Abstract
During the last years, several applications of biosurfactants with medical purposes have been reported. Biosurfactants are considered relevant molecules for applications in combating many diseases. However, their use is currently extremely limited due to their high cost in relation to that of chemical surfactants. Use of inexpensive substrates can drastically decrease its production cost. Here, twelve solid substrates were screened for the production of Bacillus subtilis SPB1 biosurfactant and the maximum yield was found with millet. A Plackett-Burman design was then used to evaluate the effects of five variables (temperature, moisture, initial pH, inoculum age, and inoculum size). Statistical analyses showed that temperature, inoculum age, and moisture content had significantly positive effect on SPB1 biosurfactant production. Their values were further optimized using a central composite design and a response surface methodology. The optimal conditions of temperature, inoculum age, and moisture content obtained under the conditions of study were 37°C, 14 h, and 88%, respectively. The evaluation of the antimicrobial activity of this compound was carried out against 11 bacteria and 8 fungi. The results demonstrated that this biosurfactant exhibited an important antimicrobial activity against microorganisms with multidrug-resistant profiles. Its activity was very effective against Staphylococcus aureus, Staphylococcus xylosus, Enterococcus faecalis, Klebsiella pneumonia, and so forth.
1. Introduction
Solid-state fermentation which involves growth of microorganisms on moist solid substrates in the absence of free flowing water, recently, has received a considerable attention of researchers due to its several advantages over the submerged fermentation [1]. Solid substrates and wastes from different origins could be treated by solid-state fermentation and useful products could be produced. Solid-state fermentation had a long history of production of traditional foods by using different organisms and is also found to have an increasing application in the production of enzymes, antibiotics, surfactants, biocides, and so forth [1]. It had been reported that in solid-state fermentation, especially in fungal one, the productions of metabolites, such as enzymes, antibiotics, organic acids, and aroma compounds, are higher than that in submerged fermentation [2].
Bacillus subtilis strains produced a broad spectrum of bioactive compounds with great potential for biotechnological and biopharmaceutical applications including surfactin, fengycin, iturin, mycosubtilins, and bacillomycins, which are amphiphilic membrane-active biosurfactants with potent antimicrobial activities [3]. These biosurfactants possessed, also, antiviral [4], antitumor [5], hemolytic [6], blood anticoagulant, and fibrinolytic [7] activities. Moreover, biosurfactants of Bacillus subtilis have numerous environmental and biotechnological applications [8] and have shown particular utility in oil recovery [9], remediation of soil contaminated by heavy metals [10], and biocontrol against phytopathogens [11] and insects [12]. The high production cost of biosurfactants has been a major concern towards restricted use in commercial applications [13, 14].
The present study aimed at the optimization of fermentation conditions in the solid substrate fermentation. We have screened twelve solid substrates for the production of B. subtilis (SPB1) biosurfactant and its antimicrobial activity was determined against multidrug-resistant bacteria. A Plackett-Burman design [15] was used to identify the most significant variables influencing biosurfactant production. A response surface methodology combined with a central composite design was then used to further optimize those variables, including temperature, inoculum age, and moisture content.
2. Materials and Methods
2.1. Microorganism
B. subtilis SPB1 (HQ392822), used through this work, was a wild type strain isolated from Tunisian soil.
Surfactant compounds were tested against a group of multiresistant bacteria isolated from clinical specimens: Pseudomonas aeruginosa, Pseudomonas stutzeri, Escherichia coli, Staphylococcus aureus, Enteroccoccus faecalis, Salmonella typhimirium, Klebsiella pneumoniae, Enterobacter cloacae, Enterobacter faecium, and Brevibacterium flavum, a group of fungi belonging to the Culture Collection of our Laboratory: Penicillium notatum, Penicillium italicum, Alternaria alternaria, Puccinia allii, Peronospora destructor, Aspergillus oryzae, Aspergillus niger, Rhizopus oryzae, and two species of yeast: Candida albicans and Saccharomyces cerevisiae. The microorganisms were maintained on LB agar slants at refrigerated temperature.
2.2. Isolation and Identification of Biosurfactant-Producing Bacteria
The screening of biosurfactants producers was performed by blood haemolysis technique as described by Master [16]. Bacteria were grown on blood agar at 37°C. The formation of the clearing zone around the colonies evidenced biosurfactant production [17].
Strain B. subtilis SPB1 used through this work was isolated from south Tunisian soil. It was identified by physiological-biochemical characterization and by 16S RNA gene ribotyping.
For the determination of 16S rDNA sequences, chromosomal DNA was extracted from B. subtilis cells using the phenol extraction method [18]. This DNA was used as template for the PCR using the universal primers Rd1 (5′AAGGAGGTGATCCAGCC3′) and Fd1 (5′AGTTTGATCCTGGCTCAG3′) and a ‘‘Gene Amp PCR System 2700” (Applied Biosystems). The 25 μL of PCR mixture contained: 0.2 mM of deoxynucleoside triphosphate, 1.32 μM concentration each primer, 0.5 U of DNA polymerase, 5 μL of 5X buffer and 1 μL (5 ng) of total DNA template. The PCR programme was 94°C for 3 min followed by 35 cycles consisting of denaturation at 94°C for 45 s, annealing at 59°C for 1 min and extension at 72°C for 2 min. Following amplification, the PCR product was purified (Promega Gel Extraction Kit, Biogène, Tunisia) and sequenced with ABI PRISMTM 3100 Genetic Analyzer (Applied Biosystems, USA).
2.3. Phylogenetic Analysis
The CLUSTALW program [18] was used to make multiple alignments between all sequences using default parameters. Dendrogram based on the sequence similarity was made using the NJPLOT program.
2.4. Inoculum and Culture Conditions
B. subtilis SPB1 strain was streaked on a nutrient agar slant [17] and incubated at 37°C. After 24 h, one loop of cells was dispensed in 3 mL of LB medium and incubated overnight at 37°C. Aliquots (0.2 mL) were used to inoculate 250 mL Erlenmeyer flasks containing 50 mL LB medium [19] and incubated in a rotatory shaker at 200 rpm and 37°C (±0.5) during 4, 11, 14, 23, 24, 31 h in accordance with the purpose. Three mL of the obtained culture were used to inoculate the production medium.
Solid-state fermentation was conducted as follows: fifteen grams of substrates (banana peels, potato peels, chick-pea flour, corn bran, corn starch, millet, soya meal, barley flour, barley bran, rice flour, cornstarch, and orange peels) were placed into a 100 mL Erlenmeyer flask, the mouth of which was stoppered with a cotton plug. Then those substrates were autoclaved twice at 120°C (1 bar) for 30 min at an interval of 8–12 h to kill the spore-forming microorganisms inhabiting the material. After autoclaving, 75 μL of KH2PO4 (1 M), 225 μL of MgSO4 (1 M), and 367 μL of deionised distilled water were aseptically added for the fortification of nutrients [20, 21], and three mL of a preculture of B. subtilis SPB1 strain grown in the LB medium was inoculated. The flasks were immersed in a water incubator for 48 h without shaking. Cultivation temperature varied from 18, 25, 28, 30, 35, 37, 42, 45, to 52°C depending on the purpose.
