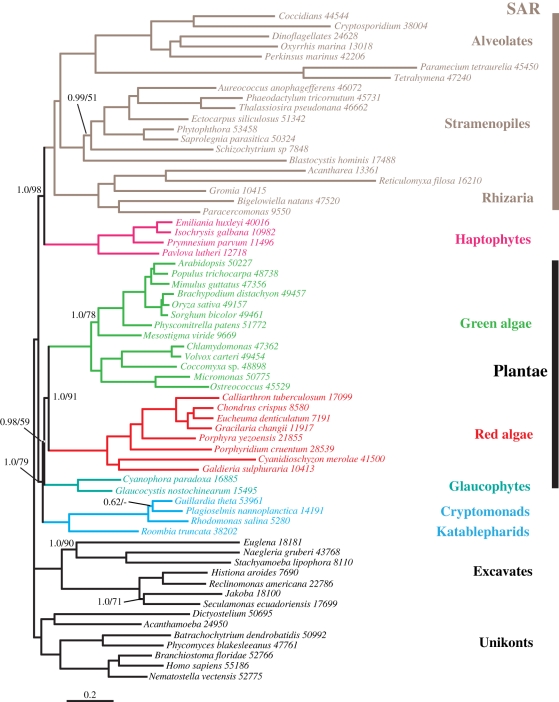
Figure 1.
Phylogeny inferred with the CAT + Γ4 model, based on the most complete taxa sampling. Support values are indicated for relevant nodes or when not maximal (PP/CAT-BP). No value at a node means support equal to 1.0 PP/100% CAT-BP. Dots mark OTUs with complete genome data available. Numbers after the OTU names are the sequence lengths. Scale bar, substitutions per position.
(b). Phylogenetic analyses
The fit on the data of two evolutionary models—the site-homogeneous LG model and the site-heterogeneous mixture CAT model—was evaluated by cross-validation (CV) as implemented in PhyloBayes v. 3.3b [40]. A learning set and a test set were generated by randomly splitting (without replacement) the original alignment into 10 replicates made of 90 per cent (50 293 amino acids) and 10 per cent (5588 amino acids) of the original sites, respectively. For each replicate, a Markov chain Monte Carlo run was then performed for a total of 5000 cycles (CAT) or 1100 cycles (LG), the lower number of cycles under LG being due to a much greater computational time per cycle. The first 1000 and 100 points were discarded as burn-in for the CAT and LG runs, respectively, and the remaining points used to compute the cross-validation log-likelihood. Bayesian inferences using the best tested model (CAT) in combination with four gamma categories for handling the rate heterogeneity across sites (Γ4) were performed in PhyloBayes v. 3.3b for all datasets. In each analysis, two independent chains were run for a minimum of 5000 cycles or until convergence of the chains was reached, removing the first 1000 cycles as burn-in and calculating the posterior consensus on the remaining trees. The Dayhoff 6 classes was used for recoding the amino acids, and analysed as above. Convergence between the chains was ascertained by examining the difference in frequency for all their bipartitions (less than 0.15 in all analyses). Bootstrap CAT proportions were computed with 100 pseudo-replicates generated with Seqboot from the Phylip v. 3.69 package [41], and run for 5000 cycles under the CAT + Γ4 model with a burn-in of 1000 cycles. For each replicate, trees were collected after the burn-in period and the resulting 100 consensus trees fed to Consense (Phylip package) to calculate the bootstrap support. Because of the extreme computational burden associated with this analysis, only two datasets could be tested with CAT bootstrap (corresponding to figures 1 and 2). The less adequate model (LG) was also evaluated for aln68 in a maximum likelihood (ML) inference using RAxML v. 7.2.8 [42]. The best ML tree was determined with the PROTGAMMA + F implementation in multiple inferences using eight randomized parsimony starting trees. Statistical support was evaluated with 100 bootstrap replicates. Fast-evolving sites were identified using the slow–fast method [43] as implemented in Slowfaster [44]. The eight most trimmed alignments were analysed with RAxML under the LG + PROTCAT + F model and 100 bootstrap replicates. The amino acid composition was visualized by assembling a 20 × 64 matrix of the frequency of each amino acids per species, and represented as a two-dimensional plot in a principal component analysis (PCA) with the R package.
Figure 2.
Phylogeny inferred with the CAT + Γ4 model, with telonemids, centrohelids and picobiliphytes removed. Support values are indicated for relevant nodes or when not maximal (PP/CAT-BP). No value at a node means support equal to 1.0 PP/100% CAT-BP. Numbers after the OTU names are the sequence lengths. Scale bar, substitutions per position.
