Abstract
Formation of cytochrome c (cyt c)/cardiolipin (CL) peroxidase complex selective towards peroxidation of polyunsaturated CLs is a pre-requisite for mitochondrial membrane permeabilization. Tyrosine residues – via the generation of tyrosyl radicals (Tyr•) - are likely reactive intermediates of the peroxidase cycle leading to CL peroxidation. We used mutants of horse heart cyt c in which each of the four Tyr residues was substituted for Phe and assessed their contribution to the peroxidase catalysis. Tyr67Phe mutation was associated with a partial loss of the oxygenase function of the cyt c/CL complex and the lowest concentration of H2O2-induced Tyr radicals in electron paramagnetic resonance (EPR) spectra. Our MS experiments directly demonstrated decreased production of CL-hydroperoxides (CL-OOH) by Tyr67Phe mutant. Similarly, oxidation of a phenolic substrate, Amplex Red, was affected to a greater extent in Tyr67Phe than in three other mutants. Tyr67Phe mutant exerted high resistance to H2O2-induced oligomerization. Measurements of Tyr fluorescence, hetero-nuclear magnetic resonance (NMR) and computer simulations position Tyr67 in close proximity to the porphyrin ring heme iron and one of the two axial heme-iron ligand residues, Met80. Thus, the highly conserved Tyr67 is a likely electron-donor (radical acceptor) in the oxygenase half-reaction of the cyt c/CL peroxidase complex.
Keywords: cytochrome c, cardiolipin, tyrosine, cardiolipin hydroperoxides, peroxidase
1. Introduction
Tunneling electrons between respiratory complexes III and IV is the major function of cytochrome c (cyt c) in normal mitochondria [1]. The newly discovered role of cyt c released into the cytosol in activating caspase cascades does not depend on its redox propensities [2]. However, recently reported participation of cyt c in selective oxidation of cardiolipin (CL) – a prerequisite for mitochondrial membranes permeabilization - heavily relies on its heme-iron related redox features [3]. A series of studies from this and other laboratories established the major stages involved in the process: 1) trans-membrane migration of CL from the inner to the outer mitochondrial membranes facilitated by phosphorylation of scramblase-3 [4-6], and possibly by mitochondrial creatine-kinase and dinucleotide phosphokinase-D [7, 8]; 2) formation of high affinity cyt c/CL complex with peroxidase activity [9]; 3) selective peroxidation of CL [10, 11]; 4) dissociation of cyt c from peroxidized CL; 5) participation of peroxidized CL in mitochondrial membrane permeabilization [3]. However, the mechanisms underlying the new catalytic identity of cyt c/CL complex have not been sufficiently characterized.
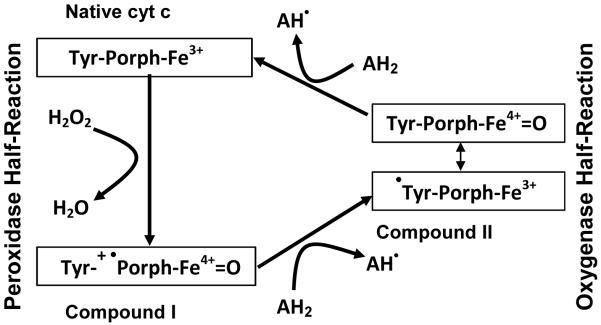
CL peroxidation includes two half-reactions catalyzed by cyt c/CL complex: 1) peroxidase half-reaction dealing with splitting of the molecules of oxidizing equivalents through homolytic and/or heterolytic mechanisms, and 2) oxygenase half-reaction leading to oxygenation of CL (Fig. 1). It has been established that H2O2 and small organic hydroperoxides (eg, tert-butyl hydroperoxide) undergo predominantly homolytic splitting on cyt c/CL complexes, while long fatty acid hydroperoxides provide for almost three orders of magnitude higher rate of the peroxidase reaction acting mainly through the heterolytic mechanism [12]. It was shown for different hemoproteins that compound I formed as a result of peroxidase half-reaction, contains two oxidizing equivalents more than the resting enzyme. One electron is removed from the iron to give the oxoferryl [Fe(IV)=O] intermediate, and a second electron is removed from the porphyrin to give a porphyrin π-cation radical. Compound I can be reduced back to the ferric enzyme by two consecutive one-electron reactions via the formation of a second enzyme intermediate, Compound II, which retains the oxoferryl group but not the porphyrin cation radical. This reduction occurs at the expense of oxidation of one of endogenous amino-acid residues (eg, tyrosine residues of cyt c) [13]. Compound II drives the oxygenase half-reaction of the cycle leading to a protein radical-driven hydrogen abstraction from a substrate, formation of a carbon-centered lipid radical and, upon addition of molecular oxygen, a peroxyl radical and an oxygenated product [14]. Our previous work indicates that tyrosyl radicals (Tyr•) are likely candidates for this role in cyt c/CL catalyzed peroxidation of polyunsaturated CLs [15] and abstraction of hydrogen from hydroxy-groups of phenolic compounds (eg, etoposide) [11]. Given that there are four Tyr residues in cyt c, we decided to more closely examine the possible role of these Tyr residues in the oxygenation of CL and phenolic compounds. To this end, we used cyt c mutants in which each of the Tyr residues was substituted for Phe. By applying MS analysis of CL peroxidation products, fluorescence characterizations of Tyr/Trp residues in cyt c and phenol oxidation products, as well as by molecular modeling and NMR spectroscopy, we concluded that Tyr67 is the prime candidate likely involved in oxygenation reactions (including CL oxygenation) while other Tyr residues remain catalytically less competent.
Figure 1. General scheme of peroxidase cycle of cyt c.
Peroxidase half-reaction utilizes hydrogen peroxide and other oxidizing equivalents to oxidize the native enzyme to compound I. Compound I can be reduced back to the ferric enzyme by two consecutive one-electron reactions via the formation of a second enzyme intermediate, Compound II, which retains the oxoferryl group but not the porphyrin cation radical. This reduction occurs at the expense of oxidation of one of endogenous amino-acid residues (eg, tyrosine residues of cyt c). Compound II drives the oxygenase half-reaction of the cycle leading to a protein radical-driven hydrogen abstraction from a substrate, formation of a carbon-centered lipid radical and, upon addition of molecular oxygen, a peroxyl radical and an oxygenated product. Porph, porphyrin ring of cyt c; Tyr, Tyrosine residues of cyt c; AH2 and AH•, substrates being oxidized and the formed radical product, respectively. The scheme is adapted from [45] with modifications.
2. Materials and methods
2.1. Reagents
Horse heart cytochrome c (cyt c , type C-7752, >95%), diethylenetriaminepentaacetic acid (DTPA), guanidine hydrochloride (GndCl), coomassie brilliant blue, n-hexane, 2-propanol, hydrogen peroxide (H2O2), and 15N isotope labeled NH4Cl were purchased from Sigma-Aldrich (St. Louis, MO). 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,1′,2,2′-tetraoleoyl-cardiolipin (TOCL) and 1,1′,2,2′-tetralinoleoyl-cardiolipin (TLCL) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). LB medium and SilverSNAP stain kit was purchased from Thermo Fisher Scientific (Rockford, IL). CM-Sepharose fast flow column was purchased from Amersham Pharmacia Biotech, Inc. (Uppsala, Sweden), Amplex Red (N-acetyl-3,7-dihydroxyphenoxazine) reagent was obtained from Molecular Probes (Eugene, OR). The plasmid pJRhrsN2 was kindly provided by Dr. Jon Rumbley, Chemistry Department, University of Minnesota (Duluth, MN).