Samples were collected at time-defined intervals for the determination of biosurfactant production yield. All experiments were performed in triplicate.
2.5. Extraction of Crude Biosurfactant
The crude biosurfactant was isolated from the cell-free broth of 24 h grown culture. The bacterial cells were removed from surfactant containing culture broth by centrifugation at 1062 g at 4°C for 20 min. The supernatant was precipitated overnight at 4°C by adding concentrated HCl to achieve a final pH of 2.0 in order to precipitate lipids and proteins [21]. Grey white pellets formed by precipitation were collected by centrifugation at 1062 g at 4°C for 20 min. The crude surfactant was lyophilized and weighted for quantification. For the extraction of biosurfactant compounds, 50 mL of chloroform-methanol (2 : 1 v/v) was added to 500 mg of the dry product and incubated in a rotatory shaker at 250 rpm, 30°C (±0.5) for 15 min. The extract was evaporated to dryness and weighted for quantification. Culture without inoculation was used as a negative control to take account possible contribution of lipids and proteins from substrates. The negative control was included in each experiment and each cultural condition. Crude biosurfactant weight was calculated as the result of subtracting the weight obtained with the negative control from that measured with the culture containing the biosurfactant producing strain. The values presented are the three determinations' average results of the two separate experiments for each cultural condition.
2.6. Antimicrobial Activity
The antimicrobial activity of the biosurfactant was tested against several Gram-positive and Gram-negative bacteria using the agar spot assay method described by Paik and et al. [22]. Each strain, used as an indicator, had been previously subcultured in its appropriate medium and temperature. A quantity of 5 mL of soft agar (0.7% agar) containing about 107 cells of indicator strain was overlaid on 2.5% LB agar plates [23]. The appearance of the inhibition zone was determined after 24 h of incubation using the appropriate medium and temperature. The values presented are the average of the results of three determinations of two separate experiments.
2.7. Determination of Emulsification Activity
Samples (0.5 mL) of cell-free supernatant were added to a screw-capped tube containing 7.5 mL of Tris-Mg (20 mM Tris HCl (pH 7.0) and 10 mM MgSO4) and 0.1 mL of kerosene. After a vigorous vortex, the tubes were allowed to sit for 1 hour. Absorbance was measured at 540 nm [24]. Emulsification activity (E.A) was defined as the measured optical density [24]. Assays were carried out in triplicates.
2.8. Determination of Bacillus subtilis SPB1 Biosurfactant Characteristics
The optimum temperature of the emulsification activity of the crude biosurfactant was determined by incubating 0.5 mL of supernatant of the culture with 7.5 mL of Tris-Mg (20 mM Tris HCl (pH 7.0) and 10 mM MgSO4) and 0.1 mL of kerosene for 1 hour at different temperatures ranging from 25 to 90°C. Thermostability of SPB1 biosurfactant was studied by the determination of the residual activity after a heat treatment during 60 min of crude biosurfactant at a range of temperatures (0, 20, 30, 40, 50, 60, 70, and 80°C). Activities were determined at 37°C. The optimum pH for biosurfactant activity was determined by incubating the supernatant in different buffers: 50 mmol L−1 citrate buffer (pH 3.0–6.0), 50 mmol L−1 phosphate buffer (pH 6.0–8.0), and 50 mmol L−1 Glycine-NaOH buffer (pH 9.0-10.0) [23]. The stability at 4°C in various pH conditions was assessed by incubating the biosurfactant at different pH and by measuring the residual activity after 24 h using the standard protocol.
2.9. Selection of a Suitable Substrate
Various industries byproducts like banana peels, potato peels, chick-pea flour, corn bran, corn flour, orange peels flour, soya meal, barley flour, barley its, rice flour, cornstarch, and millet were screened for B. subtilis SPB1 biosurfactant production. 15 g of sterilized bran were used as substrate. The substrate yielding the maximum biosurfactant production was identified and selected for further studies using response surface methodology.
2.10. Estimation of Moisture Content
Moisture content of the substrate was estimated by drying 15 g of substrate to constant weight at 105°C and the dry weight was recorded. To fix the initial moisture content of the solid medium, the substrate was soaked with the appropriate quantity of distilled water. The sample was then dried as described above, and moisture content (%) was calculated as follows [25]:
Moisture content (initial) of solid medium
| (1) |
2.11. Optimization of Bacillus subtilis SPB1 Biosurfactant Production
The optimization of B. subtilis SPB1 biosurfactant production was carried out using a statistical experimental design that consisted of two major steps. The first concerned the screening of the significant variables that affected the biosurfactant production and the second involved the further optimization of the relatively important variables.
2.12. Selection of Significant Variables by Plackett-Burman Design
Plackett-Burman design, an efficient technique for medium component optimization, was used to screen “k” variables in just “k + 1” number of experiments [15]. The Plackett-Burman design was then used to evaluate the relative importance of five parameters for B. subtilis SPB1 biosurfactant production in solid-state fermentation. In fact, this design does not consider the interaction effects among the variables and was used only to screen the important variables affecting biosurfactant production yield. The variables that were chosen for the present study were (A) temperature, (B) moisture, (C) initial pH, (D) inoculum age, and (E) inoculum size. The experimental design, with the names and actual levels of the variables, is presented in Table 1.
Table 1.
The Plackett-Burman experimental design matrix for screening culture condition factors affecting biosurfactant production by B. subtilis SPB1 strain.
| Run order | Temperature | Moisture | Initial | Inoculum size | Inoculum | Biosurfactant |
|---|---|---|---|---|---|---|
| (°C) | (%) | pH | (CFU/mL) | age (h) | mg/g | |
| A | B | C | D | E | ||
| 1 | 45 | 100 | 9 | 106 | 24 | 16.3 |
| 2 | 25 | 100 | 9 | 108 | 4 | 13.9 |
| 3 | 25 | 50 | 9 | 108 | 24 | 14.1 |
| 4 | 45 | 50 | 2 | 108 | 24 | 16.3 |
| 5 | 25 | 100 | 2 | 106 | 24 | 12.7 |
| 6 | 45 | 50 | 9 | 106 | 4 | 11.5 |
| 7 | 45 | 100 | 2 | 108 | 4 | 17.5 |
| 8 | 25 | 50 | 2 | 106 | 4 | 9.2 |
The Plackett-Burman experimental design was based on the following first order model equation:
| (2) |
where is the response (biosurfactant production yield mg/g), b0 is the model intercepts, bi is the linear coefficient, and xi is the level of the independent variable. The effects of each variable on biosurfactant production were estimated as the difference between both averages of measurements made at the higher level and at the lower level. The significance of each variable was determined via a Student's t-test.
2.13. Optimization by Response Surface Methodology
The next step in the optimization of solid-state fermentation conditions was to determine the optimum levels of the significant variables for B. subtilis SPB1 biosurfactant production. For this purpose, a response surface methodology was adopted to maximize biosurfactant production using a central composite design. The significant variables that were selected were (X1) temperature, (X2) moisture, and (X3) inoculum age. Each variable was assessed at five coded levels (−1.682, −1, 0, +1, and +1.682). A total of 24 experiments were conducted including 15 experiments for the central composite design, three supplementary experiments in the domain centre to estimate the pure error and four other experiments to check the validity of the model. The response values used in each trial were the average of three determinations of two separate experiments (Table 2).