3. Results and discussion
(a). EST sequencing of Roombia truncata and assembly of a large dataset
Katablepharids are the closest known relatives to cryptophytes [30], and one of the last hacrobian groups from which no genome-wide data are available. Katablepharid phylogenomic data are important to aid in the detection of hidden multiple substitutions that could have occurred along the branch leading to cryptophytes, a common source of non-phylogenetic signal, as well as the detection of endosymbiont-derived genes, since no plastid has been reported in katablepharids. Accordingly, we developed the first genome-wide survey of the katablepharid lineage, a transcriptome of the deep-branching R. truncata [25]. Using a fractionation method to remove this predator from its diatom prey (Navicula sp.), we isolated highly enriched R. truncata RNA and carried out cDNA sequencing using both 454 and Illumina (see the electronic supplementary material). To assess the purity of this dataset with respect to potential diatom contamination, the R. truncata transcriptome was examined by BLAST comparison against GenBank and searching for diatom-derived sequences. This analysis revealed only 14 contigs with best hits against a diatom homologue (E-value < 1e–20), specifically the centric diatom Thalassiosira pseudonana and the pennate diatom Phaeodactylum tricornudum, but none of them could be clearly assigned to be a diatom sequence.
The transcriptome was also searched for evidence of a cryptic plastid or genes derived from a lost photosynthetic endosymbiont in three ways: (i) we specifically looked by BLAST for genes related to plastid pathways known from other non-photosynthetic chromalveolates (e.g. isoprenoid, fatty acid and heme biosynthesis [45,46]); (ii) we scanned the BLAST output against Genbank for candidate plastid-derived genes by closest sequence similarity to plant, algal and/or cyanobacterial genes; and (iii) we examined the position of R. truncata in our single-gene trees that could be indicative of a red algal ancestry (see §2), as the close relationship between katablepharids and cryptophytes predicts that endosymbiotic gene transfers would be from red algae if their ancestor was plastid-bearing. Interestingly, not a single gene that could be unambiguously attributed to a cryptic plastid or derived from an ancestral plastid was identified. Altogether, the R. truncata transcriptome contained no clear evidence for an ancestral endosymbiont.
To infer a global phylogeny for eukaryotes including this new data, we developed an alignment that is based on more than twice the number of genes than the largest published phylogenomic dataset previously used to investigate similar questions [22]. Importantly, the complete genome of G. theta allowed us to concomitantly reduce the missing data for cryptophytes, resulting in an alignment characterized by 0 per cent and 14 per cent missing genes for G. theta and R. truncata, respectively, out of a total of 258 genes (55 881 amino acids). Our taxon sampling also encompasses all known other ‘hacrobian’ lineages: haptophytes, telonemids, centrohelids and the recently available picobiliphytes [22,47]. Red algae, which are another pivotal lineage for investigating plastid evolution, were also heavily sampled, with eight species included (notably the deeply diverging mesophylic P. cruentum). Finally, we used the newly sequenced genome of the rhizarian B. natans to obtain for the first time a pan-eukaryotic concatenated alignment of hundreds of genes that features at least one species with complete genome data in each major group, with the exception of glaucophytes (figure 1). This is important because each major eukaryotic group is now anchored around one or several taxa with few missing data, which increases the global phylogenetic signal contained in our alignment (see electronic supplementary material, table S1 for a detailed report on the proportion of missing data).
(b). Phylogenetic relationships of the hacrobian taxa
The complete dataset (68 OTUs, 55 881 amino acids; hereafter denoted aln68) was first analysed by ML with the LG + Γ4 model and 100 bootstrap replicates. The resulting phylogeny recovered most of the major eukaryotic groups with maximal support (electronic supplementary material, figure S1), including opisthokonts, Amoebozoa, excavates and the SAR group of stramenopiles, alveolates and Rhizaria (where alveolates and rhizarians were sisters, possibly due to the fast-evolving OTUs found in these groups). The exception, however, was Hacrobia, which was found to be polyphyletic. Specifically, this tree strongly confirmed the close relationship between katablepharids and cryptophytes with 100 per cent bootstrap (henceforth the KC group), but the KC group branched within Plantae (78% bootstrap), as did another hacrobian taxa, the picobiliphytes. The positions of the three remaining hacrobian taxa were unsupported. The general lack of support for the most ancient nodes in this tree, as opposed to robust artefactual support, can be interpreted as the result of competing signals: the genuine phylogenetic signal is diluted by equivalent non-phylogenetic signals, such as undetected homoplasy, not correctly inferred by the LG model of evolution [48]. In an attempt to counter this mutational saturation, we first conducted a site removal analysis in which the fastest-evolving sites were progressively removed from the original alignment [49]. This approach led to no improvement regarding the position of the hacrobian taxa (see electronic supplementary material, figure S2).
