Abstract
Neutralization properties of human immunodeficiency virus (HIV-1) are often defined using pseudoviruses grown in transformed cells, which are not biologically relevant HIV-1 producer cells. Little information exists on how these viruses compare to viruses produced in primary lymphocytes, particularly for globally relevant HIV-1 strains. Therefore, replication-competent chimeras encoding envelope variants from the dominant HIV-1 subtypes (A, C, and D) obtained early after infection were generated and the neutralization properties explored. Pseudoviruses generated in 293T cells were the most sensitive to antibody neutralization. Replicating viruses generated in primary lymphocytes were most resistant to neutralization by plasma antibodies and most monoclonal antibodies (b12, 4E10, 2F5, VRC01). These differences were not associated with differences in envelope content. Surprisingly, the virus source did not impact neutralization sensitivity of most viruses to PG9. These findings suggest that producer cell type has a major effect on neutralization sensitivity, but in an antibody dependent manner.
Keywords: Human Immunodeficiency Virus, Neutralizing Antibodies, Producer Cell, Pseudovirus
INTRODUCTION
A strong humoral immune response to human immunodeficiency virus type 1 (HIV-1) is considered integral to any effective vaccine response (Burton et al., 2004; Mascola and Montefiori, 2010). Thus, in vitro assays that measure HIV-1 specific neutralizing antibodies that can be used in high-throughput screening are essential. The most commonly used assay to measure HIV-1-specific neutralization employs a pseudovirus capable of a single round of infection and an indicator cell line that expresses high levels of the HIV-1 receptors [TZM-bl also named JC53BL (Li et al., 2005; Platt et al., 1998; Wei et al., 2003)]. However, at present, there is little evidence for a correlation between neutralizing antibody titers measured by the commonly used in vitro neutralization assays and protection from HIV-1 infection in humans. Thus, there is substantial interest in other assays to measure HIV-1 specific antibody activity, including other neutralization assays, such as those using peripheral blood mononuclear cells (PBMCs) (Brown et al., 2008) and antibody-dependent cellular viral inhibition (ADCVI) assays, which capture other non-neutralizing antibody activities [reviewed in (Chung et al., 2008) and (Forthal and Moog, 2009)]. Such assays require the use of replication-competent viruses, of which relatively few exist [only 30 full-length infectious viral isolates are available in the NIH AIDS Research and Reference Reagent Program (aidsreagents.org)]. Since the target of neutralizing antibodies is the HIV-1 envelope protein, one alternative approach to generate genetically diverse replication-competent viruses for these assays is to create chimeric viruses using a common full-length HIV-1 provirus and introducing diverse heterologous envelope sequences into this provirus.
A major outstanding question with regard to the use of these replication-competent chimeras is how similar is a virus generated from a chimeric provirus compared to a pseudovirus, generated by co-transfection, that contains identical genes (hereafter “isogenic”). Two studies have examined the effect on envelope expression context (either as a pseudovirus versus full-length virus) and found that this did not affect neutralization phenotype (Louder et al., 2005; Provine et al., 2009). The study by Louder and colleagues using three CXCR4 or dual-tropic subtype B envelope variants from chronic infection, reported that replication-competent chimeras and isogenic pseudoviruses exhibited equivalent neutralization sensitivity when tested against a panel of broadly neutralizing monoclonal antibodies (MAbs) (Louder et al., 2005). Similar results were observed in a study of two CCR5-using subtype A variants from chronic infection using MAbs and entry inhibitors (Provine et al., 2009).
Another relevant issue with the use of pseudoviruses for the study of neutralization sensitivity is that they are generated in transformed cells, rather than a more biologically relevant producer cell. Viruses used for single-cycle assays such as the TZM-bl assay are typically generated in human embryonic kidney (HEK) 293T cells, because it is possible to produce large amounts of virus by transient transfection in these cells (Graham et al., 1977). While a number of studies have examined the role of producer cell type on neutralization phenotype (Sawyer et al., 1994; Zhang et al., 1997), only two directly compared 293T cells and primary cells such as PBMCs as the producer cells (Louder et al., 2005; Mann et al., 2009). The study by Louder et al., demonstrated that for three CXCR4-using subtype B variants, generation of replication-competent chimeras in PBMCs significantly decreased neutralization sensitivity compared to 293T-derived virus from the same chimera (Louder et al., 2005). A separate study using clade B viral isolates from both acute and chronic infection also found that growth of virus in PBMC decreased neutralization sensitivity to the MAb 2G12, which recognizes a collection of glycan residues on the viral envelope (Mann et al., 2009; Trkola et al., 1996). However, this study used primary isolates derived by short-term co-culture of patient PBMCs rather than cloned viruses, and therefore the pseudoviruses and replication-competent viruses were not identical. Furthermore, most non-clade B viruses lack the 2G12 epitope so these findings cannot be recapitulated using most pandemic relevant proviral clones (Blish et al., 2009). Nonetheless, these studies collectively suggest that the source of virus may have a significant impact on the assessment of neutralization sensitivity.
To date there are no data examining neutralization differences of isogenic replication-competent HIV-1 and corresponding pseudovirus encoding envelope variants that represented early-stage variants of the major circulating subtypes, which are of particular interest for assessing vaccine responses and their efficacy. Therefore, we have characterized the role of envelope expression context and producer cell type for nine novel replication-competent chimeric HIV-1 isolates from the dominant circulating HIV-1 subtypes in Africa, where most new HIV-1 infections are occurring.
RESULTS
Infectivity and replication properties of chimeric viruses
Nine full-length chimeric proviruses (Table 1) were generated using a full-length proviral clone, Q23-17, that was obtained from a subtype-A infected Kenya woman early in infection (Poss and Overbaugh, 1999). The original provirus was modified to allow introduction of unique restriction sites (SmaI and XhoI) flanking the envelope gene to generate Q23XhoΔXho, and envelope sequences were introduced as described (Provine et al., 2009). A schematic of the construct used to generate these chimeras is shown in Figure 1A. The envelope variants chosen included subtype A, C, and D variants and thus represent the most common circulating strains of HIV-1 in Africa. Eight of nine of these envelopes clones were derived directly from the cells of infected individuals without culturing and all but one is from within six months of infection (range 23-142 days). Since it is not possible to identify individuals at the precise time they acquired HIV and isolate the precise viral sequence that led to their infection, the study of these variants from very early in infection provides a useful surrogate for viruses that successfully established a new infection (Table 1). Two previously generated chimeras from chronically infected transmitting mothers were also included, MG505.C2 chimera and MS208.A3 chimera (Provine et al., 2009), as well as the parental virus, Q23XhoΔXho, listed as Q23-17 env as this is the envelope gene expressed by the virus (Table 1). In all cases, the replication-competent provirus was compared to pseudovirus generated with the corresponding envelope sequences and a Q23-17 provirus deleted in env (Q23Δenv) (Poss and Overbaugh, 1999).
Table 1.