Table 2.
Design of experiment and response of the central composite experimental design for the production of biosurfactant using B. subtilis SPB1.
| Run order | X 1: temperature (°C) | X 2: moisture (%) | X 3: inoculum age (h) | Biosurfactant (mg/g)experimental values | Biosurfactant (mg/g) csalculated values |
|---|---|---|---|---|---|
| 1 | 25 | 50 | 04 | 15.22 | 15.44 |
| 2 | 45 | 50 | 04 | 12.47 | 13.15 |
| 3 | 25 | 100 | 04 | 15.40 | 15.18 |
| 4 | 45 | 100 | 04 | 17.40 | 17.14 |
| 5 | 25 | 50 | 24 | 14.90 | 15.59 |
| 6 | 45 | 50 | 24 | 14.20 | 14.88 |
| 7 | 25 | 100 | 24 | 15.85 | 15.88 |
| 8 | 45 | 100 | 24 | 19.10 | 19.42 |
| 9 | 18 | 75 | 14 | 16.20 | 15.93 |
| 10 | 52 | 75 | 14 | 17.40 | 16.99 |
| 11 | 35 | 33 | 14 | 16.58 | 15.72 |
| 12 | 35 | 117 | 14 | 19.50 | 19.32 |
| 13 | 35 | 75 | 01 | 12.50 | 12.26 |
| 14 | 35 | 75 | 31 | 14.50 | 14.30 |
| 15 | 35 | 75 | 14 | 14.30 | 14.35 |
| 16 | 35 | 75 | 14 | 12.80 | 14.35 |
| 17 | 35 | 75 | 14 | 14.91 | 14.35 |
| 18 | 35 | 75 | 14 | 13.20 | 14.35 |
| 19 | 35 | 75 | 14 | 15.80 | 14.35 |
| 20 | 35 | 75 | 14 | 14.49 | 14.35 |
| 21 | 28 | 65 | 11 | 14.42 | 14.45 |
| 22 | 42 | 69 | 11 | 14.74 | 14.12 |
| 23 | 35 | 95 | 11 | 14.48 | 15.73 |
| 24 | 35 | 75 | 23 | 15.95 | 14.59 |
2.14. Statistical Analysis and Modelling
The data obtained from the response surface methodology with regards to B. subtilis SPB1 biosurfactant production were subjected to analysis of variance (ANOVA) to check the errors and the significance of each parameter. Biosurfactant production yield was taken as a response . The data were then subjected to a multiple regression analysis to obtain an empirical model that could relate the response measured to the independent variables. The behaviour of the system was explained by the following quadratic equation:
| (3) |
where refers to the predicted response, X1, X2, X3 to the independent coded variables, b0 to the offset term, b1, b2, b3 to the linear effects, b11, b22, b33 to the squared effects and b12, b23, b13 to the interaction terms.
The statistical software package, (Nemrod-W by LPRAI Marseilles, France) [26] was used to conduct a regression analysis on the experimental data and to plot the response surface graphs. The statistical significance of the model was determined by the application of Fisher's F test [27]. The two-dimensional graphical representation of the system behaviour, called the isoresponse contour plot, was used to describe the individual and cumulative effects of the variables as well as the possible correlations existed between them.
Statistical analyses of antimicrobial activity results were performed using SPSS 17.0 for Microsoft Windows. Comparison was performed using a factorial ANOVA with subsequent post hoc test (Ryan, Einot, Gabriel, & Welsch).
3. Results
3.1. Screening of Biosurfactant-Producing Strains
Two hundred strains isolated from different locations of the south Tunisia's soil were screened for their ability to produce biosurfactants by using blood hemolysis assay [23, 28]. Several strains were selected on the basis of their high α-hemolysis activities, since the size of the clear zone developed is in proportion to the amount of the produced biosurfactant [28]. The isolate exhibiting the higher production was named SPB1. Its biochemical and molecular identification showed that it corresponded to Bacillus subtilis strain. The 16S rDNA sequence of strain SPB1 was determined and deposited in the GeneBank database under accession number HQ392822.
3.2. Antimicrobial Activity
Zones of inhibition diameter produced by different concentrations of biosurfactant were measured using agar spot assay method [21]. As shown in Table 3, biosurfactant exhibited interesting antibacterial and antifungal activities. The antimicrobial activity increased with increasing concentration of biosurfactant (P < 0.01). The compound showed higher activity against Gram-positive cocci than that of Gram-negative bacilli (P < 0.05). Its activity was very effective against Enterococcus faecalis, the halos presented a mean value of 18 mm and 20 mm when the compound was used at a concentration of 1 mg/mL and 2 mg/mL, respectively. It has a strong activity against Staphylococcus aureus. It is important to note that this strain is resistant to at least two β-lactams. Moreover, the mean halo diameter obtained with Enterobacter faecium and Brevibacterium flavum was of 13 mm, while those obtained with Escherichia coli, Pseudomonas aeruginosa, Pseudomonas stutzeri, Klebsiella pneumoniae, Enterobacter cloacae, and Salmonella typhimirium varied from 9 to 15 mm with 1 mg/mL of the compound. Indeed, it was clear that the activity against Gram-negative bacteria was lower than that obtained with Gram-positive bacteria (P < 0.0001). Furthermore, as shown in Table 3, the biosurfactant produced by B. subtilis SPB1 was also characterized by an important antifungal activity, especially, against Penicillium notatum, Penicillium italicum, and Aspergillus niger and lower activity against Rhizopus oryzae and Aspergillus oryzae. This activity was clearly negative against Alternaria alternata, Puccinia allii and Peronospora destructor. Moreover, it was clear that this compound presented an important antiyeast activity, mainly, against Candida albicans.
Table 3.
Antibacterial, antifungal, and antiyeast activities of the crude biosurfactant at different concentrations (0.5–2 g/L).
| Microorganisms | Zone of inhibition diameter (mm) | |||
|---|---|---|---|---|
| 0.5 g/L | 1 g/L | 2 g/L | ||
| Bacteria | ||||
| Staphylococcus aureus f | 19 ± 1 | 23 ± 2 | 28 ± 2 | |
| Escherichia coli d | 12 ± 1 | 15 ± 1 | 19 ± 1 | |
| Salmonella typhimirium a | 08 ± 1 | 11 ± 2 | 13 ± 2 | |
| Pseudomonas aeruginosa a | 08 ± 1 | 10 ± 1 | 12 ± 1 | |
| Enterococcus faecalis e | 14 ± 2 | 18 ± 1 | 20 ± 2 | |
| Klebsiella pneumoniae c,d | 12 ± 1 | 15 ± 1 | 18 ± 1 | |
| Enterobacter cloacae b,c | 11 ± 1 | 13 ± 1 | 15 ± 1 | |
| Enterobacter faecium b | 10 ± 1 | 13 ± 1 | 15 ± 1 | |
| Brevibacterium flavum b,c,d | 11 ± 1 | 13 ± 1 | 16 ± 1 | |
| Pseudomonas stutzeri a | 07 ± 2 | 09 ± 2 | 12 ± 2 | |
| Staphylococcus xyloxus e | 16 ± 12 | 19 ± 2 | 23 ± 2 | |
| Penicillium notatum | 09 ± 1 | 14 ± 1 | 19 ± 1 | |
|
| ||||
| Fungi | ||||
| Alternaria alternata | — | — | — | |
| Penicillium italicum | 11 ± 1 | 15 ± 1 | 20 ± 1 | |
| Puccinia allii | — | — | — | |
| Peronospora destructor | — | — | — | |
| Aspergillus oryzae | 03 ± 2 | 06 ± 2 | 09 ± 2 | |
| Aspergillus niger | 10 ± 12 | 14 ± 2 | 18 ± 2 | |
| Rhizopus oryzae | 04 ± 1 | 07 ± 1 | 09 ± 2 | |
| Candida albicans | 18 ± 2 | 21 ± 2 | 25 ± 2 | |
— = no inhibition.