2.2. Protein expression and purification
Construction of the mutant plasmids pJRhrsN2 containing Phe substitutions at each of the four Tyr positions individually and altogether was described in [16]. In addition to Tyr substitutions, the cyt c gene also carries two additional replacements, His26Asn and His33Asn associated with the higher expression yields in E. coli [17]. This mutation results in protein that is structurally and functionally similar to the wild-type [17, 18] and also with a higher peroxidase activity in the absence of CL [19]. Proteins were expressed and purified as previously reported [16, 17]. Briefly, recombinant WT and Tyr mutant proteins were expressed in E. coli by growing them either in LB medium [16] or minimal media containing 15N NH4Cl [18]. The expressed proteins were purified using a CM-Sepharose fast flow column [16-18]. Fractions with a 410/280 nm absorbance ratio > 4.0 were collected, and their purity was checked by SDS-PAGE analysis using coomassie blue staining and silver staining. Further, the purified protein was either lyophilized or stored at −80 °C after snap freezing the sample using liquid nitrogen (for NMR purposes).
2.3. Small unilamellar liposomes
Liposomes were prepared from DOPC and CL (1:1 ratio) by sonication. Individual phospholipids, stored in chloroform, were mixed and dried under nitrogen. Following this, the lipids were mixed in HEPES buffer (20 mM with 100 μM DTPA (pH 7.4)) by vortexing. Such preparations were further sonicated five times for 30s on ice using an ultrasonic probe tip sonicator (Cole-Palmer Ultrasonic Homogenizer, 20 kHz, Cole-Parmer Instrument Company,Vernon Hills, IL). Liposomes were used immediately after preparation.
2.4. Fluorescence measurements
Tryptophan and tyrosine fluorescence of cyt c in the presence of liposomes was measured using PC1 steady-state photon counting spectrofluorimeter (ISS Inc., Champaign, IL) using quartz cuvettes with volume of 50 μl. The excitation wavelengths were 275 nm and 289 nm. Fluorescence was measured in 20 mM HEPES buffer (pH7.4) with 100 μM DTPA. Before measurements cyt c (10 μM) was incubated with liposomes (TOCL/cyt c ratio 20:1). Concentration of cyt c was determined by absorbance of Soret band using UV160U spectrophotometer (Shimadzu).
2.5. Peroxidase activity measurements
Assessment of peroxidase activity with Amplex Red was performed using the fluorescence of resorufin – an oxidation product of Amplex Red - (λex - 570 nm, λem - 585 nm); 1 μM cyt c was incubated with liposomes (TOCL/cyt c ratio 20:1) for 10 min. 20 mM HEPES (pH 7.4) or PBS both containing 100 μM DTPA were used as buffers. Then 50 μM Amplex Red and 25 μM H2O2 were added, and the incubation proceeded for an additional 20 min. Fluorescence was detected by employing a “Fusion α” universal microplate analyzer and by using an excitation wavelength of 535/25 nm and an emission wavelength of 590/20 nm.
2.6. Assessment of oxidized molecular species of CL by mass spectrometry
To assess different oxidized molecular species of CL, electro-spray-ionization-liquid chromatography mass-spectrometry (LC/ESI-MS) was performed using a Dionex UltimateTM 3000 HPLC coupled online to an ESI ion source and a linear ion trap mass spectrometer (LXQ Thermo-Fisher) with the Xcalibur operating system (Thermo Fisher Scientific, San Jose, CA). CL and its oxidized molecular species were extracted by the Folch procedure [20] and separated on a normal phase column (Luna 3 μm Silica 100A, 150 × 2 mm, Phenomenex, Torrance CA) with a flow rate of 0.2 mL/min applying a gradient elution using solvents containing 5 mM CH3COONH4 (A - n-hexane : 2-propanol : water, 43:57:1 (v/v/v); and B - n-hexane : 2-propanol : water, 43:57:10 (v/v/v). Analysis of (hydroperoxy- and hydroxy-) oxidized phospholipid species was performed as described [10].
2.7. NMR spectroscopy studies
1H-15N HSQC NMR spectra of uniformly 15N isotope labeled horse heart cyt c were obtained using a ~900 MHz Bruker spectrometer. Two dimensional 1H-15N HSQC spectra of wild type cyt c were acquired using a standard HSQC pulse sequence with 64 scans in first dimension and 160 scans in the second dimension and a D1 of 1 sec. Data acquisition was carried out using Topspin Version 3.0 Software and then the spectra were processed and analyzed using NMRView, and Sparky. The control spectrum of cyt c in the absence of TOCL contained 50 μM purified wild type cyt c and 2 mM DOPC liposomes dissolved in 50 mM potassium phosphate buffer (pH 6.0) and 10% D20. Liposomes containing 250 μM TOCL and 1 mM DOPC (20:80 % ratio of total lipid) were used to obtain the spectrum in the presence of TOCL. 50 μM of cyt c from a stock solution of 2.8 mM was added to each sample.
2.8. PAGE analysis
Cyt c (8 μM) was incubated with liposomes (TOCL/cyt c ratio 20:1) in 20 mM HEPES (pH 7.4) with 100 μM DTPA for 15 min at RT. Then samples were transferred to a water bath and incubated at 37 °C for an additional 1 h in the presence of H2O2 (35 μM). Proteins were separated by 10% SDS-PAGE in Tris-glycine buffer. The gels were stained by SilverSNAP stain kit according to the manufacturer’s manual.
2.9. EPR spectra of tyrosyl radicals
Spectra were assessed after the addition of 60 μM H2O2 to 20 μM cyt c incubated with liposomes (cyt c/CL ratio 1:20) at RT for 30 sec. The samples were frozen in liquid nitrogen and EPR spectra were recorded at 77 K under the following conditions: center field, 3230 G; sweep width, 50 G; field modulation, 5 G; microwave power, 10 mW; receiver gain, 2×103; time constant, 0.1 s; time scan, 1 min. After baseline corrections, concentrations of spins were determined by double integration of EPR signals as described by Wyard [21]. The concentration of spins was related to sample concentration by recording a 100 μM Cu (EDTA) standard at 77K. Double integration of the resulting signal was used to establish a proportionality constant between spin concentrations in EPR spectra and sample concentrations. This in turn was used to calculate the concentration of spin ½ radicals in wild type and mutant forms of cyt c.
2.10. Sequence alignment and calculation of conservation
A total of 406 protein sequences corresponding to the eukaryotic cyt c family were extracted from the swissprot sequence database [22] using the criteria “family: cytochrome c family AND fragment: no NOT taxonomy: bacteria”. The extracted protein sequences were aligned using ClustalW [23] to obtain the multiple sequence alignment (MSA). After deleting the gaps containing unaligned areas from the MSA, a sequence conservation/consensus plot was created using the weblogo tool [24].