Characterization of Envelope Variants
| Original envelope variant namea | Name used here | Clade | Length of infectionb | Virus sourcec | Tierd | Reference |
|---|---|---|---|---|---|---|
| Q23-17 env | Q23-17 env | A | 142 dpi | Blood, Adult | 1b | (Long et al., 2002; Poss and Overbaugh, 1999) |
| Q168.b23 | Q168.b23 | A | 23 dpi | Blood, Adult | 1b | (Long et al., 2002) |
| Q461.e2 | Q461.e2 | A | 28 dpi | Blood, Adult | 2 | (Long et al., 2002) |
| MG505.W0M.ENV.C2 | MG505.C2 | A | Chronic | Blood, Adult | 2 | (Wu et al., 2006) |
| MS208.W6B.ENV.A3 | MS208.A3 | A | Chronic | Breast milk | 2 | (Rainwater et al., 2007) |
| BJ613.W6M.ENV.E1 | BJ613.E1 | A | Week 6 | Blood, Infant | 2 | (Wu et al., 2006) |
| BS208.W6M.ENV.B1 | BS208.B1 | A | Week 6 | Blood, Infant | 2 | (Wu et al., 2006) |
| QB099.391M.ENV.B1 | QB099.B1 | C | 391 dpi | Blood, Adult | 2 | (Blish et al., 2009) |
| QC406.70M.ENV.F3 | QC406.F3 | C | 70 dpi | Blood, Adult | 2 | (Blish et al., 2009) |
| BJ412.W6M.ENV.S3 | BJ412.S3 | C | Week 6 | Blood, Infant | 2 | (Wu et al., 2006) |
| QA013.70I.ENV.M12 | QA013.M12 | D | 70 dpi | Primary isolate, Adult | 2 | (Blish et al., 2009) |
| QD435.100M.ENV.A4 | QD435.A4 | D | 100 dpi | Blood, Adult | 2 | (Blish et al., 2009) |
The original nomenclature for the published envelope, and used for the sequence designation in Genbank (http://www.ncbi.nlm.nih.gov/genbank/). The longer envelope designation is truncated as indicated, and this abbreviated version is used throughout the text and figures.
For adults, this estimate is based on published data and methods for estimating time of infection as described (Lavreys et al., 2004). For infants (BJ412, BJ613, and BS208), the time post infection is the first time point where the infant was HIV-1 positive after birth as described (Wu et al., 2006).
Blood = isolated directly from PBMCs from the individual, either adult or infant as denoted; Breast milk = isolated from breast milk cells; Primary isolate = isolated after short-term culture in PBMCs from an adult individual.
The Tier designation was estimated based on comparison with viruses that have been described in prior studies (Seaman et al., 2010), as defined in the Materials and Methods.
FIG 1. Generation and characterization of chimeric proviruses encoding diverse envelope variants.
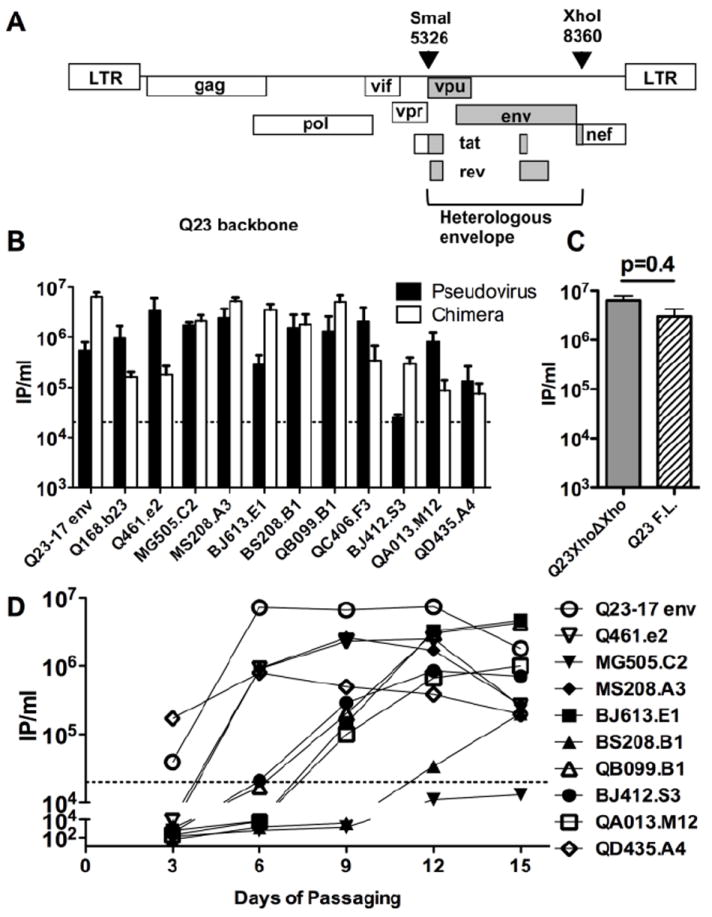
(A) Schematic of the chimeric proviruses derived from Q23XhoΔXho. Restriction sites used to introduce envelope sequences, SmaI and XhoI, are shown. The numbering refers to the 5’ (SmaI) and 3’ (xhoI) most extents of the restriction sites based on the Q23-17 genome (Genbank ID: AF004885). The portions of the chimeras that are derived from Q23XhoΔXho (LTRs, gag, pol, vif, and vpr) are shown in white boxes, those derived from the insert sequences (vpu, env, and rev) are shown in hatched boxes. The tat and nef genes are chimeras of the two viral strains. (B) Infectious titer of replication-competent chimeras compared to isogenic pseudoviruses generated in 293T cells. IP/ml refers to infectious particles per ml (IP/ml) determined using TZM-bl reporter cells. Black bars indicate the IP/ml for the pseudovirus while white bars indicate the IP/ml of the virus derived from the chimeric provirus; the envelope of each pair tested is shown at the bottom of the graph. The data represent the results of at least two separate experiments (each performed in duplicate); mean and SEM are shown. Dashed line denotes minimum IP/ml needed to perform the neutralization assay. (C) Infectious titer of the Q23XhoΔXho compared to the parental virus, Q23-17, which is the wild-type Q23 viral provirus prior to manipulation of the XhoI site for cloning (Provine et al., 2009). (D) Infection of PBMCs with the chimeric viruses generated in 293T cells. The envelope insert of each virus is listed. PBMC were infected with 293T-derived replication-competent chimera at an MOI of 0.01. The graph shows the IP/ml (infectious particles per ml determined in TZM-bl reporter cells) in cell-culture supernatant versus days post-infection. The symbol corresponding to the viruses tested is shown to the right of the graph. Representative data from one of two PBMC infections are shown. Dashed line denotes minimum IP/ml needed to perform the neutralization assay.
The viral titers of the isogenic pseudoviruses and replication-competent chimeras generated by transfection of HEK 293T cells were generally high, ranging from 2×104 to as high as 6×107 infectious particles per ml (Fig 1B). However, there were virus specific differences in the relative titer of the pseudovirus compared to the isogenic replication-competent chimera. On one extreme, the replication-competent chimeras of BJ412.S3 and BJ613.E1 as well as the parental Q23-17 env produced approximately 10-times as many infectious particles as the pseudovirus system, despite transfection of equal amounts of DNA (Fig 1B). In contrast, the pseudovirus systems of QA013.M12, Q168.B23, and Q461.e2 all produced at least 10-times as many infectious particles as the isogenic chimera system (Fig 1B).