Complex models of evolution that detect multiple substitutions at sites with better accuracy than classical models such as LG have been shown to be more powerful for investigating difficult phylogenetic questions [50]. In fact, a CV test showed that the site-heterogeneous CAT + Γ4 model fitted our alignment significantly better than the site-homogeneous LG + Γ4 model used in the ML reconstruction, with a scored average over 10 replicates of 6215 + 145, and, accordingly, Bayesian inferences with the CAT + Γ4 model were used in all subsequent analyses of our data. When applied to the same alignment (aln68), the CAT + Γ4 model provided a similar picture overall, but with several critical differences (figure 1). The KC group remained sister to the Plantae and picobiliphytes supported by 1.0 PP and 57 per cent CAT-BP, with picobiliphytes again closer to Plantae with affinities to glaucophytes (0.92 PP; 50% CAT-BP). The remaining hacrobians—haptophytes, centrohelids and telonemids—branched with the SAR group (1.0 PP; 51% CAT-BP), this time with telonemids possibly the closest lineage to SAR. It is also worth noting that, within SAR, the CAT + Γ4 model recovered alveolates and stramenopiles together with maximal support. This difference compared with the LG + Γ4 model is probably due to better prediction of homoplasic positions that have accumulated on the fast-evolving alveolates and rhizarians.
(c). How robust is this new topology?
The case for a monophyletic Hacrobia has mostly rested on the photosynthetic haptophytes and cryptophytes [25], so to test their possible polyphyly suggested in figure 1 we removed taxa in several permutations. The rationale behind this is twofold: removing unstable or deviating taxa might help to identify artefactual groupings and at the same time improve the resolution across the tree in general. We first removed the three heterotrophic hacrobian lineages (telonemids, centrohelids and picobiliphytes) that were represented by a single member in our dataset and failed to show clear evolutionary affinities in figure 1. In this analysis (figure 2), the KC group remained closely related to plants but the CAT bootstrap for this relationship increased to 79 per cent. More strikingly, the haptophytes–SAR grouping received near-maximal support (1.0 PP; 98% CAT-BP), which demonstrates the negative effect of the three removed lineages on the general stability of the tree while providing a clear evolutionary framework for haptophytes.
Second, the addition of katablepharids to a phylogenomic dataset and its strong grouping with cryptophytes created the opportunity to test the position of the KC group by alternatively removing one of its members, which is of interest since cryptophytes branched with haptophytes in previous analyses of smaller datasets without katablepharids [22,24]. The removal of R. truncata led to no topological change, suggesting that the polyphyly of hacrobians was not simply due to adding katablepharid data (electronic supplementary material, figure S3). However, the converse did not hold: when cryptophytes (and picobiliphytes) were removed, R. truncata branched specifically with glaucophytes (electronic supplementary material, figure S4, 0.98 PP). One model violation that may be responsible for this grouping is amino acid compositional heterogeneity. To verify that this was not biasing our inference, we measured the compositional deviation of each taxon in a posterior predictive test (electronic supplementary material, table S2 and figure S4). Interestingly, among the taxa that did not significantly deviate from the global empirical frequencies were R. truncata and the glaucophyte Glaucocystis nostochinearum, while the second glaucophyte, Cyanophora paradoxa, was only slightly below the 5 per cent threshold. The amino acid composition of this dataset was also visualized as a two-dimensional plot in a PCA (electronic supplementary material, figure S5), altogether demonstrating that the composition of R. truncata and both glaucophytes are not similar. However, when the amino acids were recoded into six biochemically similar categories and the resulting alignment analysed as above, the R. truncata–glaucophyte association was not recovered (electronic supplementary material, figure S6). Data recoding have been used to weaken possible compositional biases [48], so this analysis suggests that this association may be due in part to compositional heterogeneity undetected in the PCA.