3. RESULTS
3.1. Structure-based prediction of tyrosine involvement in radical formation
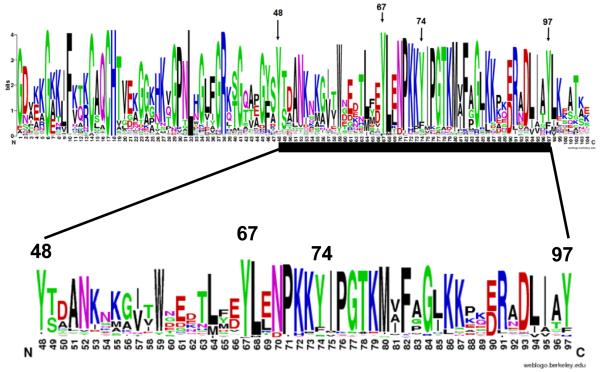
To obtain initial insights into the possible involvement of Tyr residues in catalysis of oxygenation reactions, we performed computational structural analysis of the relative locations of four Tyr with respect to the heme. The positions of the four Tyr residues (Tyr48, Tyr67, Tyr74 and Tyr97) in the native cyt c structure are shown in (Fig. 2). The distances between the −OH groups of these Tyr to the closest atom in the heme chromophore and to its iron are provided in (Table 1). The −OH groups of buried Tyr48 and Tyr67 residues are within 4 Å of the heme moiety and are thus much closer as compared to the surface-accessible Tyr74 and Tyr97. However, only Tyr67 lies in close proximity to the iron co-ordination site. Further, we examined the sequence conservation of the Tyr residues at positions 48, 67, 74 and 97 in cyt c family. We performed multiple sequence alignment of all the protein sequences that belong to ‘cyt c family’ and generated the sequence conservation plot (Fig. 3). Of the four tyrosines compared, Tyr67 is one the most highly conserved residues in cyt c, similar to residues His18, Cys13 and Cys17. The only mutation that occurred at this position was tyrosine to phenylalanine (frequency <10%). Other tyrosines, Tyr48, Tyr74 and Tyr97 in horse heart cyt c are significantly less conserved compared to Tyr67. Thus, the distance analysis along with sequence conservation is compatible with the involvement of Tyr67 in the oxygenase catalytic activity.
Figure 2. Location of tyrosines in cyt c with respect to the heme ligand.
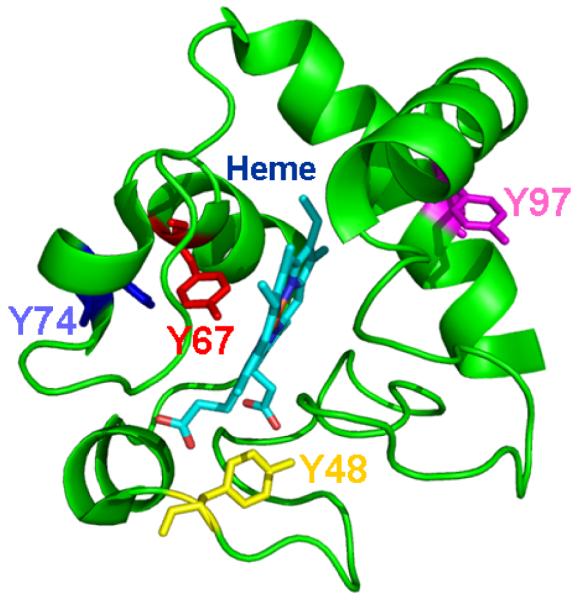
The x-ray crystal structure of horse heart cyt c (pdbid: 1HRC) was used to generate the image. Cyt c is represented as a cartoon and the heme moiety is rendered as sticks. Tyrosines 48, 67, 74 and 97 are also rendered as sticks and colored yellow, red, blue and magenta, respectively.
Table 1.
Distance measurements from the −OH groups of tyrosines in cyt c to Heme (row 1) and Iron (row 2). The distances reported for heme are those to the closest atom, which is listed in brackets.
| Tyr 48 | Tyr 67 | Tyr 74 | Tyr 97 | |
|---|---|---|---|---|
| Heme | 2.7Å (O1A) |
3.3Å (C2A) |
10.3 Å (O2A) |
10.1 Å (CMB) |
| Iron | 9.3 Å | 4.6 Å | 14.7 Å | 13.8 Å |
Figure 3. Sequence conservation in cyt c family.
Sequences corresponding to the eukaryotic cyt c family were downloaded from swissprot [22], aligned with ClustalW [23] and displayed using weblogo [24]. The total height of the stack at each position corresponds to the sequence conservation at that position, represented in bits. The height of different symbols with in a stack at a particular position corresponds to the relative frequency of that amino acid in the sequence.
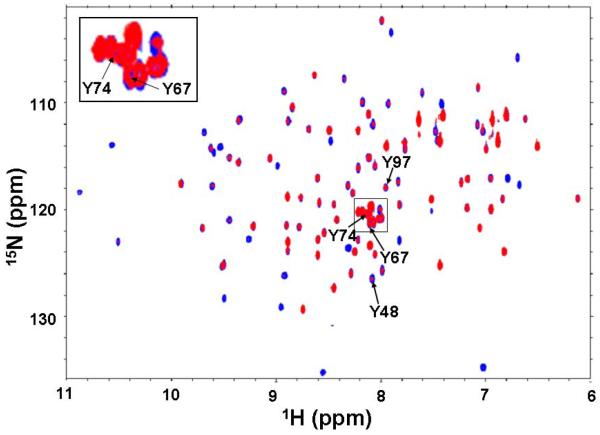
Because the oxygenase activity towards polyunsaturated CL requires CL binding to cyt c, ideally, we would have conducted the above distance measurements in the structure of the complex between cyt c and CL. However, since this structure is not known, we conducted NMR spectroscopic measurements to gauge the amount of change each tyrosine might experience upon CL binding. 1H, 15N heteronuclear single quantum coherence (HSQC) spectra correlate protons bound to nitrogen atoms and allow detection of backbone NH groups in proteins. Their signals are sensitive to the local environment experienced by each 1H, 15N pair and can thus be used to detect changes upon CL binding. The 1H, 15N-HSQC spectra of cyt c alone (blue) and in the presence of CL (red) are superimposed in (Fig. 4), and the positions of the signals arising from the four Tyr residues are labeled. There was no significant change in chemical shifts corresponding to any of the tyrosines - Tyr48 (15N - 126.48 ppm, 1H - 8.07 ppm), Tyr67 (15N - 121.24 ppm, 1H - 8.10 ppm), Tyr74 (15N - 120.37 ppm, 1H – 8.11 ppm), and Tyr97 (15N – 117.93 ppm, 1H – 7.94 ppm) when comparing the spectra in the presence and absence of TOCL. Signal intensities corresponding to Tyr48 in particular, and to a lesser extent of Tyr97 decreased while those of Tyr67 and Tyr74 remained essentially unchanged. This strongly indicates that there are possible changes in the dynamics of Tyr, but the structure remains likely unaltered, in particular for the tyrosine closest to the heme iron, Tyr67, whose signal intensity remains unaltered. We conclude that the prediction of tyrosine involvement in oxygenase activity based on the native cyt c structure remains valid for the CL-bound state.
Figure 4. NMR spectra of cyt c.
Overlay of two-dimensional HSQC spectra of wild type horse heart cyt c in the absence (colored blue) and presence (colored red) of TOCL. The peaks corresponding to the different Tyr residues are indicated by arrows and labeled accordingly. The area of the most crowded region in the spectrum, highlighted in a box is shown as an expanded inset for clarity purposes. A total of 50 μM wild type cyt c dissolved in 50 mM potassium phosphate (pH 6.0) was used to acquire each spectrum. The TOCL/cyt c ratio was kept at 5:1 in the red spectrum and DOPC to TOCL was maintained at a percent ratio of 80:20 in total lipid content.