To test whether the modification of the Q23-17 provirus, by mutation of a XhoI site to facilitate chimera creation, had any impact on replication properties, we compared replication of the parental Q23-17 virus and the modified Q23XhoΔXho, which differs at one amino acid in Nef. There was no significant difference in titer between virus generated from the Q23-17 proviral HIV-1 clone and the Q23XhoΔXho clone (Fig 1C; p=0.4, Mann-Whitney U-test) suggesting that the modification of the cloning sites in Q23-17 to generate a unique XhoI site did not alter the replication properties. Additionally, Q23XhoΔXho was compared to full-length Q23-17 in all subsequent experiments and no notable difference between the two constructs was found (data not shown). The Q23-17 env chimeric virus (Q23XhoΔXho) produced the highest titer of virus (6.4×107 infectious particles per ml), but there were a number of chimeras that produced similar levels of infectious virus (Fig 1B). These results demonstrate that introduction of a heterologous envelope gene does not generally impair virus replication.
The chimeras were also assessed for their ability to establish spreading infection in human PBMCs in culture. Nine of the 12 replication-competent chimeras replicated to high titer in PMBCs, with a titer of >105 infectious particles by day nine (Fig 1D). In most cases, viral titer increased until day 12, but had diminished by day 15, presumably due to infected cell death. The titers of two viruses, QC406.F3 chimera and Q168.B23 chimera, dropped below detection by day 6 (data not shown). A third virus, MG505.C2 chimera established low level persistent infection over the 15 days in culture (Fig 1D). However, the titer never reached the 2×104 IP/ml needed to perform subsequent experiments (dashed horizontal line, Fig 1D). These three viruses were therefore excluded from subsequent studies utilizing PBMC-derived viruses. Several viruses replicated to equivalent levels as Q23-17 env (Fig 1D), again suggesting that introduction of a heterologous envelope does not necessarily negatively affect virus replication capacity.
Both envelope expression context and producer cell type affect sensitivity of HIV-1 to plasma antibody neutralization
Pooled plasma was used to examine the affect of the virus source on neutralization sensitivity against HIV-1 specific antibodies in plasma from HIV-1 infected individuals. The plasma was from HIV-1-infected Kenyans, and presumably includes individuals infected with subtype A, C, and D, which are the common subtypes in this region (Neilson et al., 1999). This plasma pool was tested against the pseudoviruses, isogenic 293T-derived chimeras, and the PMBC-derived chimeras. The chimeras generated in 293T cells exhibited a 1.8-fold decrease in sensitivity to neutralization compared to the pseudoviruses when tested against the HIV-1 positive plasma pool (Fig 2; Supplemental Table 1). The difference in IC50s (reciprocal dilution of plasma required to neutralize 50% of the virus) between the two groups was statistically significant (p=0.05). For ten of the viruses, the differences in IC50 values between pseudovirus and chimera were approximately two-fold or less. However, two isolates were dramatically more resistant when generated as 293T-derived chimeras. 293T-derived Q461.e2 chimera was approximately four-fold more resistant to neutralization than the matched pseudovirus, and 293T-derived QA013.M12 chimera was approximately 16-fold more resistant. These results suggest that virus generated from full-length replication-competent virus is generally more resistant to neutralization that an isogenic pseudovirus, although the magnitude of the difference is small. However, isolated specific differences can be several-fold greater than the median difference observed between the groups.
FIG 2. Comparison of virus source on neutralization sensitivity to an HIV-1 positive plasma pool.
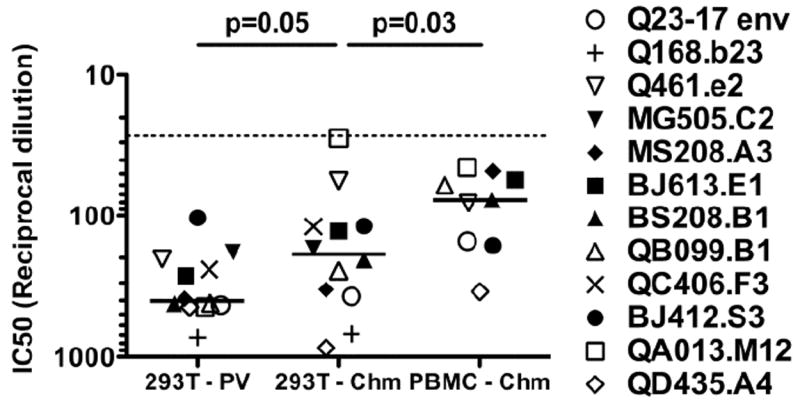
The source of the virus is shown on the x-axis: 293T-PV denotes pseudovirus obtained by transfecting 293T cells; 293T-Chm denotes virus obtained by transfecting 293T cells with a chimeric provirus; PBMC-Chm denotes virus obtained by infecting PBMCs with 293T-derived chimeric virus. The envelope of the various virus types is indicated to the right with the corresponding symbol. In the case of the chimeric viruses, the indicated designated envelope corresponds to the chimeric provirus in Fig. 1D. The IC50 (50% inhibitory concentration) is shown on the y-axis, and each symbol represents the mean IC50 of that virus calculated from a minimum of three experiments, each performed in triplicate. If a sample failed to reach 50% neutralization or was neutralized greater than 50% at the greatest dilution tested, the IC50 was set as the highest or lowest dilution of plasma tested, respectively. Dashed line denotes highest concentration of plasma tested. Solid line represents the median IC50 of each group. The P-value, shown above, was calculated by Wilcoxon signed-rank test. The HIV-1 positive plasma pool was obtained by pooling plasma from 31 HIV-1 positive individuals from Kenya.
Two other groups had noted decreased neutralization sensitivity for subtype B HIV-1 grown in PBMCs compared to 293T cells (Louder et al., 2005; Mann et al., 2009). To determine if growth of subtype A, C, and D viruses in PBMC had a similar affect on neutralization susceptibility as the subtype B variants, nine replication-competent chimeras that were capable of growth in PBMCs (all but MG505.C2 chimera, QC406.F3 chimera, and Q168.B23 chimera) were generated in 293T cells and in PBMCs and compared. When the effect of producer cell type was examined, a 2.7-fold reduction in median neutralization sensitivity of the PBMC-derived chimera was observed when compared to the 293T-derived chimera (Fig 2; p=0.03). Not all viruses were more resistant to neutralization when grown in PBMCs compared to 293T cells; three chimeras were slightly more sensitive to neutralization (BJ412.S3, QA013.M12, and Q461.e2). Overall the magnitude of differences between 293T-derived and PBMC-derived isolates was not variable from one isolate to the next. Only MS208.A3 exhibited an IC50 that was greater than four-fold different between the 293T-derived and PBMC-derived chimeras. Replication-competent virus grown in PBMC was five-fold more resistant to neutralization than pseudovirus generated in 293T cells. In sum, these data demonstrate that producer cell type has a major effect on neutralization sensitivity of isolates to plasma from HIV-1-infected individuals.