Another characteristic that may account for the grouping of katablepharids and glaucophytes is their relatively slow rate of evolution—in a tree hypothetically rooted between unikonts and bikonts, R. truncata and glaucophytes indeed displayed the shortest branches (electronic supplementary material, figure S4 and table S3). It has been shown that sequences with lower-than-average rates can artificially group together based on shared ancestral positions lost from other taxa, an artefact known as ‘short-branch exclusion’ [51]. Thus, it was important to confirm that this pattern can be overcome when either branch leading to R. truncata or glaucophytes is interrupted by faster-evolving lineages. Reintroducing cryptophytes to this dataset appeared to reduce the attraction between R. truncata and glaucophytes, so that the KC group branched as sister to Plantae (electronic supplementary material, figure S7). Reintroducing picobiliphytes appeared to have the same effect by breaking the short branch leading to glaucophytes, and also showed R. truncata as sister to Plantae (electronic supplementary material, figure S8).
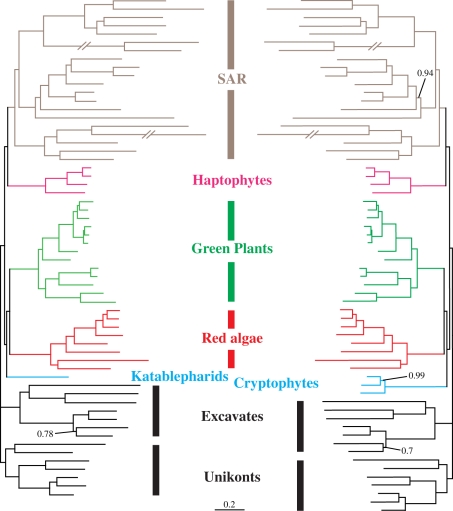
To evaluate whether the relationship between the KC group and Plantae is itself a result of the artefactual attraction between R. truncata and glaucophytes, we tested the consistency of the inferred relationships after the removal of glaucophytes from three datasets: from that shown in figure 1, and two alignments corresponding to figure 2 but with cryptophytes or katablepharids alternately removed. The first dataset met the expectations: the KC group branched with red algae + green plants (and picobiliphytes), distantly related to haptophytes, centrohelids and telonemids (electronic supplementary material, figure S9). Similarly, with only cryptophytes or R. truncata included in the absence of glaucophytes, we recovered fully resolved trees in which either was sister to red algae and green plants, again distantly related from haptophytes (figure 3). This illustrates the general consistency of the dataset throughout the taxa removal experiments, by showing both plastid-lacking (katablepharids) and photosynthetic (cryptophytes) representatives occupying the very same phylogenetic position.
Figure 3.
Phylogenies inferred with the CAT + Γ4 model, based on the dataset shown in figure 2, but with glaucophytes and either cryptophytes (right) or katablepharids (left) removed. Support values are indicated when not maximal (PP). No value at a node means support equal to 1.0 PP. All branches are drawn to scale, except the branches leading to Ciliates and Foraminifera, which were shortened for practical reasons. Scale bar, substitutions per position.
(d). Implication of a polyphyletic Hacrobia on plastid evolution
Overall, two significant conclusions emerge from this work. First, after more than doubling the amount of data, our analyses show that nuclear gene data do not support the Hacrobia hypothesis. Second, the phylogenetic position of a major eukaryotic lineage, haptophytes, is now robustly inferred to be closely related to the SAR group. This is the strongest case, receiving nearly maximal support when the more unstable lineages were removed (figure 2), which is noteworthy because this is the largest and most ecologically significant of all the hacrobian lineages. The relationship between the KC group and the Plantae is unfortunately less solid, yet is highly consistent in all datasets analysed. Importantly, regardless of where this group will finally branch, these analyses suggest that a monophyletic cryptophyte–haptophyte host lineage is unlikely. Telonemids, centrohelids and picobiliphytes remain of uncertain evolutionary origin, and sequencing more from related species is now crucial.
This result contrasts with some plastid phylogenies that showed the monophyly of photosynthetic hacrobians [13], but is congruent with others where stramenopiles and haptophytes branched together [8,14,52]. Regardless of which plastid phylogeny is correct, however, they all favour a unique secondary endosymbiosis giving rise to plastids in hacrobians and SAR taxa (because these plastids are monophyletic to the exclusion of the red algal plastids). Moreover, the monophyly of hacrobian plastids is also supported by their shared possession of a horizontally transferred ribosomal protein-encoding gene rpl36, which is found in no other plastid lineages [19]. Squaring this plastid data with that of their hosts has long been the challenge to sorting out the history of red algal plastids. In general, the evolution of the host lineages has proven not so much contradictory as difficult to resolve [22,33,53]. Now, however, our analysis of the largest collection of host-derived data with the most sophisticated models converged towards a scenario at the very least inconsistent with a simple vertical inheritance of secondary red plastids in hacrobians, and by extension in chromalveolates as a whole.