Different letters indicates the difference between two means is statistically significant (P < 0.05).
3.3. Screening of Various Substrates and Optimization of Incubation Time
Biosurfactant production by B. subtilis SPB1 was studied with different solid substrates supplemented with nutrient medium. The studies on the production of SPB1 biosurfactant using various solid substrates were carried for 72 h. In the case of potato peels, chick-pea flour, corn bran, corn flour, millet, soya bean meal, barley flour, and orange peels, the maximum biosurfactant production was observed on the first 24 h (Figure 1), whereas in case of banana peels, barley its, rice flour and corn starch it was noted on the third day. Among all the solid substrates used for our study, the highest biosurfactant production was found using millet. It was evaluated to 14 mg of biosurfactant per g of substrate after 24 h of culture.
Figure 1.
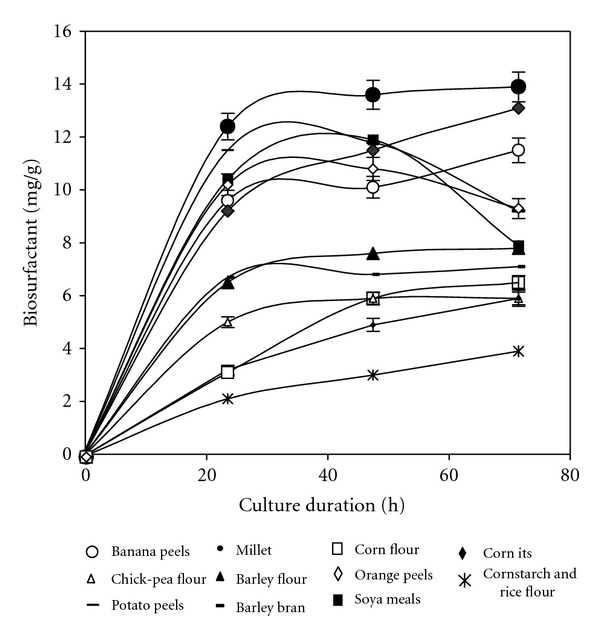
Evaluation of biosurfactant production by B. subtilis SPB1 strain during 72 h of fermentation using different cheap substrates.
When varying millet concentrations, it was clear that the optimal production (14.5 mg of biosurfactant per g of substrate) was obtained when using only 10 g of dehydrated substrate (data not shown). The corresponding emulsification activity of the produced biosurfactant was 1.55.
3.4. Screening of Culture Condition Factors Affecting Biosurfactant Production
The purpose of the first optimization step is to identify which factors, representing culture conditions, have significant effect on B. subtilis SPB1 biosurfactant production. The Placket-Burman design with height rows was shown in Table 1. The last column indicated the measured responses. The adequacy of the model was calculated, and the variables evidencing statistically significant effects were screened via Student's t-test for ANOVA (Table 4). Factors evidencing P values of less than 0.05 were considered to have significant effects on the response and were, therefore, selected for further optimization studies. Among the variables screened, temperature, moisture, and inoculum age were identified as the most significant variables influencing biosurfactant production by B. subtilis SPB1. The values of the other variables were statistically insignificant and were, therefore, not considered in the subsequent analysis. The optimum levels of the three significant variables selected were further determined by performing a central composite design.
Table 4.
Estimated effect, regression coefficient, and corresponding t and P values for surfactin production in eight variable Plackett-Burman design experiments.
| Code | Estimate coefficient | F. inflation | Standard deviation | t value | P value |
|---|---|---|---|---|---|
| Intercept | 13.937 | 0.238 | 58.36 | <0.01*** | |
| A | 1.463 | 1.00 | 0.238 | 6.12 | * |
| B | 1.162 | 1.00 | 0.238 | 4.87 | * |
| C | 0.013 | 1.00 | 0.238 | 0.05 | 93.3 |
| D | 1.512 | 1.00 | 0.238 | 6.33 | * |
| E | 0.912 | 1.00 | 0.238 | 3.82 | 6.2 |
***P < 0.01 (very significant).
*(Significant).
3.5. Optimization of Selected Variables Using Response Surface Methodology
The central composite experimental design was applied to find out the optimum conditions for B. subtilis SPB1 biosurfactant production through the determination of the optimum levels of the significant factors temperature (X1), moisture (X2), and inoculums (X3). The results obtained were subjected to an ANOVA to determine the significant differences. As shown in Table 2, there was a considerable variation in SPB1 biosurfactant production yield which was heavily depended on the levels of the three independent variables in the medium. In fact, biosurfactant production yield varied from 12.47 mg/g (run 2) to 19.50 mg/g (run 12).
By applying a least-squares method to the experimental data, the following second-order polynomial equation was found to adequately explain the biosurfactant production yield by considering only the significant terms (Table 5):
Table 5.
Estimate regression coefficients for biosurfactant production using data in coded units.
| Number | Estimate coefficient | F. inflation | Standard deviation | t experimental | Significance |
|---|---|---|---|---|---|
| b 0 | 14.356 | 0.334 | 43.02 | *** | |
| b 1 | 0.313 | 1.00 | 0.250 | 1.26 | *** |
| b 2 | 1.070 | 1.00 | 0.250 | 4.29 | *** |
| b 3 | 0.607 | 1.00 | 0.250 | 2.43 | *** |
| b 1-1 | 0.746 | 1.01 | 0.243 | 3.07 | *** |
| b 2-2 | 1.119 | 1.01 | 0.243 | 4.61 | *** |
| b 3-3 | −0.379 | 1.01 | 0.242 | −1.57 | NS |
| b 1-2 | 1.064 | 1.00 | 0.334 | 3.18 | *** |
| b 1-3 | 0.396 | 1.00 | 0.336 | 1.18 | *** |
| b 2-3 | 0.138 | 1.00 | 0.336 | 0.41 | NS |
(***): Significant at the level 99.9%; (NS): nonsignificant at the level 95%.
| (4) |
The fit of the model was evaluated by the coefficient of determination R2, which was 0.983, indicating that 98.3% of the variability in the response could be explained by the model. The predicted R2 of 0.983 was in reasonable agreement with the adjusted R2 of 0.941 indicating that the regression model could be used to analyze trends of responses. The closer the value of R (multiple correlation coefficient) to 1, the better the correlation between the observed and the predicted values is [29]. The coefficient of variation CV indicates the degree of accuracy with which the treatments are compared. Usually, the higher the value of CV, the lower the reliability of the experiment is. In this experiment, a lower value (4.61) indicates higher reliability.