Producer cell type, not envelope expression context, affects sensitivity to neutralizing MAbs of moderate potency
The neutralization sensitivities of the twelve isogenic pairs of pseudoviruses and replication-competent chimeras were compared using three moderately potent HIV-1 specific MAbs that were tested in prior studies against pseudoviruses generated with the envelope variants used here (Blish et al., 2009; Blish et al., 2008; Rainwater et al., 2007; Wu et al., 2006). Of note, the envelopes under study exhibited a range of sensitivities to these MAbs, which included two (2F5 and 4E10) that target adjacent conserved epitopes in the membrane proximal external region (MPER) of gp41 (Muster et al., 1993; Zwick et al., 2001) and one (IgG1 MAb b12), which targets a discontinuous epitope overlapping the CD4 binding pocket (Burton et al., 1994). When the pseudoviruses and chimeras generated in 293T cells were compared using the three MAbs there was no significant difference in the distributions of IC50s between the two groups (Fig 3A, b12: p=0.3; Fig 3B, 4E10: p=0.85; Fig 3C, 2F5: p=0.16). The IC50s of the pseudoviruses were in good agreement with the published results (Blish et al., 2009; Blish et al., 2008; Rainwater et al., 2007; Wu et al., 2006), when available (data not shown).
FIG 3. Effect of virus source on neutralization phenotype to the first generation broadly neutralizing monoclonal antibodies.
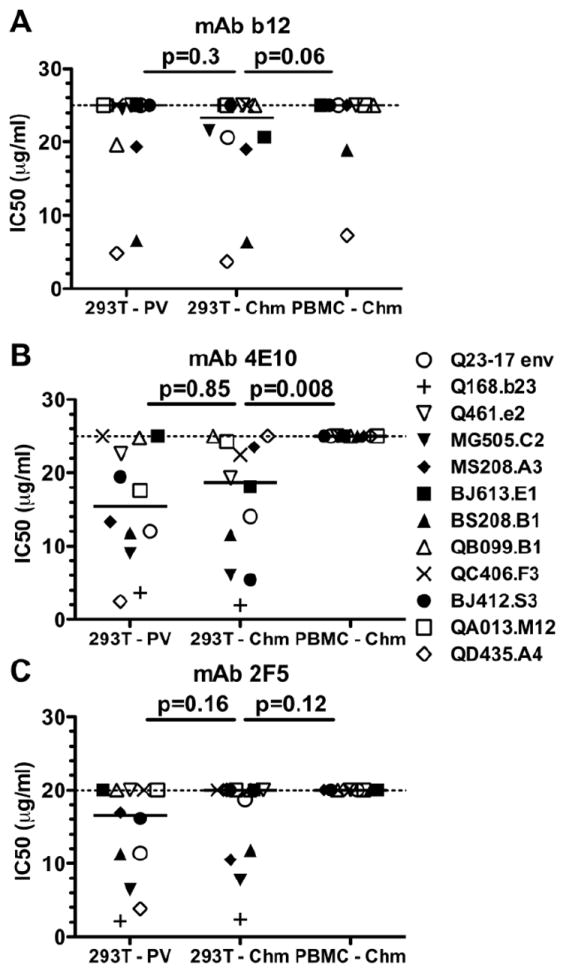
The layout for this figure is as described in the legend to Figure 2, with results for different monoclonal antibodies shown for each panel. (A) Viruses tested against the CD4 binding site directed MAb IgG1 b12 (Burton et al., 1994). (B) Viruses tested against the gp41 directed MAb 4E10 (Zwick et al., 2001). (C) Viruses tested against the gp41 directed MAb 2F5 (Muster et al., 1993). Each symbol represents the mean IC50 of the virus indicated by the envelope it contains to the right, calculated from a minimum of two experiments, performed in triplicate.
While no significant overall differences between IC50s of the 293T-derived pseudoviruses and 293T-derived chimeras were observed with these three MAbs, it is interesting to note that individual viral variants were not always functionally equivalent when produced either as a pseudovirus or a chimera. Specifically, the QD435.A4 envelope was >10-fold more sensitive to neutralization in the context of a pseudovirus compared to a chimera when tested against the MAbs 4E10 and 2F5 (Fig 3B and 3C, open diamond; Supplemental Table 2), but was equally sensitive to b12 (Fig 3A; Supplemental Table 2). Against 4E10 the chimera failed to reach 50% neutralization, whereas the pseudovirus was neutralized 50% at <2.5μg/ml. Against 2F5, the pseudovirus was sensitive to neutralization (IC50 of 3.8μg/ml) while the chimera was neutralized <50% at the highest concentration of antibody tested. In contrast, 293T-derived BJ412.S3 chimera (Fig 3B, filled circle; Supplemental Table 2) was approximately four-fold more sensitive the neutralization by 4E10 than the corresponding pseudovirus. These results demonstrate that while pseudovirus and replication-competent chimera generated in 293T cells are generally comparable, there are some virus-specific differences, as was also observed by Louder et al. (Louder et al., 2005).
To determine if the decreased sensitivity of PBMC-derived virus observed for plasma antibodies was also observed when MAbs were used, 293T-derived and PBMC-derived chimeras were compared. The PBMC-derived chimera was more resistant to neutralization than the 293T-derived chimera for all three MAbs tested (Fig 3; Supplemental Table 2). While only 4E10 was found to require significantly higher MAb levels to achieve neutralization of PBMC virus (Fig 3B; p=0.008), in all cases the PBMC-derived virus was more resistant to neutralization than analogous virus produced in 293T cells. In fact, every PBMC-derived chimera variant failed to reach 50% neutralization at the highest concentration of antibody tested when 4E10 and 2F5 were used (Fig 3B and 3C). When b12 was used, four of the variants that were neutralized when grown in 293T cells were no longer neutralized when grown in PBMCs, and the other two viruses, BS208.B1 chimera and QD435.A4 chimera, were more resistant (Fig 3A). A limitation of these findings was the relatively poor neutralizing potency of b12, 4E10, and 2F5 against the viruses employed. Hence, statistical significance was unachievable, despite a trend in increased neutralization resistance, with b12 and 2F5 because more than 50% of the chimeras were not neutralized, even when grown in 293T cells (Fig 3A and 3C).
In general, virus derived after PBMC passage was also not neutralized by a combination of the three MAbs (tested at a combined concentration of 50μg/ml total MAb). The exception was QD435.A4, which exhibited moderate sensitivity to the combined MAbs (IC50 19μg/ml; data not shown). These findings support the previous reports that short-term growth in PBMCs confers increased resistance to neutralization by monoclonal antibodies compared to virus derived by transient transfection of 293T cells (Louder et al., 2005; Mann et al., 2009).
The effect of producer cell on neutralization by highly potent MAbs, VRC01 and PG9
Two MAbs, VRC01 and PG9, that exhibit previously unprecedented neutralization breadth and potency have recently been described (Walker et al., 2009; Wu et al., 2010). VRC01 is a CD4 binding site directed antibody (Wu et al., 2010). PG9 is directed against the variable loops of gp120, and its target is thought to be a quaternary structure formed by the envelope trimer that includes glycans (Walker et al., 2009). Given the notable differences in neutralization sensitivity between 293T-derived versus PBMC-derived virus with plasma and the other MAbs, we examined the neutralization potential of VRC01 and PG9 against viruses generated in these two cell types. In the case of VRC01, PBMC-derived chimeras were more resistant to neutralization than 293T-derived chimeras (Fig 4A; p=0.0078). Both the median IC50 of the PBMC-derived chimera was approximately 6-fold greater than the 293T-derived chimera (Supplemental Table 3) and each isolate was more neutralization resistant when generated in PBMCs as compared to 293T cells. These findings are consistent with the observations made using the other less broadly neutralizing antibodies.