There are two obvious possibilities to explain the discrepancy between the host and plastid data. The first is simply that one set of phylogenies is misleading. The monophyly of chromalveolate plastids might be due to very poor red algal or haptophyte and cryptophyte sampling, as well as the deceiving behaviour of alveolate plastid data making it mostly unusable in phylogenies. Conversely, the polyphyly of hacrobians based on nuclear data might also be the result of limited taxon sampling or model misspecifications. The other possibility, however, is that the plastids are genuinely monophyletic and the hosts are not, a situation that could have arisen if higher-order endosymbioses took place, as predicted by the phylogenomic falsification of the chromalveolate hypothesis [33] or the recent study of endoplasmic reticulum-associated degradation (ERAD) components implicated in protein import across plastid membranes [54]. In this latter study, however, the host ERAD copies point to a common origin for haptophytes and cryptophytes, a scenario that we did not recover here. Yet any models that reconcile plastid and host data by means of additional layers of endosymbiosis should explain several observations. On the plastid side, these include: monophyletic hacrobian plastids (in particular, the shared rpl36 transfer in hacrobians), monophyletic alveolate and stramenopile plastids, and likely monophyletic chromalveolate plastids as a whole. On the host side, these now include: monophyletic SAR and haptophyte hosts, and the possibly independent origin of the KC host component. The emerging central difficulties with the most simple version of the chromalveolate hypothesis are therefore all related to the KC group, which should serve to refocus our efforts on the evolution of this lineage.
While the topology of the host lineages may complicate some aspects of plastid distribution in the ways noted earlier, it can also simplify others. In particular, many lineages of chromalveolates lack plastids, but in some alveolates where plastid ancestry can be reasonably inferred, evidence for this ancestry has been found in their nuclear genomes (e.g. Perkinsus and Oxyrrhis [6,45]). Many hacrobian lineages also lack plastids, but based on the host phylogeny described here at least some of these are not necessarily secondarily plastid-lacking; instead their ancestors may have never possessed the capacity to use light to produce energy. For example, we found no molecular evidence in the R. truncata transcriptome for a cryptic plastid or plastid-derived genes, and similarly no such genes were unambiguously identified in the metagenome of picobiliphytes [47], suggesting the ancestral state of the KC clade (and picobiliphytes) was non-photosynthetic.
Developing a robust integrative theory for plastid evolution conciliating plastid and host data could come from expanding taxon sampling in two ways. First, adding more sampling to sparsely sampled groups, including host data from new KC lineages, glaucophytes, telonemids, centrohelids and haptophytes, and plastid data from red algae. Second, new lineages with potentially intermediate positions can be very important, as the R. truncata transcriptome shows, and Palpitomonas or Rappemonads are already identified candidates [28,32]. We expect that several such new lineages with close affinities to ‘hacrobian’ taxa will be found, and it remains to be seen how such data will affect the larger picture of plastid evolution.
Acknowledgements
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (227301), and by a grant from the Tula Foundation to the Centre for Microbial Diversity and Evolution. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under contract no. DE-AC02-05CH11231. We thank J. M. Archibald, M. W. Gray, G. I. McFadden and C. E. Lane for project contributions to the sequencing of the nuclear genome of G. theta and B. natans. We thank Thierry Heger for sharing R scripts for performing the principal component analysis. F.B. thanks the Swiss National Science Foundation for a prospective researcher post-doctoral fellowship. P.J.K. is a Fellow of the Canadian Institute for Advanced Research.