3.2. Fluorescence of tyrosines and tryptophan
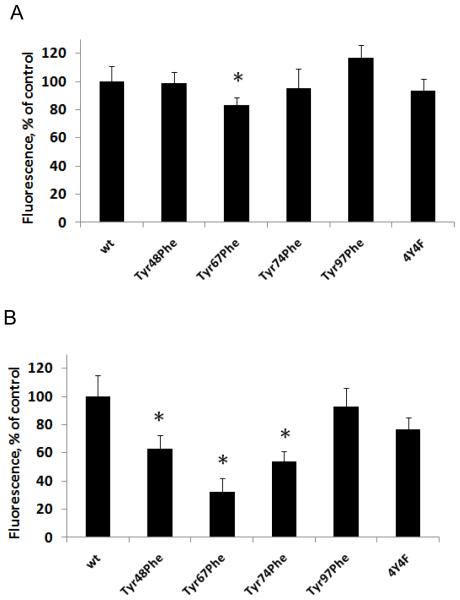
To experimentally validate the predicted proximity of Tyr67 to the heme in the CL-bound state and oxygenase catalysis, we employed measurements of intrinsic UV-fluorescence. In mammalian cyt c, both Tyr- and Trp-residues can potentially contribute to UV-fluorescence. In heme iron-deficient cyt c or in the presence of strong denaturants such as high concentrations of GndCl, fluorescence from Tyr and Trp residues is observed [25], while in native cyt c, the fluorescence is almost completely quenched via the energy transfer mechanism between these amino acid residues and the heme group. Binding of TOCL (in the form of TOCL/DOPC liposomes) to cyt c leads to the appearance of a weak but detectable Tyr/Trp fluorescence response from the protein (Fig. 5A). To resolve the contribution of the four different Tyr and the Trp to the overall UV fluorescence response from cyt c/TOCL complexes, we studied the fluorescence of Tyr mutants of cyt c.
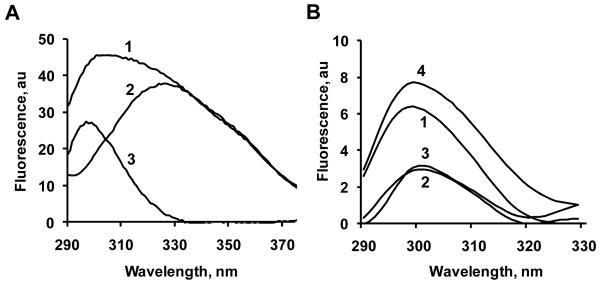
Figure 5. Fluorescence spectra of wild type and mutant forms of horse heart cyt c. Assessments of contribution of different Tyr residues in total tyrosine fluorescence of cyt c.
A – Typical fluorescence emission spectra of complexes of TOCL with wild type cyt c and 4Y4F mutant (λex 275 nm) (1-wt cyt c, 2-4Y4F mutant, 3-spectrum of Tyr residues of wild type cyt c). The spectra were obtained in 20 mM HEPES buffer (pH 7.4) with 100 μM DTPA using 10 μM of cyt c and TOCL/cyt c ratio 20:1.
B. - Fluorescence spectra of cyt c tyrosines calculated by the “subtraction” approach (1-Tyr48, 2-Tyr67, 3-Tyr74, 4-Tyr97). The fluorescence spectra after subtraction were smoothened using Gaussian convolution method provided by PeakFit v4.12.
In aqueous solutions, the fluorescence excitation maxima for Tyr are at shorter wavelengths (270 and 275 nm) than for Trp (280 and 289 nm) while the emission maxima of Tyr and Trp are at 303 and 350 nm, respectively. For cyt c/TOCL complexes, the emission spectrum of wild type cyt c contained features characteristic of Tyr and Trp fluorescence. Expectedly, only one maximum - corresponding to Trp fluorescence - was detectable for the 4Y4F mutant lacking all four Tyr residues (Fig. 5A). Accordingly, the emission spectra obtained for the complex of cyt c of 4Y4F mutant with TOCL using excitation at 275 nm (max for Tyr) and at 289 nm (max for Trp) differed in the magnitudes but had only one emission maximum. Based on these data, subtraction of 4YF4 spectrum from that of wild type cyt c yielded the spectrum corresponding to fluorescence of all four Tyr without interference from the Trp residue. This procedure has also been performed for each of the Tyr mutants to yield fluorescence spectra of three (out of total four) Tyr residues. Finally, by subtracting the spectra for the sum of three Tyr residues from the spectra obtained for all four Tyr residues, we obtained the spectra for individual Tyr residues. This protocol has been performed and the spectra for each of the four Tyr cyt c mutants were obtained in the presence of CL (Fig. 5A, B). The intensities of fluorescence from different Tyr were not equal and decreased in the following order: Tyr97>Tyr48>Tyr74=Tyr67. The fact that Tyr67’s fluorescence quenching was maximal (compared to other Tyr residues) suggests - in line with the computer modeling results - that Tyr67 is located in closest proximity to the heme-moiety in the cyt c/CL complex thus pointing to its possible involvement in the catalysis of oxygenase reactions.
3.3 Peroxidase activity of cyt c mutants
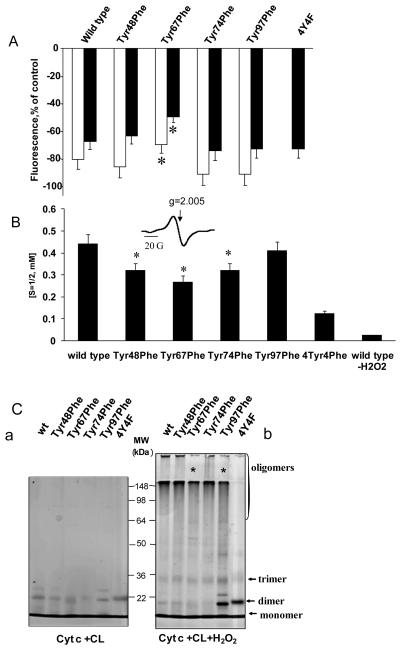
To experimentally assess the role of different Tyr residues in the oxidation reactions catalyzed by cyt c/TOCL complexes, we compared the efficiency of the mutants in oxidation of a prototypical phenolic substrate, Amplex Red, to its product, resorufin [26]. The Tyr67Phe mutant demonstrated a significantly decreased peroxidase activity as compared to other Tyr mutants (Fig. 6A, B). When the reaction was performed in PBS buffer, we also found that substitution of Tyr residues for Phe had inhibitory effect on peroxidase activity of two cyt c mutants – Tyr48Phe and Tyr74Phe (Fig. 6B). Experiments performed in 20 mM HEPES buffer, indicated that Tyr67Phe was the only mutant that demonstrated decreased peroxidase activity) (Fig. 6A). These results are compatible with the involvement of Tyr67 in catalysis of oxidative reactions towards a prototypical phenolic substrate, Amplex Red.
Figure 6. Peroxidase activity of complexes of cyt c with TOCL (wild type and mutated cyt c) measured by the fluorescence of resorufin (an Amplex Red oxidation product).
Incubation conditions: concentration of cyt c was 1 μM, TOCL/cyt c ratio was 20:1, 50 μM Amplex Red and 25 μM H2O2. Buffer-20 mM HEPES (pH 7.4), 100 μM DTPA (A), or PBS , 100 μM DTPA (B),*p<0.01 vs. wild-type cyt c.