FIG 4. Neutralization potency of VRC01 and PG9 against 293T-derived versus PBMC-derived replication-competent chimeras.
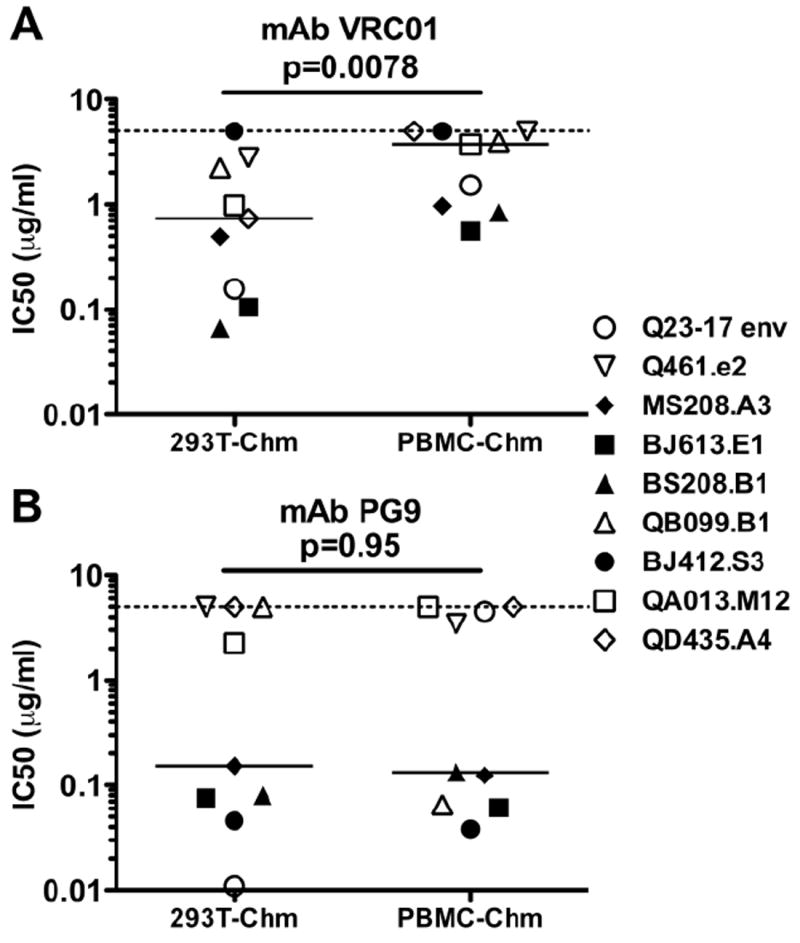
The layout for this figure is similar to that described in the legend to Figure 2. (A) Viruses tested against PG9 (Walker et al., 2009). (B) Viruses tested against VRC01 (Wu et al., 2010). Each symbol represents the mean IC50 of the virus indicated to the right, calculated from a minimum of two experiments, performed in triplicate.
Surprisingly, there was no difference in the neutralization sensitivity of PBMC-versus 293T-derived viruses using the MAb PG9. The median IC50s of 293T-derived chimeras and PBMC-derived chimeras were comparable when PG9 was used (Fig 4B; p=0.95). For seven virus pairs, there was a less than three-fold difference in neutralization sensitivity between viruses obtained from the two growth conditions. However, one of the chimeric viruses, QB099.B1, was approximately 77-fold more sensitive to neutralization when grown in PBMCs compared to 293T cells (Supplemental Table 3). In contrast, Q23-17 env chimera displayed neutralization characteristics more comparable to what was observed with the other antibodies. The 293T-derived chimera displayed an IC50 of 0.011μg/ml, whereas the PBMC-derived chimera had an IC50 of 4.50μg/ml, a more than 400-fold decrease in neutralization sensitivity for the PBMC-derived virus (Supplemental Table 3). Thus, for the majority of MAbs tested (b12, 4E10, 2F5, and VRC01), PBMC-derived chimeras displayed increased neutralization resistance compared to 293T-derived chimeras, with only PG9 displaying comparable potency between the two growth conditions overall.
Envelope levels in PBMC- versus 293T-derived viruses
We performed Western blot analyses to determine if differences in neutralization sensitivity between viruses derived from 293T versus PBMC were related to differences in Envelope levels. The results of these experiments are summarized in Fig. 5. Overall, there were no differences in the amount of Envelope cleavage between viruses derived from the two cell populations (Fig. 5A). To estimate the relative Envelope density of the different viruses, Envelope levels were calculated in relation to p24 levels. There was no statistically significant difference in the ratio of total Envelope (gp120 plus gp160)/p24 between viruses generated in 293T versus PBMC viruses (p=0.11; Fig 5B). However, PBMC-derived viruses had a trend towards significantly higher gp120/p24 ratios compared to 293T-derived viruses (p=0.079; Fig. 5C). The magnitude of this difference was small, less than 2-fold overall. Interestingly, there were some viruses with a greater than 2-fold difference in gp120/p24 levels, such as QA013 (3-fold greater expression on PBMC-derived virus) and QB099 (3.6-fold lower expression on PBMC-derived virus). However, these differences in gp120/p24 ratio were not reflected by consistent differences in neutralization patterns. For example, for QA013, the PBMC-derived virus was more resistant to VRC01 and PG9 than its 293T-derived counterpart, but displayed no differences (less than 2-fold) in sensitivity to the polyclonal plasma. In comparison, with QB099, the PBMC-derived virus showed similar sensitivity to VRC01 (less than 2-fold difference in IC50), increased sensitivity to PG9 compared to 293T-derived virus, but four-fold decreased sensitivity to the polyclonal plasma pool. Overall, there was no correlation between Envelope levels, as measured by either total Env/p24 or gp120/p24, and neutralization sensitivity, as defined by IC50 values, to either the plasma pool or the MAbs PG9 and VRC01 (Supplemental Figure 1).
FIG 5. Quantification of Envelope processing and Envelope incorporation of 293T-derived and PBMC-derived replication competent chimeras.