References
- 1.Gould S. B., Waller R. F., McFadden G. I. 2008. Plastid evolution. Annu. Rev. Plant. Biol. 59, 491–517 10.1146/annurev.arplant.59.032607.092915 (doi:10.1146/annurev.arplant.59.032607.092915) [DOI] [PubMed] [Google Scholar]
- 2.Keeling P. J. 2004. Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 91, 1481–1493 10.3732/ajb.91.10.1481 (doi:10.3732/ajb.91.10.1481) [DOI] [PubMed] [Google Scholar]
- 3.Reyes-Prieto A., Weber A. P., Bhattacharya D. 2007. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168 10.1146/annurev.genet.41.110306.130134 (doi:10.1146/annurev.genet.41.110306.130134) [DOI] [PubMed] [Google Scholar]
- 4.Palmer J. D., Soltis D. E., Chase M. W. 2004. The plant tree of life: an overview and some points of view. Am. J. Bot. 91, 1437–1445 10.3732/ajb.91.10.1437 (doi:10.3732/ajb.91.10.1437) [DOI] [PubMed] [Google Scholar]
- 5.Howe C., Barbrook A., Nisbet R., Lockhart P., Larkum A. W. 2008. The origin of plastids. Proc. R. Soc. B 363, 2675–2685 10.1098/rstb.2008.0050 (doi:10.1098/rstb.2008.0050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowack E. C., Melkonian M., Glöckner G. 2008. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418 10.1016/j.cub.2008.02.051 (doi:10.1016/j.cub.2008.02.051) [DOI] [PubMed] [Google Scholar]
- 7.Archibald J. M. 2009. The puzzle of plastid evolution. Curr. Biol. 19, R81–R88 10.1016/j.cub.2008.11.067 (doi:10.1016/j.cub.2008.11.067) [DOI] [PubMed] [Google Scholar]
- 8.Rogers M. B., Gilson P. R., Su V., McFadden G. I., Keeling P. J. 2007. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol. Biol. Evol. 24, 54–62 10.1093/molbev/msl129 (doi:10.1093/molbev/msl129) [DOI] [PubMed] [Google Scholar]
- 9.Keeling P. J. 2009. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 56, 1–8 10.1111/j.1550-7408.2008.00371.x (doi:10.1111/j.1550-7408.2008.00371.x) [DOI] [PubMed] [Google Scholar]
- 10.Lane C. E., Archibald J. M. 2008. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol. Evol. 23, 268–275 10.1016/j.tree.2008.02.004 (doi:10.1016/j.tree.2008.02.004) [DOI] [PubMed] [Google Scholar]
- 11.Cavalier-Smith T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 46, 347–366 [DOI] [PubMed] [Google Scholar]
- 12.Cavalier-Smith T. 2000. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5, 174–182 [DOI] [PubMed] [Google Scholar]
- 13.Janouskovec J., Horak A., Obornik M., Lukes J., Keeling P. J. 2010. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl Acad. Sci. USA 107, 10 949–10 954 10.1073/pnas.1003335107 (doi:10.1073/pnas.1003335107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon H. S., Hackett J. D., Pinto G., Bhattacharya D. 2002. The single, ancient origin of chromist plastids. Proc. Natl Acad. Sci. USA 99, 15 507–15 512 10.1073/pnas.242379899 (doi:10.1073/pnas.242379899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fast N. M., Kissinger J. C., Roos D. S., Keeling P. J. 2001. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 18, 418–426 [DOI] [PubMed] [Google Scholar]
- 16.Harper J. T., Keeling P. J. 2003. Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol. Biol. Evol. 20, 1730–1735 10.1093/molbev/msg195 (doi:10.1093/molbev/msg195) [DOI] [PubMed] [Google Scholar]
- 17.Patron N. J., Rogers M. B., Keeling P. J. 2004. Gene replacement of fructose-1,6-bisphosphate aldolase supports the hypothesis of a single photosynthetic ancestor of chromalveolates. Eukaryot. Cell 3, 1169–1175 10.1128/EC.3.5.1169-1175.2004 (doi:10.1128/EC.3.5.1169-1175.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teich R., Zauner S., Baurain D., Brinkmann H., Petersen J. 2007. Origin and distribution of Calvin cycle fructose and sedoheptulose bisphosphatases in plantae and complex algae: a single secondary origin of complex red plastids and subsequent propagation via tertiary endosymbioses. Protist 158, 263–276 10.1016/j.protis.2006.12.004 (doi:10.1016/j.protis.2006.12.004) [DOI] [PubMed] [Google Scholar]