Activation of cyt c into a peroxidase can be achieved by highly oxidizable polyunsaturated CLs (such as tetralinoleoyl-cardiolipin, TLCL) as well as by non-oxidizable saturated CLs (eg, tetramyristoyl-cardiolipin, TMCL) and/or mono-unsaturated CL (tetraoleoyl-cardiolipin, TOCL). In the latter cases and in the absence of other oxidizable exogenous (eg, phenolic) substrates, the oxidizing potential of cyt c/CL complexes is re-directed towards oxidation of its own Tyr and Trp residues resulting in their oxidative modification and, commonly, dimerization/oligomerization [11, 27, 28]. These oxidative events can be monitored by the disappearance of characteristic fluorescence of Tyr and Trp residues as well as by the formation of protein immobilized radicals, most commonly (Tyr•) [29, 30]. To further assess the involvement of Tyr residues in the oxidation reaction towards the protein amino acids, we performed measurements of protein fluorescence and formation of (Tyr•) in the presence of H2O2.
Treatment of cyt c/TOCL complexes with H2O2 decreased both Tyr and Trp fluorescence. The Tyr67Phe mutant exhibited the lowest fluorescence decrease (for both Tyr and Trp fluorescence) as compared to any other mutant – supporting the conclusion that this mutant has the lowest peroxidase activity of all the mutants studied again emphasizing the catalytic competence of Tyr67 (Fig. 7A).
Figure 7. H2O2-dependent changes in Tyr and Trp fluorescence, production of protein-derived radicals and oligomerization of wild type and mutated forms of cyt c.
A.- H2O2-dependent decrease of tyrosine (λem 303 nm) and tryptophan (λem 325 nm) fluorescence of wild type and mutated cyt c in complexes with TOCL (λex 275nm) (open bars, Tyr fluorescence, %; filled black bars, Trp fluorescence, %). *p<0.01 vs wild-type cyt c.
B.- Production of protein-derived radicals as a result of incubation of of wild type and mutated cyt c in complexes with TOCL with H2O2. The inset shows a typical low-temperature EPR spectrum of protein-derived radicals formed after the addition of H2O2 to incubated liposomes cyt c.
C.- Silver-stained SDS PAGE (10%) of untreated complexes of cyt c mutants with CL (a) and H2O2–treated cyt c/TOCL complexes (wild type and mutated forms of cyt c) (b). Cyt c (8 μM) was incubated with liposomes (TOCL/cyt c ratio 20:1) in 20 mM HEPES (pH 7.4) for 1h in the presence of H2O2 (35 μM) and 100 μM DTPA. Lower contents of oligomers in Tyr67Phe and Tyr97Phe mutants are marked by asterisks. Lane 1, recombinant wild type cyt c (wt); Lane 2, Tyr48Phe cyt c mutant (Tyr48Phe); Lane 3, Tyr67Phe cyt c mutant (Tyr67Phe); Lane 4, Tyr74Phe cyt c mutant (Tyr74Phe); Lane 5, Tyr97Phe cyt c mutant (Tyr97Phe); Lane 6, 4Tyr4Phe cyt c mutant lacking all Tyr residues (4Y4F).
To assess the ability of different cyt c mutants to facilitate the H2O2-dependent production of Tyr• we employed low temperature EPR spectroscopy that has been successfully used in studies of peroxidase activity of cyt c [15, 30, 31]. Typical low-temperature EPR spectra representing the characteristic signal of Tyr• with a peak-to-trough width of ~16 G and a g factor of ~2.005 were obtained from all complexes of cyt c mutants with TOCL pre-incubated in the presence of H2O2 (Fig. 7B). We further assessed the concentrations of H2O2-induced Tyr• in different cyt c mutants. Interestingly a pronounced EPR signal was still present in the 4Tyr4Phe mutant suggesting that, in the absence of all four Tyr residues, electron withdrawal from another amino acid residue took place. This is in line with previous findings on other hemoproteins (cyclooxygenase, hemoglobin) that have documented the formation of radicals of amino acids different from Tyr, such as Cys, Trp and Met, during peroxidase reactions [32, 33]. We further employed the “subtraction protocol” (similar to the described above for the assessments of the contribution of individual Tyr residues in the fluorescence of cyt c) and evaluated the contribution of each of four Tyr residues in the EPR signals from Tyr•. The concentration of protein-immobilized (Tyr•) radicals formed from different participating Tyr increased in the following order: Tyr97<Tyr48=Tyr74<Tyr67 - in accord with the lowest ability of Tyr67Phe mutant to oxidize Amplex Red.
We further used SDS-PAGE to detect the formation of dityrosine-cross-links and protein oligomerization – resulting from the recombination of protein-immobilized Tyr• radicals [27] - in mutant cyt c/TOCL complexes treated with H2O2. SDS-PAGE analysis revealed accumulation of protein oligomers for wild type cyt c and all mutants with substituted single Tyr residues. Tyr67Phe and Tyr97Phe displayed lower amounts of oligomers than other mutants. Interestingly, accumulation of dimers and trimers (but not high molecular weight oligomers) was observed for Tyr97Phe/CL complex treated with H2O2. Expectedly, the Y4F4 mutant lacking Tyr residues was minimally sensitive to H2O2 as revealed by the absence of high molecular weight oligomers and accumulation of only relatively small amounts of dimers and trimers (Fig. 7C).
3.4 Oxidation of cardiolipin in the presence of H2O2
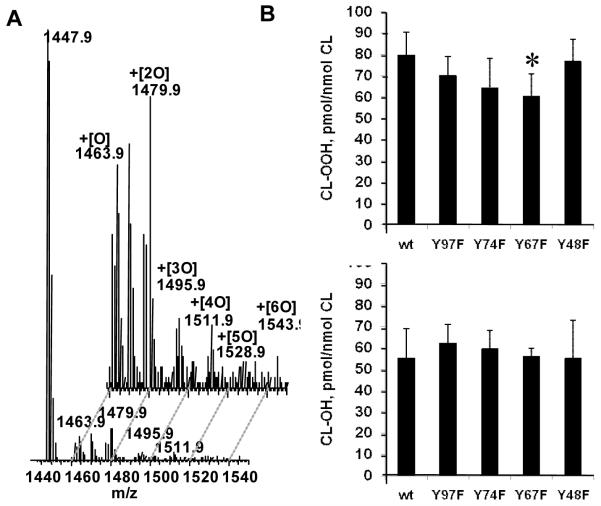
To evaluate the contribution of Tyr residues in CL oxidation, we utilized readily oxidizable polyunsaturated TLCL and conducted ESI-LC-MS assessments of its oxygenation by wild type and mutant forms of cyt c in the presence of H2O2. A typical full ESI-LC-MS spectrum of TLCL oxidized by wild type cyt c is presented in Fig. 8A. Molecular species of mono-, di- and tri-hydroperoxy modified TLCL represented by molecular ions with m/z 1479.9, 1511.9 and 1543.9, respectively, were detected in the mass spectrum (along with a molecular ion with m/z 1448 from non-oxidized TLCL). In addition, molecular ions of mono-, di- and tri-hydroxy molecular species of TLCL with m/z 1463.9, 1495.9 and 1528.9 were also present in the spectra (Fig. 8A). Qualitatively, similar TLCL oxidation products were detected in the spectra obtained from all four cyt c mutants (Tyr97Phe, Tyr74Phe, Tyr67Phe, Tyr48Phe). However, the relative intensity of molecular ions with m/z 1479.9 corresponding to mono-hydroperoxy species of TLCL formed by Tyr67Phe mutant was significantly lower than that produced from TLCL by wild type cyt c. No significant differences in relative intensities of these molecular ions were detected in the presence of other cyt c mutants. Consequently, quantitative assessment of TLCL oxidation products showed that a lower amount of CL-OOH was formed when TLCL was incubated with Tyr67Phe mutant (60.4 ± 10.6 pmol/nmol CL) compared to wild type cyt c (79.8 ± 10.7 pmol/nmol CL) (Fig. 8Ba). No differences in accumulation of hydroxy-molecular species of CL incubated in the presence of wild-type and Tyr67Phe or other Tyr97Phe, Tyr74Phe, Tyr48Phe mutants were observed (Fig. 8Bb). Thus, substitution of Tyr67 for Phe resulted in a decreased oxygenase activity of cyt c towards TLCL.