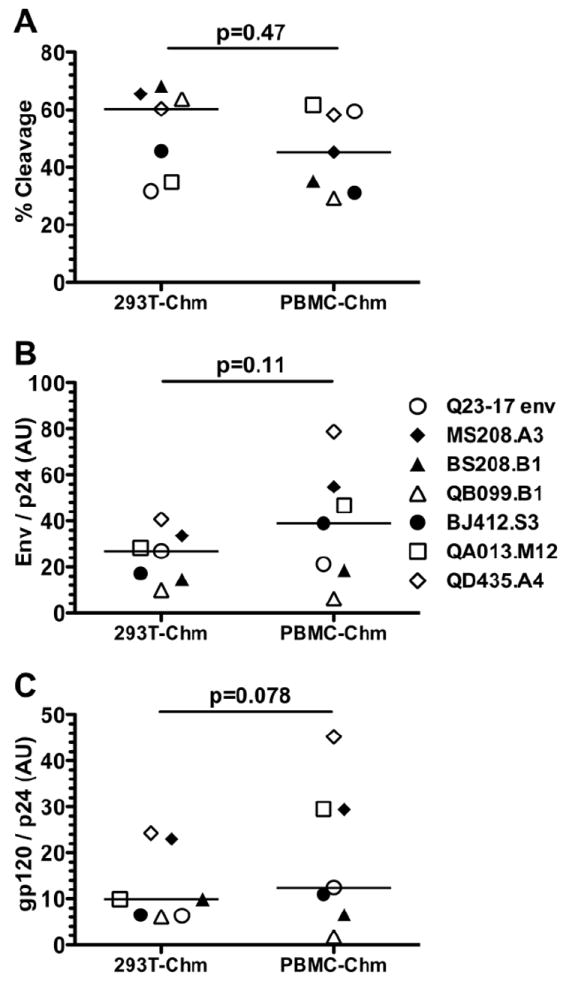
The source of the virus is shown on the x-axis: 293T-Chm denotes virus obtained by transfecting 293T cells with a chimeric provirus; PBMC-Chm denotes virus obtained by infecting PBMCs with 293T-derived chimeric virus. The envelope of the various virus types is indicated to the right with the corresponding symbol. Quantitative Western blotting was performed to determine the amount of gp120, gp160, and p24 (see Materials and Methods). (A) The % cleavage was calculated as integrated intensity (i.i.) of gp120 / (i.i. gp120 + i.i. gp160). (B) The total Env / p24 was calculated as (i.i. gp120 + i.i. gp160) / i.i. p24. (C) Amount of gp120 / p24 was calculated as the integrated i.i. gp120 / i.i. p24. The P-value, shown above each figure, was calculated by Wilcoxon signed-rank test. Each symbol represents average data from two experiments.
DISCUSSION
The findings of this study demonstrate that most viruses generated in lymphocyte cultures are more resistant to antibody neutralization than virus generated by transfection of 293T cells; this is consistent with previous studies utilizing clade B viruses (Louder et al., 2005; Mann et al., 2009). Thus, the current assays that rely on 293T-derived virus likely overestimate the true potency of the antibody under study. This can be seen as a positive feature of such assays, as it allows better detection of neutralizing antibody responses with less antibody. However, when considering the true potency of the antibody- that is the amount needed to neutralize virus produced by the typical target cells for HIV-1 in vivo- it is important to keep in mind that viruses in vivo may be significantly less sensitive to neutralization than the test virus. Unfortunately, it is not possible to directly calculate such a difference for any given virus or antibody, because there is considerable variation in the producer cell effect depending on the antibody as well as the specific virus. Here we found there is a significant producer cell effect with some broadly neutralizing MAbs such as VRC01, b12, 2F5, and 4E10, as well as pooled plasma from HIV-1 infected individuals, but this was not true with another very broad and potent MAb, PG9. However, given that the median difference in IC50 between the 293T-derived and PBMC-derived viruses was less than a five-fold difference, the inflated Ab potency determined using 293T-derived viruses is likely to be only several-fold in most cases. Interestingly, some viruses from lymphocytes were more resistant to one antibody but not another, while others were more resistant to all antibodies, suggesting there is likely no one feature of lymphocyte-derived viruses that explains their neutralization resistance.
Isogenic pseudoviruses and replication-competent chimeras generated in the same 293T producer cell were generally equivalent in neutralization sensitivity to the MAbs tested. This finding is consistent with a previous study using clade B viruses from chronic infection (Louder et al., 2005). Here, we observed that the chimeras generated in 293T cells were most resistant to antibodies in plasma for HIV-1-infected individuals than the corresponding pseudovirus, although the magnitude of the difference is small (~2-fold). For both MAbs and plasma, isolate specific variation was observed.
There are multiple non-exclusive explanations for the decreased neutralization sensitivity of the PMBC-derived compared to 293T-derived chimeras. Several studies have demonstrated that changes in the glycosylation of envelope can alter neutralization sensitivity (Duenas-Decamp et al., 2008; Utachee et al., 2010). A study comparing the effect of PBMCs versus macrophages as producer cell on neutralization sensitivity found macrophage-derived viruses were more resistant to neutralization than PBMC-derived viruses, and this was correlated with different glycosylation between the two cell types (Willey et al., 1996). Furthermore, a study examining the effect of producer cell type on amount and type of glycans found significant differences in gp120 glycosylation when produced in different cell lines, including 293T cells and Jurkat cells (Raska et al., 2010). These differences in glycosylation were shown to alter binding of IgG purified from HIV-1 infected individuals to gp120. If differences in glycan structure are important in driving differences between neutralization sensitivity of viruses made in 293T cells and PBMCs, then the results with PG9 are somewhat surprising. Glycans on Env have been shown to be critical for neutralization of virus by PG9 (Walker et al., 2009), and therefore changes in virus growth that alter the properties of envelope glycans would have been predicted to have the greatest impact on PG9 recognition, as observed for another glycan-dependent MAb, 2G12 (Mann et al., 2009). We did observe two viruses where the producer cell had a dramatic affect on PG9 recognition, in one case, the PBMC-derived virus was more neutralization sensitive and in the other, the opposite was true. These variants may hold clues to the features of viruses from different sources that drive neutralizing antibody recognition.
We examined whether the total amount of envelope, or the form of envelope present on the virion correlated with neutralization sensitivity. There was no overall correlation between envelope levels and neutralization sensitivity among the viruses tested, suggesting that the inherent antigenic properties of individual viral variants, not envelope density on the virion, is the major determinant of neutralization sensitivity, as observed in prior studies (Blish et al., 2008).
While the neutralization profile may be largely defined by nature of the viral variant itself, for an individual viral variant, differences in envelope density due to producer cell type could nonetheless contribute to the observed differences in neutralization susceptibility of a PBMC-derived virus compared to its 293T-derived counterpart. We found that the ratio of gp120 to p24 was higher for PBMC-derived viruses than for 293T viruses, suggesting a higher gp120 density for viruses generated in PBMCs. This is consistent with a prior study of three subtype B viruses which showed relatively few envelope spikes on 293T-derived chimeras (between one and three) compared to PBMC-derived virus [(an estimated 12 and 19 spikes;(Louder et al., 2005)]. The magnitude of the differences observed here for viruses from diverse subtypes was much more modest, overall less than two-fold. Moreover, virus pairs that showed the greatest differences in gp120/p24 levels based on producer cell type did not show a corresponding consistent change in neutralization sensitivity. Indeed, there are studies that suggest a relationship increased envelope levels of PBMC-derived viruses correlates with increased neutralization resistance (Blay et al., 2007) and others that do not (Provine et al., 2009). Thus, it is possible that Envelope density may contribute to the decreased neutralization sensitivity of the PBMC-derived viruses for some viral variants, but this is not generally the case. The fact that not all viruses or virus/antibody combinations are affected by producer cell also suggests that the basis for these differences in neutralization sensitivity due to producer cell may be complex.