- 19.Rice D. W., Palmer J. D. 2006. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 4, 31. 10.1186/1741-7007-4-31 (doi:10.1186/1741-7007-4-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burki F., Shalchian-Tabrizi K., Minge M. A., Skjaeveland A., Nikolaev S. I., Jakobsen K. S., Pawlowski J. 2007. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 2, e790. 10.1371/journal.pone.0000790 (doi:10.1371/journal.pone.0000790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackett J. D., Yoon H. S., Li S., Reyes-Prieto A., Rümmele S. E., Bhattacharya D. 2007. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol. 24, 1702–1713 10.1093/molbev/msm089 (doi:10.1093/molbev/msm089) [DOI] [PubMed] [Google Scholar]
- 22.Burki F., et al. 2009. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, telonemia and centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol. Evol. 1, 231–238 10.1093/gbe/evp022 (doi:10.1093/gbe/evp022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burki F., Shalchian-Tabrizi K., Pawlowski J. 2008. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol. Lett. 4, 366–369 10.1098/rsbl.2008.0224 (doi:10.1098/rsbl.2008.0224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patron N. J., Inagaki Y., Keeling P. J. 2007. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr. Biol. 17, 887–891 10.1016/j.cub.2007.03.069 (doi:10.1016/j.cub.2007.03.069) [DOI] [PubMed] [Google Scholar]
- 25.Okamoto N., Chantangsi C., Horak A., Leander B. S., Keeling P. J. 2009. Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the hacrobia taxon nov. PLoS ONE 4, e7080. 10.1371/journal.pone.0007080 (doi:10.1371/journal.pone.0007080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavalier-Smith T., von der Heyden S. 2007. Molecular phylogeny, scale evolution and taxonomy of centrohelid heliozoa. Mol. Phylogenet. Evol. 44, 1186–1203 10.1016/j.ympev.2007.04.019 (doi:10.1016/j.ympev.2007.04.019) [DOI] [PubMed] [Google Scholar]
- 27.Cuvelier M. L., Ortiz A., Kim E., Moehlig H., Richardson D. E., Heidelberg J. F., Archibald J. M., Worden A. Z. 2008. Widespread distribution of a unique marine protistan lineage. Environ. Microbiol. 10, 1621–1634 10.1111/j.1462-2920.2008.01580.x (doi:10.1111/j.1462-2920.2008.01580.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E., Harrison J. W., Sudek S., Jones M. D., Wilcox H. M., Richards T. A., Worden A. Z., Archibald J. M. 2011. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc. Natl Acad. Sci. USA 108, 1496–1500 10.1073/pnas.1018259108 (doi:10.1073/pnas.1018259108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Not F., Valentin K., Romari K., Lovejoy C., Massana R., Töbe K., Vaulot D., Medlin L. K. 2007. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science 315, 253–255 10.1126/science.1136264 (doi:10.1126/science.1136264) [DOI] [PubMed] [Google Scholar]
- 30.Okamoto N., Inouye I. 2005. The katablepharids are a distant sister group of the cryptophyta: a proposal for Katablepharidophyta divisio nova/Kathablepharida phylum novum based on SSU rDNA and beta-tubulin phylogeny. Protist 156, 163–179 10.1016/j.protis.2004.12.003 (doi:10.1016/j.protis.2004.12.003) [DOI] [PubMed] [Google Scholar]
- 31.Shalchian-Tabrizi K., et al. 2006. Telonemia, a new protist phylum with affinity to chromist lineages. Proc. R. Soc. B 273, 1833–1842 10.1098/rspb.2006.3515 (doi:10.1098/rspb.2006.3515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yabuki A., Inagaki Y., Ishida K. 2010. Palpitomonas bilix gen. et sp. nov.: a novel deep-branching heterotroph possibly related to archaeplastida or hacrobia. Protist 161, 523–538 10.1016/j.protis.2010.03.001 (doi:10.1016/j.protis.2010.03.001) [DOI] [PubMed] [Google Scholar]
- 33.Baurain D., et al. 2010. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes and stramenopiles. Mol. Biol. Evol. 27, 1698–1709 10.1093/molbev/msq059 (doi:10.1093/molbev/msq059) [DOI] [PubMed] [Google Scholar]
- 34.Bodyl A., Stiller J. W., Mackiewicz P. 2009. Chromalveolate plastids: direct descent or multiple endosymbioses? Trends Ecol. Evol. 24, 119–121 10.1016/j.tree.2008.11.003 (doi:10.1016/j.tree.2008.11.003) [DOI] [PubMed] [Google Scholar]
- 35.Dorrell R. G., Smith A. G. 2011. Do red and green make brown? Perspectives on plastid acquisitions within chromalveolates. Eukaryot. Cell 10, 856–868 10.1128/EC.00326-10 (doi:10.1128/EC.00326-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez-Puerta M. V., Delwiche C. F. 2008. A hypothesis for plastid evolution in chromalveolates. J. Phycol. 44, 1097–1107 10.1111/j.1529-8817.2008.00559.x (doi:10.1111/j.1529-8817.2008.00559.x) [DOI] [PubMed] [Google Scholar]