The statistical significance of (4) was checked by Fischer's F test. The results from this test as well as those from ANOVA yielded a very low P value, thus indicating that the model was highly significant and reliable (Table 6). The Student t distribution and the parameter estimate are given in Table 4. When the magnitude of the t test value is large and the P value is small, this indicates that the corresponding coefficient is highly significant [30]. As far as the current study is concerned, the estimated parameters and the corresponding P values suggest that, among all the independent variables, X1 (temperature), X2 (moisture), and X3 (inoculum age) had a significant effect on SPB1 biosurfactant production.
Table 6.
ANOVA analysis for response surface quadratic model.
| Sum of squares | df | Mean square | F value | P value | Significance | |
|---|---|---|---|---|---|---|
| Prob > F | ||||||
| Regression | 63.6369 | 9 | 7.0708 | 7.7595 | <0.001 | *** |
| Residual | 12.7574 | 14 | 0.9112 | |||
| Lack of fit | 6.6542 | 9 | 0.7394 | 0.6057 | 0.0758 | NS |
| Pure error | 6.1032 | 5 | 1.2206 | |||
|
| ||||||
| Total | 76.3943 | 23 | ||||
(NS): nonsignificant at the level 95%.
The effect of the interaction of various physicochemical parameters on biosurfactant production by B. subtilis was investigated by plotting the response surface curves against any two independent variables while keeping the third independent variable at the “0” level. Thus three response surfaces were obtained by considering all the possible combinations. The interactive roles of temperature, moisture, and inoculum on biosurfactant production by B. subtilis SPB1 are illustrated in the three-dimensional curves of the calculated response surface shown in Figure 2. These plots are helpful in studying the effects of the factors' variation in the studied field and consequently, in determining the optimal experimental conditions [31].
Figure 2.
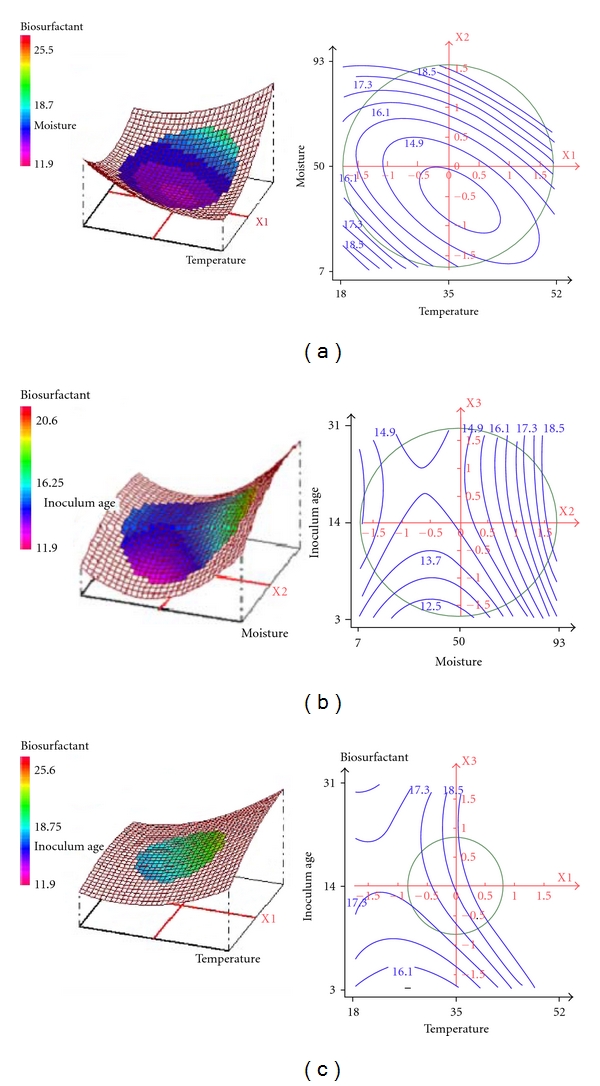
Effect of physical factors on the production of biosurfactant: response surface plot (left) and its contour plot (right) of interaction between (a) moisture and temperature, (b) inoculum age and moisture, (c) inoculum age and temperature.
The 3D response surface curves and their respective contour plots provided information about the interaction between two parameters and allowing an easy prediction and interpretation of the optimum experimental conditions.
Figure 2(a) indicated an interaction between moisture and temperature level. However a decrease was observed in SPB1 biosurfactant production at low moisture level percentage. It showed that moisture content (93%) was required with 35°C temperature for high yield of the response due to the maximum microbial activity which might be related to a balance between temperature activity for oxygen penetration and water availability [31].
Figure 2(b), showing the response surface plot, indicated no significant interaction between moisture and inoculum age. It depicted that a linear increase in SPB1 biosurfactant production was observed when moisture was increased up to 93%. However, for moisture middle value, it sharply declined.
Figure 2(c) showed an interaction between temperature and inoculum age indicating that a maximum biosurfactant production was observed when temperature was at 35°C and inoculum age was at 25 h.
3.6. Validation of the Model
The model was verified for the three variables within the design space. Four combinations of production conditions were selected by the software and were then tested for B. subtilis SPB1 biosurfactant production (Table 2). The experimental values of biosurfactant production that were determined were found to be in a good agreement with those that were statistically predicted by the model (R2 = 0.983), thus confirming the authenticity and reliability of the model. In addition, the average error (difference between observed and predicted value) was close to zero, indicating the absence of bias in the predictions made by the model.
3.7. Validation of the Optimum Points of the Factors
The response surfaces graphs showed the optimal levels of variables for biosurfactant production yield. The value of X1 (Temperature), X2 (Moisture), and X3 (inoculum age) were found to be 37°C, 88%, and 14 h, respectively. The biosurfactant production yield was 20.8 mg/g while the predicted value was 19.15 mg/g ± 1.36. This yield was very important compared to that reported in earlier studies. It is the highest reported to date for B. subtilis species in solid-state fermentation.
3.8. Time Course of Surfactin Production by B. subtilis SPB1
The time course of the biosurfactant production by B. subtilis SPB1 for both the optimized and the unoptimized media are shown in Figure 3. In the optimum conditions, the biosynthesis of the biosurfactant started since the first hour of culture and reached a maximum after 24 hours. The maximum of the biosurfactant production obtained under unoptimized conditions was only 12.5 after 24 h. By optimising the medium composition and the culture conditions, the production of SPB1 biosurfactant was enhanced from 12.5 mg/g to 20.8 mg/g.
Figure 3.
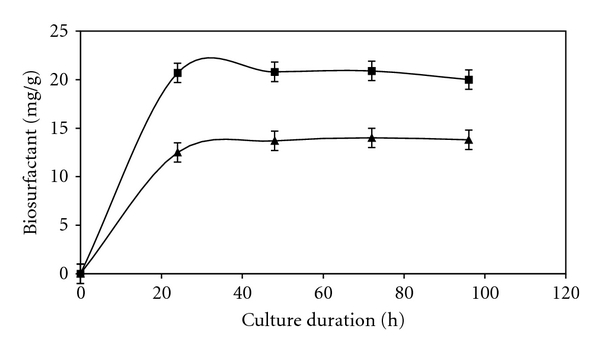
Kinetics of surfactin production by Bacillus subtilis: (▲) surfactin production (mg/g) before optimization; (■) surfactin production after optimization.