As part of this study, we have developed nine novel replication-competent chimeras comprising envelope variants from the most common circulating HIV-1 subtypes, A, C, and D. Six are from early in infection in adults who acquired HIV-1 through sexual contact and three are from recently infected infants. These proviral clones produce high titer virus when transfected into 293T cells, and nine of the 12 chimeras tested established a high level infection of human PBMCs. Thus, these chimeric proviruses should be useful for examining the potential of interventions such as vaccines and microbicides to inhibit globally relevant viruses produced in primary cells. Since these viruses are replication-competent, they can also be employed in a range of cell types, including organ cultures and other models used to study early events in HIV-1 infection.
In conclusion, in this study of a novel panel of globally relevant HIV-1 envelope variants, we found that producer cell type has a dramatic effect on neutralization sensitivity. Pseudoviruses, which are employed in the most commonly used HIV-1 neutralization assays, are the most sensitive form of the virus, and thus may overestimate the potency of most HIV-1 antibodies. Lymphocyte-derived viruses were less sensitive to neutralization by the MAbs b12, 4E10, 2F5, and VRC01, as well as polyclonal plasma antibodies. However, it is notable that the producer cell did not effect neutralization by PG9 in most cases. Thus, these findings suggest complex determinants of neutralization sensitivity that are significantly affected by the cellular origin of the virus.
MATERIALS AND METHODS
HIV-1 plasmids
The HIV-1 envelopes used in this study have all been previously described. Three, BJ412.W6M.ENV.S3, BJ613.W6M.ENV.E1, and BS208.W6M.ENV.B1 are from recently infected Kenyan infants and two, MG505.W0M.ENV.C2 and MS208.W6B.ENV.A3 are from chronically infected mothers in the same study (Rainwater et al., 2007; Wu et al., 2006). Seven are envelope clones derived from recently infected Kenyan women, QA013.70I.ENV.M12, QB099.391M.ENV.B1, QC406.70M.ENV.F3, QD435.100M.ENV.A4, Q168.b23, Q461.e2, and Q23-17 env (Blish et al., 2009; Long et al., 2002). The envelope clone names are abbreviated in the paper to include only the subject ID and the clone number; for example, QA013.70I.ENV.M12 is designated QA013.M12. Q23Δenv was generated from a clade A HIV-1 proviral genome (Q23-17) (Poss and Overbaugh, 1999) by engineering an out-of-frame deletion in env (Long et al., 2002). Full-length chimeras were generated from the parental Q23-17 infectious proviral clone (Genbank ID: AF004885), as previously described (Provine et al., 2009). Briefly, a derivative of Q23-17 (Q23XhoΔXho) that was engineered to encode a XhoI site in nef (A to G transition at base 8358 resulting in an amino acid change of glutamine to glutamic acid) was digested with SmaI and XhoI, gel purified and ligated to a SmaI/XhoI digested fragment encoding the envelope genes of interest (the envelope clones under study were amplified with primers that contained the desired SmaI and XhoI sites – see references above for source of virus and primers employed) The resulting chimeric provirus was verified by restriction site digestion and sequence analysis of the whole envelope gene and flanking Q23 genomic sequence.
Virus preparation in 293T cells
Viruses were generated by transfection of 293T cells using FuGENE 6 Transfection Reagent (Roche, Mannheim, Germany). In all instances, 293T cells were plated at 2×106 cells in a T-75 flask. After one day, 4μg of total plasmid DNA was incubated with 388μl DMEM and 12μl of FuGENE, incubated at RT for 15 minutes, and added dropwise to the cells. The pseudoviruses were generated by cotransfection of the Q23Δenv plasmid and respective env plasmid at a 1:1 molar ratio; for the replication-competent viruses, 4μg of plasmid was transfected. After 48 hours, virus was harvested, cellular debris was removed by filtration through a 0.22μm filter (Steriflip, Millipore Corporation, Billerica, MA), and the viral supernatant was stored at -80°C for future use.
Virus preparation in peripheral blood mononuclear cells
PBMCs from HIV-1 negative donors were isolated as previously described (Mascola et al., 2002) and resuspended in complete RPMI (RPMI + 10% FBS (fetal bovine serum) +1% L-Glutamine + 1% PSF (Penicillin, Streptomycin, and Fungizone) +10U/ml interleukin 2 (IL-2) +10μg/ml phytohemagglutinin (PHA). After ~48hrs at 37°C, cells were pelleted, washed in PBS, and resuspended in complete RPMI plus IL-2 without PHA at a concentration of 2×106 cells/ml and returned to 37°C. After a further 48 hour incubation, 2×107 PBMCs at 6.7×106 cells/ml were infected with the virus derived from transfected 293T cells, at a multiplicity of infection of 0.01. After one hour incubation at 37°C, cells were gently pelleted by centrifugation at 400×g for 5min at room temperature, supernatant was removed and the cells were returned to a concentration of 2×106 cells/ml. Every 72 hours, cells were pelleted, supernatant was removed, aliquoted and frozen at -80°C. The PBMCs were subsequently resuspended at 2×106 cells/ml for additional culture over a course of 15 days.
Determination of viral infectivity
The infectious titer of viruses generated by transfection of 293T cells or infection of PBMCs was determined by infecting TZM-bl cells (Platt et al., 1998; Wei et al., 2003) and enumerating blue foci as described (Vodicka et al., 1997). Briefly, 100μl of serially diluted virus supernatant was added to 2×104 TZM-bl cells in 500μl of complete DMEM media (DMEM +10% FBS, 1% L-Glutamine, and 1% PSF) + 0.6μg of DEAE-Dextran, and incubated at 37°C. After 48 hours the supernatant was removed and 500μl of a 2.7% formaldehyde and 0.8% gluteraldehyde solution in PBS was added to fix the cells. After a maximum of 10 minutes the fix solution was removed and the cells were washed three times in PBS. The PBS was subsequently removed and 200μl of the staining solution (0.004M Potassium Ferrocyanide, 0.004M Potassium Ferricyanide, 0.002M MgCl2, and 0.4mg/ml X-gal) was added. The cells were stained for 50 minutes at 37°C after which time the staining solution was removed, cells were washed in PBS, and wells that contained between 30-300 blue foci were counted. The number of infectious particles per ml was then calculated based on the dilution. Each experiment was performed in duplicate, at least twice using two separate transfections.
Tier designation
The Tier designation for these envelope variants was estimated using the data and methods of Seaman et al (Seaman et al., 2010). It was not possible to directly compare the Tier cutoffs defined by Seaman with the IC50s of viruses used here because different plasma were used to define the IC50 values. However, because some of the envelope variants from our studies were analyzed in the prior study and determined to be Tier 2 viruses (Seaman et al., 2010), we were able to compare the relative sensitivity of these known Tier 2 variants versus the ones studied here using IC50s that were determined using the same pooled plasma (Blish et al., 2007a; Blish et al., 2009). This comparison was used to estimate the Tier designation.