Figure 8. Oxidation of TLCL by cyt c.
Cyt c (5 μM) was incubated with liposomes (with total of 250 μM phospholipids) containing 20% TLCL, in the presence of H2O2 (100 μM) in 20 mM HEPES, pH 7.4, containing 100 μM DTPA for 10 min at 37 °C. At the end of incubation lipids were extracted and resolved by LC-ESI-MS.
A. - Typical ESI-LC-MS spectra of TLCL incubated in the presence of wild type cyt c.
B. - Accumulation of hydroperoxy- (a) and hydroxy- (b) molecular species of TLCL formed in the presence of either wild-type cyt c or different cyt c mutants. Data are expressed as means ± SD, n =4 for wild type and n= 6 for cyt c mutants, *p<0.03 vs. wild-type cyt c. wt-wild type, Y48F-Tyr48Phe, Y67F-Tyr67Phe, Y74F-Tyr74Phe, Y97F-Tyr97Phe.
DISCUSSION
Native cyt c exhibits a very weak peroxidase activity due to the engagement of all six of the heme-iron coordination bonds in interactions with the porphyrin ring and two distal axial ligands His18 and Met80, respectively [34]. Protein unfolding that includes a loosening/loss of the coordination of heme-iron by the distal ligand Met80 [9, 28, 35, 36], such as has been observed upon interactions of cyt c with anionic phospholipids, particularly CL, results in increased accessibility of catalytic iron to small molecules - H2O2, organic peroxides. This confers increased peroxidase activity, as is observed in cyt c/CL complexes. Several previous studies have proposed that two types of interactions - electrostatic and hydrophobic – participate in the formation of cyt c complex with CL. While the electrostatic interaction is mainly driven by the charges between the protein and phosphate groups of CL, the hydrophobic interaction involves the insertion of the lipid acyl chain in a hydrophobic channel present in the cyt c. In line with this, solutions with high ionic strength (eg, high concentration of NaCl) interfere with the cyt c/CL complex formation through the disruption of the electrostatic forces [34]. We and others have shown that the phosphate ions in the phosphate buffer compete with cardiolipin for electrostatic binding with positively charged Lys residues on cyt c [9, 36]. Further, the NMR studies from our group also suggest that the interaction of CL with cyt c is more effective in Hepes buffer compared to phosphate buffer(Yanamala, unpublished data). Thus, the presence of phosphate ions/high salt concentrations significantly change the peroxidase activity of cyt c by attenuating cyt c/CL interactions and preventing the unfolding of the protein by CL.
In current study, in the absence of CL, commercial preparations of wild type cyt c (purchased from Sigma) exhibit very low peroxidase activity. In contrast, recombinant wild type cyt c used in our studies has detectable peroxidase activity even in the absence of CL. A likely reason for this is the presence of two additional replacements in the protein (His26Asn and His33Asn). These replacements were introduced to increase the expression yield of cyt c in E. coli [17,18]. Especially, His33 was indicated to form a covalent attachment with heme by forming a non-native histidine ligand in unfolded cyt c, replacing Met80. Replacing a His with an Asn at either position 26/33 may interfere with the catalysis of peroxidase reaction of cyt c [34].
Similar to several other peroxidases (eg, prostaglandin synthase) cyt c/CL complexes entertain two activities – 1) peroxidase that includes splitting of a peroxide and formation of the primary reactive intermediates and 2) oxygenase that involves the formation of protein immobilized radical intermediates, frequently Tyr radicals, abstraction of hydrogen and addition of oxygen to the oxidizable substrate. The addition of molecular oxygen to a lipid carbon-centered radical yields a peroxyl radical and, finally, a molecular product, hydroperoxide [37]. In fact, the intermediate production of Tyr radicals in cyt c/CL complexes has been documented [15, 28]. In this work, we assessed the relative contribution of one or more of the four conserved Tyr residues in cyt c to the overall peroxidase catalysis including both activities.
Several lines of evidence presented here imply the primary involvement of Tyr67 as compared to all other tyrosines in the oxygenase function of the cyt c/CL complex. Our MS experiments directly demonstrated decreased production of CL-OOH (but not CL-OH) by Tyr67Phe mutant but not by other Tyr mutants of cyt c. Notably, CL-OOH may be formed by the addition of molecular oxygen to carbon-centered radical of TLCL generated via Tyr•-driven hydrogen abstraction. In contrast, the reduction of CL-OOH to CL-OH – in the peroxidase half-cycle of the reaction – was unaffected by the Tyr67Phe substitution. Similarly, formation of protein-immobilized (Tyr•) radicals and oxidation of a phenolic substrate, Amplex Red, was maximally affected in Tyr67Phe mutant. Further, the Tyr67Phe mutant exerted high resistance to H2O2-induced oligomerization in the absence of oxidation substrates.
Noteworthy, there are differences between the covalent oligomerization of cyt c found in our experiments and previously reported non-covalent oligomers of cyt c [38]. Non-covalent oligomers of cyt c can be formed due to unfolding of the protein by exposure to high temperature or treatments with low concentrations of SDS (0.1%) or ethanol. Using native gel electrophoresis we also documented the formation of cyt c oligomers after it’s unfolding by tetraoleoyl-cardiolipin (TOCL) [3, 9]. In contrast, here we detected the formation of covalent oligomers that retained their aggregated state during SDS-PAGE. Prior to SDS electrophoresis, the samples were boiled in the presence of 2% SDS, a treatment known to disrupt non-covalent interactions. It is tempting to speculate that the covalent cyt c oligomers were produced, most likely, due to the formation of dityrosine cross-links generated by the recombination of protein-immobilized Tyr• radicals.
Finally, our structural studies of cyt c/CL complexes, including Tyr fluorescence, hetero-nuclear NMR, and computer simulations, positioned Tyr 67 in close proximity to the porphyrin ring, heme iron and one of the two axial heme-iron ligand residues, Met80. This structural proximity may favor the generation of Tyr• from Tyr67 as opposed to Tyr48, Tyr74 and Tyr97. It is also possible that - similar to other peroxidases - further transfer of the radical reaction with the involvement of other radical forming residues occurs in the cyt c/CL complexes. This radical-transfer function of Tyr67 may explain why other Tyr mutants (Tyr74Phe and Tyr97Phe) did not demonstrate significant decrease of the catalytic activity. Our data are compatible with the findings that mutation [39], nitration [40, 41] or iodination [42] of Tyr67 drastically alters various heme-linked properties, suggesting that this residue plays an important role in heme ligation and catalytic activity. Finally, Tyr67 has recently also been shown to mediate the disruption of the interaction between heme-iron and Met80 in response to nitration in the more distant Tyr74 via a network of interactions mediated by the mobile Ω loop [16].