3.9. B. subtilis SPB1 Biosurfactant Characteristics
The thermoactivity and thermostability of the crude biosurfactant from B. subtilis are shown in Figure 4(a). The optimum of emulsification activity was obtained with temperatures lower than 37°C. This bioemulsifier retained more than 45% and 25% of its activity when incubated at 60 and 80°C, respectively. Moreover, up to 60°C, it was completely stable and retained more than 80% and 50% of its activity after incubation during 60 min at 70 and 80°C, respectively. The optimum of biosurfactant activity was shown for pH ranging from 5 to 9 and B. subtilis SPB1 biosurfactant was very stable over a considerable pH range from 2 to 9 (Figure 4(b)).
Figure 4.
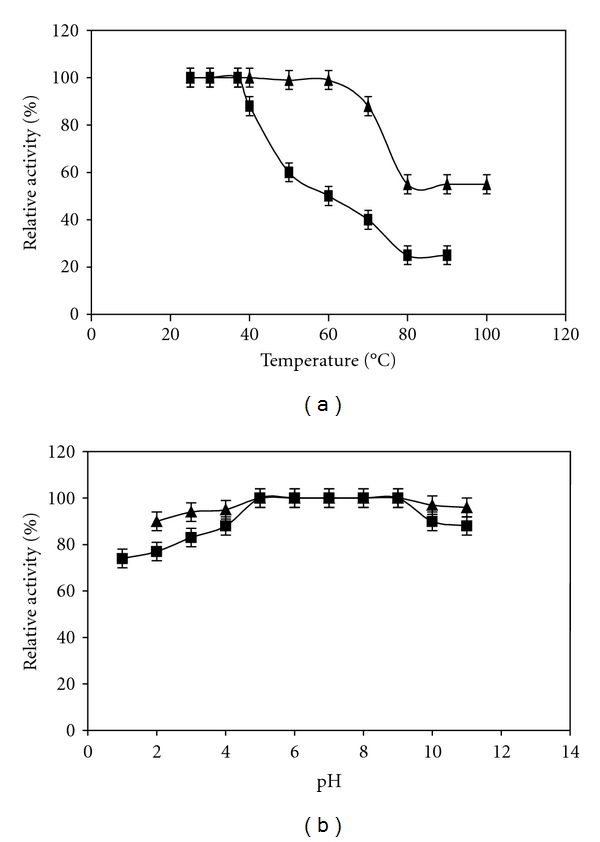
Effect of temperature (a) and pH (b) on the activity (■) and the stability (▲) of the crude biosurfactant produced by Bacillus subtilis SPB1.
4. Discussion
The production of surface active compounds, or biosurfactants, by microorganisms has been a subject of increasing interest in recent years, especially due to their potential applications in biotechnological and biopharmaceutical domains. Hemolytic activity has been used for the isolation of lipopeptide biosurfactant [32] and rhamnolipids [33]. The hydrophilic part of biosurfactant (the cationic part) is proposed to initiate electrostatic interaction with the negatively charged component of the membrane [34]. Biosurfactant producing capacity in liquid medium was found to be associated with hemolytic activity [35]. Thus, hemolytic activity appears to be a good screening criterion for surfactant producing strains [35]. Therefore, using such strategy, B. subtilis SPB1 strain was selected as the highest biosurfactant producer strain based on the size of the created hemolysis zone [23]. Several biosurfactants which exhibit antimicrobial activity against various microorganisms have been previously described. They include surfactin and iturin produced by B. subtilis strains [3], rhamnolipids from Pseudomonas species [36], mannosylerythritol lipids from Candida antarctica [37], and biosurfactants produced by some fungi [37]. Moreover, there are few reports about the antimicrobial activity of biosurfactants isolated from lactobacilli; only biosurfactants obtained from S. thermophilus A and L. lactis 53 showed significant antimicrobial activity against several bacterial and yeast strains isolated from explanted voice prostheses [38]. The biosurfactant isolated in the present study exhibited a broad spectrum of action, including antimicrobial activity against microorganisms with multidrug-resistant profiles. The compound showed higher activity against Gram-positive cocci than against Gram-negative bacilli. Its activity was very important against Enterococcus faecalis. These results are of great interest, since these microorganisms have natural resistance to aztreonam, co-trimoxazole, cephalosporins, chloramphenicol, and clindamycin and low sensibility to aminoglycosides and penicillin G as discussed by Furtado et al. [39]. The antimicrobial activity of the compounds was also considerable against Staphylococcus aureus which is known to be resistant to at least two β-lactams. Moreover, it was clear that the activity against Gram-negative bacteria was lower when compared to Gram-positive bacteria. Singh and Cameotra [40] have also observed that the lipopeptide N1, produced by B. subtilis C1, was active against several microorganisms, especially S. aureus and Mycobacterium sp. Recently, interest in the potential industrial application of B. subtilis biosurfactants in medicine has increased. This is partially because their antiadhesive activity against a variety of pathogens was indicative of their potential use in the protection of biomaterials [41]. Furthermore, the biosurfactant produced by B. subtilis SPB1 was also characterized by an important antifungal activity, especially, against Penicillium notatum, Penicillium italicum, and Aspergillus niger and lower activity against Rhizopus oryzae and Aspergillus oryzae. This activity was clearly negative against Alternaria alternata, Puccinia allii, and Peronospora destructor. Moreover, it was clear that this compound presented also an important antiyeast activity, mainly, against Candida albicans.