- 37.Douglas S. E., et al. 2001. The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091–1096 10.1038/35074092 (doi:10.1038/35074092) [DOI] [PubMed] [Google Scholar]
- 38.Frada M., Probert I., Allen M. J., Wilson W. H., de Vargas C. 2008. The ‘Cheshire Cat’ escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc. Natl Acad. Sci. USA 105, 15 944–15 949. 10.1073/pnas.0807707105 (doi:10.1073/pnas.0807707105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burki F., Kudryavtsev A., Matz M. V., Aglyamova G. V., Bulman S., Fiers M., Keeling P. J., Pawlowski J. 2010. Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol. Biol. 10, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lartillot N., Lepage T., Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 10.1093/bioinformatics/btp368 (doi:10.1093/bioinformatics/btp368) [DOI] [PubMed] [Google Scholar]
- 41.Felsenstein J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5, 164–166 [Google Scholar]
- 42.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 43.Brinkmann H., Philippe H. 1999. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol. Biol. Evol. 16, 817–825 [DOI] [PubMed] [Google Scholar]
- 44.Kostka M., Uzlikova M., Cepicka I., Flegr J. 2007. SlowFaster, a user-friendly program for slow–fast analysis and its application on phylogeny of Blastocystis. BMC Bioinformatics 9, 341. 10.1186/1471-2105-9-341 (doi:10.1186/1471-2105-9-341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuzaki M., Kuroiwa H., Kuroiwa T., Kita K., Nozaki H. 2008. A cryptic algal group unveiled: a plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol. Biol. Evol. 25, 1167–1179 10.1093/molbev/msn064 (doi:10.1093/molbev/msn064) [DOI] [PubMed] [Google Scholar]
- 46.Slamovits C. H., Keeling P. J. 2008. Plastid-derived genes in the nonphotosynthetic alveolate Oxyrrhis marina. Mol. Biol. Evol. 25, 1297–1306 10.1093/molbev/msn075 (doi:10.1093/molbev/msn075) [DOI] [PubMed] [Google Scholar]
- 47.Yoon H., Price D., Stepanauskas R., Rajah V., Sieracki M., Wilson W., Yang E., Duffy S., Bhattacharya D. 2011. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332, 714–717 10.1126/science.1203163 (doi:10.1126/science.1203163) [DOI] [PubMed] [Google Scholar]
- 48.Philippe H., Brinkmann H., Lavrov D., Littlewood D., Manuel M., Worheide G., Baurain D. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9, e1000602. 10.1371/journal.pbio.1000602 (doi:10.1371/journal.pbio.1000602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Ezpeleta N., Brinkmann H., Roure B., Lartillot N., Lang B. F., Philippe H. 2007. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 56, 389–399 10.1080/10635150701397643 (doi:10.1080/10635150701397643) [DOI] [PubMed] [Google Scholar]
- 50.Lartillot N., Philippe H. 2008. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Proc. R. Soc. B 363, 1463–1472 10.1098/rstb.2007.2236 (doi:10.1098/rstb.2007.2236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiller J. W., Harrell L. 2005. The largest subunit of RNA polymerase II from the Glaucocystophyta: functional constraint and short-branch exclusion in deep eukaryotic phylogeny. BMC Evol. Biol. 5, 71. 10.1186/1471-2148-5-71 (doi:10.1186/1471-2148-5-71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan H., Parks N., Kozera C., Curtis B. A., Parsons B. J., Bowman S., Archibald J. M. 2007. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol. Biol. Evol. 24, 1832–1842 10.1093/molbev/msm101 (doi:10.1093/molbev/msm101) [DOI] [PubMed] [Google Scholar]
- 53.Harper J. T., Waanders E., Keeling P. J. 2005. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. Syst. Evol. Micr. 55, 487–496 10.1099/ijs.0.63216-0 (doi:10.1099/ijs.0.63216-0) [DOI] [PubMed] [Google Scholar]
- 54.Felsner G., Sommer M. S., Gruenheit N., Hempel F., Moog D., Zauner S., Martin W., Maier U. G. 2011. ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane. Genome Biol. Evol. 3, 140–150 10.1093/gbe/evq074 (doi:10.1093/gbe/evq074) [DOI] [PMC free article] [PubMed] [Google Scholar]