Our data are also consistent with the previously described “Markov-process” nature of cyt c unfolding and the identified sequentially unfolding units that get involved in the process with increasing concentrations of denaturants. The core of structural stability and the next most stable unit encompasses the upper half of the protein shown in Fig. 2, and includes Tyr67. Furthermore, recent molecular dynamics simulations support an active role of Tyr67 in the stability of cyt c [43]. Finally, Tyr67 is amongst the most highly conserved residues in cyt c, similar to the residues involved directly in heme coordination (Fig. 3). The only mutation permitted at this site is Phe. This is in contrast to other Tyr residues that are significantly less conserved.
In summary, we have identified a novel role for the highly conserved Tyr67 in serving as the primary electron-donor (radical acceptor) in the oxygenase half-reaction of the cyt c/CL peroxidase complex. These findings may be useful for the discovery of novel specific inhibitors of peroxidation reaction catalyzed by cyt c/CL complexes based on their positioning at the Tyr67-heme-iron interface and the ability to act as competitive donors of electron for the Tyr phenoxyl radicals. Among physiologically relevant reductants, interesting candidates may be sterically hindered phenols with sufficiently high reducing redox potentials, such as homologues of vitamin E (~−0.5V [44]). Regulators of this type may display a pronounced anti-apoptotic potential by inhibiting peroxidation of polyunsaturated CLs.
Research Highlights.
-
>
Cyt c forms peroxidase complexes with cardiolipin (CL).
-
>
Tyrosyl radicals are reactive intermediates of cyt c/CL peroxidase.
-
>
The complexes catalyze CL peroxidation.
-
>
Tyr 67 is closest to the heme-iron of cyt c.
-
>
Tyr67 mutant displayed weakest EPR radical signals and lowest CL peroxidation.
-
>
Tyr 67 is a likely electron-donor in the oxygenase half-reaction of cyt c/CL complex.
Acknowledgments
This study was supported by NIH: HL70755, HL094488; U19 AIO68021; by NIOSH OH008282, Grant RFFI 08-04-01074-a.
Abbreviations
- CL
cardiolipin
- TOCL
1,1′,2,2′-tetraoleoylcardiolipin
- TLCL
1,1′,2,2′-tetralinoleoylcardiolipin
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- cyt c
cytochrome c
- wt
wild type
- Tyr•
tyrosyl radicals
- GndCl
guanidine hydrochloride
- DTPA
diethylenetriaminepentaacetic acid
- H2O2
hydrogen peroxide
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- PBS
phosphate buffered saline
- Amplex Red
N-acetyl-3,7-dihydroxyphenoxazine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nelson DL, Cox MM, Lehninger A. In: Lehninger Principles of Biochemistry. 4th ed. Freeman WH, editor. New York: 2004. [Google Scholar]
- [2].Kluck RM, Martin SJ, Hoffman BM, Zhou JS, Green DR, Newmeyer DD. Cytochrome c activation of CPP32-like proteolysis plays a critical role in a Xenopus cell-free apoptosis system. The EMBO journal. 1997;16:4639–4649. doi: 10.1093/emboj/16.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nature chemical biology. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- [4].He Y, Liu J, Grossman D, Durrant D, Sweatman T, Lothstein L, Epand RF, Epand RM, Lee RM. Phosphorylation of mitochondrial phospholipid scramblase 3 by protein kinase C-delta induces its activation and facilitates mitochondrial targeting of tBid. Journal of cellular biochemistry. 2007;101:1210–1221. doi: 10.1002/jcb.21243. [DOI] [PubMed] [Google Scholar]
- [5].Liu J, Chen J, Dai Q, Lee RM. Phospholipid scramblase 3 is the mitochondrial target of protein kinase C delta-induced apoptosis. Cancer research. 2003;63:1153–1156. [PubMed] [Google Scholar]
- [6].Liu J, Dai Q, Chen J, Durrant D, Freeman A, Liu T, Grossman D, Lee RM. Phospholipid scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol Cancer Res. 2003;1:892–902. [PubMed] [Google Scholar]
- [7].Lacombe ML, Tokarska-Schlattner M, Epand RF, Boissan M, Epand RM, Schlattner U. Interaction of NDPK-D with cardiolipin-containing membranes: Structural basis and implications for mitochondrial physiology. Biochimie. 2009;91:779–783. doi: 10.1016/j.biochi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- [8].Schlattner U, Tokarska-Schlattner M, Ramirez S, Bruckner A, Kay L, Polge C, Epand RF, Lee RM, Lacombe ML, Epand RM. Mitochondrial kinases and their molecular interaction with cardiolipin. Biochimica et biophysica acta. 2009;1788:2032–2047. doi: 10.1016/j.bbamem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- [9].Belikova NA, Vladimirov YA, Osipov AN, Kapralov AA, Tyurin VA, Potapovich MV, Basova LV, Peterson J, Kurnikov IV, Kagan VE. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tyurin VA, Tyurina YY, Kochanek PM, Hamilton R, DeKosky ST, Greenberger JS, Bayir H, Kagan VE. Oxidative lipidomics of programmed cell death. Methods in enzymology. 2008;442:375–393. doi: 10.1016/S0076-6879(08)01419-5. [DOI] [PubMed] [Google Scholar]
- [11].Tyurina YY, Kini V, Tyurin VA, Vlasova, Jiang J, Kapralov AA, Belikova NA, Yalowich JC, Kurnikov IV, Kagan VE. Mechanisms of cardiolipin oxidation by cytochrome c: relevance to pro- and antiapoptotic functions of etoposide. Molecular pharmacology. 2006;70:706–717. doi: 10.1124/mol.106.022731. [DOI] [PubMed] [Google Scholar]
- [12].Belikova NA, Tyurina YY, Borisenko G, Tyurin V, Samhan Arias AK, Yanamala N, Furtmuller PG, Klein-Seetharaman J, Obinger C, Kagan VE. Heterolytic reduction of fatty acid hydroperoxides by cytochrome c/cardiolipin complexes: antioxidant function in mitochondria. Journal of the American Chemical Society. 2009;131:11288–11289. doi: 10.1021/ja904343c. [DOI] [PubMed] [Google Scholar]
- [13].Svistunenko DA. Reaction of haem containing proteins and enzymes with hydroperoxides: the radical view. Biochimica et biophysica acta. 2005;1707:127–155. doi: 10.1016/j.bbabio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- [14].Duggan KC, Musee J, Marnett LJ. Peroxidase active site activity assay. Methods in molecular biology (Clifton, N.J. 644:55–65. doi: 10.1007/978-1-59745-364-6_6. [DOI] [PubMed] [Google Scholar]
- [15].Vlasova, Tyurin VA, Kapralov AA, Kurnikov IV, Osipov AN, Potapovich MV, Stoyanovsky DA, Kagan VE. Nitric oxide inhibits peroxidase activity of cytochrome c.cardiolipin complex and blocks cardiolipin oxidation. J Biol Chem. 2006;281:14554–14562. doi: 10.1074/jbc.M509507200. [DOI] [PubMed] [Google Scholar]
- [16].Abriata LA, Cassina A, Tortora V, Marin M, Souza JM, Castro L, Vila AJ, Radi R. Nitration of solvent-exposed tyrosine 74 on cytochrome c triggers heme iron-methionine 80 bond disruption. Nuclear magnetic resonance and optical spectroscopy studies. J Biol Chem. 2009;284:17–26. doi: 10.1074/jbc.M807203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rumbley JN, Hoang L, Englander SW. Recombinant equine cytochrome c in Escherichia coli: high-level expression, characterization, and folding and assembly mutants. Biochemistry. 2002;41:13894–13901. doi: 10.1021/bi026543y. [DOI] [PubMed] [Google Scholar]