Despite possessing many commercially attractive properties and clear advantages compared with their synthetic counterparts, the production of microbial surfactants on a commercial scale has not been realized because of their low yields and high production costs. In light of the economic constraints associated with biosurfactant production, essentially two basic strategies have been explored to a greater extent and have been reported to be effective in substantially increasing the production of biosurfactants. They corresponded, respectively, to the use of cheaper and waste substrates to lower the initial raw material costs involved in the process and the development of efficient bioprocesses, including optimization of the culture conditions for maximum biosurfactant production. Maximizing productivity or minimizing production costs demands the use of process optimization strategies that involve multiple factors. The classical method of medium optimization is laborious, time consuming, and does not guarantee the determination of the optimal conditions for metabolite production. To tackle this problem and make the optimization process easier, a statistical optimization strategy based on response surface methodology has been used by various investigators. This method was successfully used to determine the optimum media, inoculum and environmental conditions for the enhanced biosurfactant production by B. subtilis [42]. These optimization methods would help the industry to design the best media containing cheaper substrates and to use the most favourable environmental conditions for improved biosurfactant production. There were many reports on the production of lipopeptide biosurfactants by B. subtilis species in solid-state fermentation [43, 44]. Biosurfactant production was 9.26 mg/g when B. subtilis S3 was cultivated for five days in solid-state fermentation containing high gluten flour and rice bran [45]. B. subtilis RB14-CS produced 14 mg/g dry solid material after four days of fermentation using soybean crude residue (okara) [46]. After that, response surface methodology was applied to predict the optimum amounts of the carbon and nitrogen source for biosurfactant production by B. subtilis in solid-state fermentation [47], in that the maximum production yield was 15.591 mg/g under optimized condition. In the present study, B. subtilis SPB1 biosurfactant production yield was increased significantly through application of the solid-state fermentation and the response surface methodology. The central composite design exploited in the present study enabled us to investigate the culture conditions that support biosurfactant overproduction. A high degree of similarity was observed between the predicted and experimental values that reflected the accuracy and applicability of the response surface methodology to optimise the process for biosurfactant production. The maximum production was achieved at temperature of 37°C, moisture of 88%, and inoculum age of 14 h. Excess or diminution in one of these values may cause a disruption in the corresponding production yield. The yield of the biosurfactant (20.8 mg/g) produced by B. subtilis SPB1 reported in this study is the highest reported to date for B. subtilis species in solid-state fermentation. In addition, the use of millet as a substrate in solid-state fermentation for biosurfactants production by B. subtilis is unique. By increasing the biosurfactant yield via this experimental design approach, the production cost would markedly be reduced, enhancing feasibility of commercial application of this powerful biosurfactant. The properties of the crude biosurfactant were also studied. This bioemulsifier was very stable over a considerable pH and temperature range. The result was in accordance with those reported by Horowitz et al. [48] showing that biosurfactant from Bacillus licheniformis 86 was stable in the temperatures range of 25–120°C. Therefore, the stability of this biosurfactant over a wide range of pH and after heat treatment indicates that it preserves its activity at extreme conditions, which is an extremely interesting feature in view of its potential use in detergent industries and bioremediation of hydrocarbons.
Conflict of Interests
The authors have declared no conflict of interests.
Acknowledgments
This work has been supported by grants from ‘‘Tunisian Ministry of Higher Education, Scientific Research and Technology,” The authors would like to express their gratitude to Professor Faiza Ghribi, English professor at Schooling Plus Language Teaching and Training Center for carefully proofreading and editing the paper of the present study.
References
- 1.Martins S, Mussatto SI, Martínez-Avila G, Montañez-Saenz J, Aguilar CN, Teixeira JA. Bioactive phenolic compounds: production and extraction by solid-state fermentation. Biotechnology Advances. 2011;29(3):365–373. doi: 10.1016/j.biotechadv.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Holker U, Lenz J. Solid-state fermentation—are there any biotechnological advantages? Current Opinion in Microbiology. 2005;8(3):301–306. doi: 10.1016/j.mib.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Sriram MI, Kalishwaralal K, Deepak V, Gracerosepat R, Srisakthi K, Gurunathan S. Biofilm inhibition and antimicrobial action of lipopeptide biosurfactant produced by heavy metal tolerant strain Bacillus cereus NK1. Colloids and Surfaces B. 2011;85(2):174–181. doi: 10.1016/j.colsurfb.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Henry G, Deleu M, Jourdan E, Thonart P, Ongena M. The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cellular Microbiology. 2011;13(11):1824–1837. doi: 10.1111/j.1462-5822.2011.01664.x. [DOI] [PubMed] [Google Scholar]
- 5.Cameotra SS, Makkar RS. Recent applications of biosurfactants as biological and immunological molecules. Current Opinion in Microbiology. 2004;7(3):262–266. doi: 10.1016/j.mib.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi T, Hasumi K. Enhancement of plasminogen activation by surfactin C: augmentation of fibrinolysis in vitro and in vivo. Biochimica et Biophysica Acta. 2002;1596(2):234–245. doi: 10.1016/s0167-4838(02)00221-2. [DOI] [PubMed] [Google Scholar]
- 7.Kim SD, Park SK, Cho JY, et al. Surfactin C inhibits platelet aggregation. Journal of Pharmacy and Pharmacology. 2006;58(6):867–870. doi: 10.1211/jpp.58.6.0018. [DOI] [PubMed] [Google Scholar]
- 8.Banat IM, Franzetti A, Gandolfi I, et al. Microbial biosurfactants production, applications and future potential. Applied Microbiology and Biotechnology. 2010;87(2):427–444. doi: 10.1007/s00253-010-2589-0. [DOI] [PubMed] [Google Scholar]
- 9.Youssef N, Simpson DR, Duncan KE, et al. In-situ biosurfactant production by Bacillus strains injected into a limestone petroleum reservoir. Applied and Environmental Microbiology. 2007;73(4):1239–1247. doi: 10.1128/AEM.02264-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulligan CN. Environmental applications for biosurfactants. Environmental Pollution. 2005;133(2):183–198. doi: 10.1016/j.envpol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Touré Y, Ongena M, Jacques P, Guiro A, Thonart P. Role of lipopeptides produced by Bacillus subtilis GA1 in the reduction of grey mould disease caused by Botrytis cinerea on apple. Journal of Applied Microbiology. 2004;96(5):1151–1160. doi: 10.1111/j.1365-2672.2004.02252.x. [DOI] [PubMed] [Google Scholar]
- 12.Assie LK, Deleu M, Arnaud L, et al. Insecticide activity of surfactins and iturins from a biopesticide Bacillus subtilis Cohn (S499 strain) Mededelingen. 2002;67(3):647–655. [PubMed] [Google Scholar]
- 13.Rahman PKSM, Gakpe E. Production, characterisation and applications of biosurfactants. Biotechnology. 2008;7(2):360–370. [Google Scholar]
- 14.Yeh MS, Wei YH, Chang JS. Bioreactor design for enhanced carrier-assisted surfactin production with Bacillus subtilis. Process Biochemistry. 2006;41(8):1799–1805. [Google Scholar]
- 15.Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946;33(4):305–325. [Google Scholar]
- 16.Master RN. Nitrate reduction. In: Isenberg HD, editor. Clinical Microbiology Procedure Handbook. Washington, DC, USA: ASM Press; 1996. [Google Scholar]
- 17.Sambrouk J, Fritch EF, Maniatis T. Molecular Cloning: A laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghribi D, Zouari N, Trigui W, Jaoua S. Use of sea water as salts source in starch- and soya bean-based media, for the production of Bacillus thuringiensis bioinsecticides. Process Biochemistry. 2007;42(3):374–378. [Google Scholar]
- 20.Ohno A, Ano T, Shoda M. Production of a lipopeptide antibiotic surfactin with recombinant Bacillus subtilis. Biotechnology Letters. 1992;14(12):1165–1168. doi: 10.1002/bit.260470212. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee S, Das P, Sen R. Towards commercial production of microbial surfactants. Trends in Biotechnology. 2006;24(11):509–515. doi: 10.1016/j.tibtech.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Paik HD, Bae SS, Park SH, Pan JG. Identification and partial characterisation of tochicin, a bacteriocin produced by Bacillus thuringiensis subsp. tochigiensis. Journal of Industrial Microbiology and Biotechnology. 1997;19(4):294–298. doi: 10.1038/sj.jim.2900462. [DOI] [PubMed] [Google Scholar]
- 23.Kamoun F, Mejdoub H, Aouissaoui H, Reinbolt J, Hammami A, Jaoua S. Purification, amino acid sequence and characterization of bacthuricin F4, a new bacteriocin produced by Bacillus thuringiensis. Journal of Applied Microbiology. 2005;98(4):881–888. doi: 10.1111/j.1365-2672.2004.02513.x. [DOI] [PubMed] [Google Scholar]
- 24.Patel RM, Desai AJ. Surface-active properties of rhamnolipids from Pseudomonas aeruginosa GS3. Journal of Basic Microbiology. 1997;37(4):281–286. doi: 10.1002/jobm.3620370407. [DOI] [PubMed] [Google Scholar]
- 25.Kunamneni A, Kumar KS, Singh S. Response surface methodological approach to optimize the nutritional parameters for enhanced production of α-amylase in solid state fermentation by Thermomyces lanuginosus. African Journal of Biotechnology. 2005;4(7):708–716. [Google Scholar]
- 26.Mathieu D, Nony J, Phan-Tan-Luu R, Nemrod W. New Efficient Methodology for Research Using Optimal Design (NEMROD) Software. Marseilles, France: LPRAI; 2000. [Google Scholar]
- 27.Nair SR, Panda T. Statistical optimization of medium components for improved synthesis of pectinase by Aspergillus niger. Bioprocess Engineering. 1997;16(3):169–173. [Google Scholar]
- 28.Carrera P, Cosmina P, Grandi G. Method of producing surfactin with the use of mutant of Bacillus subtilis. US Patant no. 5227294, 1993.
- 29.Sayyad SA, Panda BP, Javed S, Ali M. Optimization of nutrient parameters for lovastatin production by Monascus purpureus MTCC 369 under submerged fermentation using response surface methodology. Applied Microbiology and Biotechnology. 2007;73(5):1054–1058. doi: 10.1007/s00253-006-0577-1. [DOI] [PubMed] [Google Scholar]
- 30.Karthikeyan RS, Rakshit SK, Baradarajan A. Optimization of batch fermentation conditions for dextran production. Bioprocess Engineering. 1996;15(5):247–251. [Google Scholar]
- 31.Kammoun R, Naili B, Bejar S. Application of a statistical design to the optimization of parameters and culture medium for α-amylase production by Aspergillus oryzae CBS 819.72 grown on gruel (wheat grinding by-product) Bioresource Technology. 2008;99(13):5602–5609. doi: 10.1016/j.biortech.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 32.Nitschke M, Pastore GM. Production and properties of a surfactant obtained from Bacillus subtilis grown on cassava wastewater. Bioresource Technology. 2006;97(2):336–341. doi: 10.1016/j.biortech.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 33.Iqbal S, Khalid ZM, Malik KA. Enhanced biodegradation and emulsification of crude oil and hyperproduction of biosurfactants by a gamma ray-induced mutant of Pseudomonas aeruginosa. Letters in Applied Microbiology. 1995;21(3):176–179. doi: 10.1111/j.1472-765x.1995.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 34.Pag U, Oedenkoven M, Papo N, Oren Z, Shai Y, Sahl HG. In vitro activity and mode of action of diastereomeric antimicrobial peptides against bacterial clinical isolates. Journal of Antimicrobial Chemotherapy. 2004;53(2):230–239. doi: 10.1093/jac/dkh083. [DOI] [PubMed] [Google Scholar]
- 35.Carrillo PG, Mardaraz C, Pitta-Alvarez SI, Giulietti AM. Isolation and selection of biosurfactant-producing bacteria. World Journal of Microbiology and Biotechnology. 1996;12(1):82–84. doi: 10.1007/BF00327807. [DOI] [PubMed] [Google Scholar]
- 36.Benincasa M, Abalos A, Oliveira I, Manresa A. Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa LBI from soapstock. Antonie van Leeuwenhoek. 2004;85(1):1–8. doi: 10.1023/B:ANTO.0000020148.45523.41. [DOI] [PubMed] [Google Scholar]
- 37.Meylheuc T, Methivier C, Renault M, Herry JM, Pradier CM, Bellon-Fontaine MN. Adsorption on stainless steel surfaces of biosurfactants produced by gram-negative and gram-positive bacteria: consequence on the bioadhesive behavior of Listeria monocytogenes. Colloids and Surfaces B. 2006;52(2):128–137. doi: 10.1016/j.colsurfb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigues L, Banat IM, Teixeira J, Oliveira R. Biosurfactants: potential applications in medicine. Journal of Antimicrobial Chemotherapy. 2006;57(4):609–618. doi: 10.1093/jac/dkl024. [DOI] [PubMed] [Google Scholar]
- 39.Furtado GHC, Martins ST, Coutinho AP, Soares GMM, Wey SB, Medeiros EAS. Incidência de Enterococcus resistente á vancomicina em um hospital universitário no Brasil. Revista de Saúde Pública. 2005;39:41–46. doi: 10.1590/s0034-89102005000100006. [DOI] [PubMed] [Google Scholar]
- 40.Singh P, Cameotra SS. Potential applications of microbial surfactants in biomedical sciences. Trends in Biotechnology. 2004;22(3):142–146. doi: 10.1016/j.tibtech.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 41.Walencka E, Ró alska S, Sadowska B, Ró alska B. The influence of Lactobacillus acidophilus-derived surfactants on staphylococcal adhesion and biofilm formation. Folia Microbiologica. 2008;53(1):61–66. doi: 10.1007/s12223-008-0009-y. [DOI] [PubMed] [Google Scholar]
- 42.Sen R, Swaminathan T. Response surface modeling and optimization to elucidate and analyze the effects of inoculum age and size on surfactin production. Biochemical Engineering Journal. 2004;21(2):141–148. [Google Scholar]
- 43.Akihiro O, Takashi A, Makoto S. Production of the antifungal peptide antibiotic, iturin by Bacillus subtitilis NB22 in solid state fermentation. Journal of Fermentation and Bioengineering. 1993;75:23–27. [Google Scholar]
- 44.Ohno A, Ano T, Shoda M. Production of a lipopeptide antibiotic, surfactin, by recombinant Bacillus subtilis in solid state fermentation. Biotechnology and Bioengineering. 1995;47(2):209–214. doi: 10.1002/bit.260470212. [DOI] [PubMed] [Google Scholar]
- 45.Kuo CY. Optimization of cultivation conditions for iturin A production by Bacillus subtilis using solid state fermentation. Taiwan: Da-Yeh University; 2006. M.S. thesis. [Google Scholar]
- 46.Mizumoto S, Hirai M, Shoda M. Production of lipopeptide antibiotic iturin A using soybean curd residue cultivated with Bacillus subtilis in solid-state fermentation. Applied Microbiology and Biotechnology. 2006;72(5):869–875. doi: 10.1007/s00253-006-0389-3. [DOI] [PubMed] [Google Scholar]
- 47.Mizumoto S, Shoda M. Medium optimization of antifungal lipopeptide, iturin A, production by Bacillus subtilis in solid-state fermentation by response surface methodology. Applied Microbiology and Biotechnology. 2007;76(1):101–108. doi: 10.1007/s00253-007-0994-9. [DOI] [PubMed] [Google Scholar]
- 48.Horowitz S, Gilbert JN, Griffin WM. Isolation and characterization of a surfactant produced by Bacillus licheniformis 86. Journal of Industrial Microbiology. 1990;6(4):243–248. [Google Scholar]