- [18].Liu W, Rumbley J, Englander SW, Wand AJ. Backbone and side-chain heteronuclear resonance assignments and hyperfine NMR shifts in horse cytochrome c. Protein Sci. 2003;12:2104–2108. doi: 10.1110/ps.03211303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pecina P, Borisenko GG, Belikova NA, Tyurina Y, Pecinova A, Lee I, Samhan-Arias AK, Przyklenk K, Kagan VE, Huttemann M. Phosphomimetic Substitution of Cytochrome c Tyrosine 48 Decreases Respiration and Binding to Cardiolipin, and Abolishes Ability to Trigger Downstream Caspase Activation. Biochemistry. doi: 10.1021/bi100486s. [DOI] [PubMed] [Google Scholar]
- [20].Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- [21].Wyard SJ. Double integration of electron spin resonance spectra. J. Sci. Instrum. 1965;42:769–770. [Google Scholar]
- [22].Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic acids research. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Thompson JD, Gibson TJ, Higgins DG, Baxevanis Andreas D. Current protocols in bioinformatics. 2002. Multiple sequence alignment using ClustalW and ClustalX; p. 3. editoral board. [et al, Chapter 2. Unit 2. [DOI] [PubMed] [Google Scholar]
- [24].Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Strottmann JM, Stellwagen A, Bryant C, Stellwagen E. Spectral studies of horse heart porphyrin cytochrome c. J Biol Chem. 1984;259:6931–6936. [PubMed] [Google Scholar]
- [26].Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Analytical biochemistry. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- [27].Chen YR, Mason RP. Mechanism in the reaction of cytochrome c oxidase with organic hydroperoxides: an ESR spin-trapping investigation. The Biochemical journal. 2002;365:461–469. doi: 10.1042/BJ20020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kapralov AA, Kurnikov IV, Vlasova, Belikova NA, Tyurin VA, Basova LV, Zhao Q, Tyurina YY, Jiang J, Bayir H, Vladimirov YA, Kagan VE. The hierarchy of structural transitions induced in cytochrome c by anionic phospholipids determines its peroxidase activation and selective peroxidation during apoptosis in cells. Biochemistry. 2007;46:14232–14244. doi: 10.1021/bi701237b. [DOI] [PubMed] [Google Scholar]
- [29].Ehrenshaft M, Mason RP. Protein radical formation on thyroid peroxidase during turnover as detected by immuno-spin trapping. Free Radic Biol Med. 2006;41:422–430. doi: 10.1016/j.freeradbiomed.2006.02.023. [DOI] [PubMed] [Google Scholar]
- [30].Qian SY, Chen YR, Deterding LJ, Fann YC, Chignell CF, Tomer KB, Mason RP. Identification of protein-derived tyrosyl radical in the reaction of cytochrome c and hydrogen peroxide: characterization by ESR spin-trapping, HPLC and MS. The Biochemical journal. 2002;363:281–288. doi: 10.1042/0264-6021:3630281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen YR, Chen CL, Chen W, Zweier JL, Augusto O, Radi R, Mason RP. Formation of protein tyrosine ortho-semiquinone radical and nitrotyrosine from cytochrome c-derived tyrosyl radical. J Biol Chem. 2004;279:18054–18062. doi: 10.1074/jbc.M307706200. [DOI] [PubMed] [Google Scholar]
- [32].Marnett LJ. Cyclooxygenase mechanisms. Curr Opin Chem Biol. 2000;4:545–552. doi: 10.1016/s1367-5931(00)00130-7. [DOI] [PubMed] [Google Scholar]
- [33].Jia Y, Buehler PW, Boykins RA, Venable RM, Alayash AI. Structural basis of peroxide-mediated changes in human hemoglobin: a novel oxidative pathway. J Biol Chem. 2007;282:4894–4907. doi: 10.1074/jbc.M609955200. [DOI] [PubMed] [Google Scholar]
- [34].Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang J, Stoyanovsky DA, Wipf P, Kochanek PM, Greenberger JS, Pitt B, Shvedova AA, Borisenko G. Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic Biol Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sanghera N, Pinheiro TJ. Unfolding and refolding of cytochrome c driven by the interaction with lipid micelles. Protein Sci. 2000;9:1194–1202. doi: 10.1110/ps.9.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sinibaldi F, Fiorucci L, Patriarca A, Lauceri R, Ferri T, Coletta M, Santucci R. Insights into cytochrome c-cardiolipin interaction. Role played by ionic strength. Biochemistry. 2008;47:6928–6935. doi: 10.1021/bi800048v. [DOI] [PubMed] [Google Scholar]
- [37].Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. Journal of lipid research. 1998;39:1529–1542. [PubMed] [Google Scholar]
- [38].Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, Sugiura Y, Kataoka M, Higuchi Y. Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12854–12859. doi: 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Luntz TL, Schejter A, Garber EA, Margoliash E. Structural significance of an internal water molecule studied by site-directed mutagenesis of tyrosine-67 in rat cytochrome c. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:3524–3528. doi: 10.1073/pnas.86.10.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cassina AM, Hodara R, Souza JM, Thomson L, Castro L, Ischiropoulos H, Freeman BA, Radi R. Cytochrome c nitration by peroxynitrite. J Biol Chem. 2000;275:21409–21415. doi: 10.1074/jbc.M909978199. [DOI] [PubMed] [Google Scholar]
- [41].Rodriguez-Roldan V, Garcia-Heredia JM, Navarro JA, Rosa MA, Hervas M. Effect of Nitration on the Physicochemical and Kinetic Features of Wild-Type and Monotyrosine Mutants of Human Respiratory Cytochrome c. Biochemistry. 2008 doi: 10.1021/bi800910v. [DOI] [PubMed] [Google Scholar]
- [42].Rinaldi TA, Tersariol IL, Dyszy FH, Prado FM, Nascimento OR, Di Mascio P, Nantes IL. Protonation of two adjacent tyrosine residues influences the reduction of cytochrome c by diphenylacetaldehyde: a possible mechanism to select the reducer agent of heme iron. Free Radic Biol Med. 2004;36:802–810. doi: 10.1016/j.freeradbiomed.2003.12.002. [DOI] [PubMed] [Google Scholar]
- [43].Singh SR, Prakash S, Vasu V, Karunakaran C. Conformational flexibility decreased due to Y67F and F82H mutations in cytochrome c: molecular dynamics simulation studies. Journal of molecular graphics & modelling. 2009;28:270–277. doi: 10.1016/j.jmgm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- [44].Kagan VE, Kuzmenko AI, Shvedova AA, Kisin ER, Tyurina YY, Yalowich JC. Myeloperoxidase-catalyzed phenoxyl radicals of vitamin E homologue, 2,2,5,7,8-pentamethyl- 6-hydroxychromane, do not induce oxidative stress in live HL-60 cells. Biochemical and biophysical research communications. 2000;270:1086–1092. doi: 10.1006/bbrc.2000.2564. [DOI] [PubMed] [Google Scholar]
- [45].Furtmuller PG, Jantschko W, Zederbauer M, Jakopitsch C, Arnhold J, Obinger C. Kinetics of interconversion of redox intermediates of lactoperoxidase, eosinophil peroxidase and myeloperoxidase. Jpn J Infect Dis. 2004;57:S30–31. [PubMed] [Google Scholar]