Neutralization assay
Neutralization assays were performed using TZM-bl cell targets as previously described (Blay et al., 2007; Wu et al., 2006). The monoclonal antibodies 2F5 and 4E10 (acquired from the AIDS Research and Reference Reagent Program, National Institutes of Health) and IgG1 b12 (kindly provided by Dennis Burton) were used at a maximum concentration of 20, 25, and 25μg/ml, respectively. The monoclonal antibodies PG9 (kindly provided by Dennis Burton and the IAVI NAC) and VRC01 (kindly provided by John Mascola and the VRC team) were used at a maximum concentration of 5μg/ml. An HIV positive plasma pool, generated from 31 HIV-1-infected women from Kenya who had CD4 counts >500 cells/ml, was used at a starting dilution of 1:27. Each antibody or plasma was serially diluted two-fold and incubated with 500 infectious particles based on the titer in TZM-bl cells as described above. For viruses where an IC50 could not be determined for PG9 and VRC01, the MAbs were diluted five-fold. The antibody and virus were incubated for 1 hr, after which time, 104 TZM-bl cells and 10μg/ml DEAE-Dextran were added to each well. After 48h, luciferase activity was subsequently measured by a Thermo Scientific luminometer (Waltham, MA) using the Galacto-Lite Plus kit per the manufacturer’s instructions (Applied Biosystems, Foster City, CA). At each dilution, percent neutralization was calculated as [(value for virus only - value for cell only) - (value for antibody - value for cell only)] / (value for virus only - value for cell only). The 50% inhibitory concentration was calculated by interpolation of the linear regression of the neutralization curves converted to a logarithmic scale, performed in Microsoft Excel (Microsoft, Redmond, WA). In cases where the 50% inhibitory concentration was greater or less than the concentrations tested the value was reported as the maximum or minimum concentration/dilution, respectively. All experiments using monoclonal antibodies were performed twice, in triplicate, using virus generated by two independent transfections, and all results that did not agree within 2.5-fold were repeated two further times. The average IC50 was determined using all replicates. All neutralization experiments using the HIV-1 positive plasma pool were performed three times, in triplicate, using virus from at least two transfections.
Western blots
Purified pseudoviral preparations were prepared for Western blotting as previously described (Blish et al., 2007b; Lovelace et al., 2011). Briefly, pseudoviral supernatants were pelleted by microcentrifugation and resuspended in lysis buffer [1% Triton X-100 (Sigma)/PBS]. Samples were resolved on a 4–12% gradient Bis-Tris polyacrylamide gel (Invitrogen), followed by electrotransfer to a nitrocellulose membrane. A rabbit polyclonal antisera to HIV-1 Env (Doria-Rose et al., 2005) and mouse anti-p24 (cat. no. 4121; NIH AIDS Research and Reference Reagent Program) were used as a primary antibodies, with IRDye700DX-conjugated goat-anti-rabbit IgG and IRDye800DX-conjugated goat-anti-mouse IgG as secondary antibodies (Rockland Immunochemicals), respectively. Western blotting and protein quantification was performed using the Odyssey infrared imagining system (LI-COR Biosciences). Percent cleavage was calculated as the signal due to gp120 over the signal due to total envelope protein (gp120 plus gp160). Amount of total Env/p24 was calculated as (gp160 plus gp120) / p24 and gp120 / p24 was calculated by quantification of the gp120 and p24 bands. Western blot analysis was performed twice and the average values for the two experiments are presented in Fig 5.
Supplementary Material
The y-axis denotes IC50 for each viral isolate as calculated in Figure 2 for the HIV plasma pool or Figure 4 for the MAbs PG9 and VRC01. The x-axis is either the total Env / p24 or gp120 / p24 as calculated in Figure 5. The cell type used to produce each virus is listed vertically on the left of the figure, and the antibody/plasma used for the neutralization data is shown at the top of the figure. The R2 and p values were calculated by Pearson’s linear regresion analysis, and the line of best fit is shown on each graph.
Supplemental Table 1. 50% inhibitory concentration of HIV-1 positive plasma pool against 293T-derived pseudoviruses and chimeras and PBMC-derived chimeras
Supplemental Table 2. 50% inhibitory concentration of neutralizing antibodies b12, 4E10, and 2F5 against 293T-derived pseudoviruses and chimeras and PBMC-derived chimeras
Supplemental Table 3. 50% inhibitory concentration of broadly neutralizing antibodies VRC01 and PG9 against 293T-derived chimeras and PBMC-derived chimeras
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blay W, Kasprzyk T, Misher L, Richardson B, Haigwood N. Mutations in envelope gp120 can impact proteolytic processing of the gp160 precursor and thereby affect neutralization sensitivity of human immunodeficiency virus type 1 pseudoviruses. Journal of Virology. 2007;81:13037–13049. doi: 10.1128/JVI.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blish C, Blay W, Haigwood N, Overbaugh J. Transmission of HIV-1 in the face of neutralizing antibodies. Current HIV research. 2007a;5:578–587. doi: 10.2174/157016207782418461. [DOI] [PubMed] [Google Scholar]
- Blish CA, Jalalian-Lechak Z, Rainwater S, Nguyen M-A, Dogan OC, Overbaugh J. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. Journal of Virology. 2009;83:7783–7788. doi: 10.1128/JVI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blish CA, Nedellec R, Mandaliya K, Mosier DE, Overbaugh J. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS. 2007b;21:693–702. doi: 10.1097/QAD.0b013e32805e8727. [DOI] [PubMed] [Google Scholar]
- Blish CA, Nguyen M-A, Overbaugh J. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Medicine. 2008;5:0091–0103. doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B, Wieczorek L, Sanders-Buell E, Rosa Borges A, Robb M, Birx D, Michael N, McCutchan F, Polonis V. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology. 2008;375:529–538. doi: 10.1016/j.virol.2008.02.022. [DOI] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nature Immunology. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Chung A, Rollman E, Johansson S, Kent SJ, Stratov I. The utility of ADCC responses in HIV infection. Current HIV research. 2008;6:515–519. doi: 10.2174/157016208786501472. [DOI] [PubMed] [Google Scholar]
- Doria-Rose N, Learn G, Rodrigo A, Nickle D, Li F, Mahalanabis M, Hensel M, McLaughlin S, Edmonson P, Montefiori D, Barnett S, Haigwood N, Mullins J. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. Journal of Virology. 2005;79:11214–11224. doi: 10.1128/JVI.79.17.11214-11224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. Journal of Virology. 2008;82:5807–5814. doi: 10.1128/JVI.02585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forthal DN, Moog C. Fc receptor-mediated antiviral antibodies. Current opinion in HIV and AIDS. 2009;4:388–393. doi: 10.1097/COH.0b013e32832f0a89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. The Journal of general virology. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Lavreys L, Baeten JM, Kreiss JK, Richardson BA, Chohan BH, Hassan W, Panteleeff DD, Mandaliya K, Ndinya-Achola JO, Overbaugh J. Injectable contraceptive use and genital ulcer disease during the early phase of HIV-1 infection increase plasma virus load in women. The Journal of infectious diseases. 2004;189:303–311. doi: 10.1086/380974. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. Journal of Virology. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EM, Rainwater SMJ, Lavreys L, Mandaliya K, Overbaugh J. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Research and Human Retroviruses. 2002;18:567–576. doi: 10.1089/088922202753747914. [DOI] [PubMed] [Google Scholar]
- Louder MK, Sambor A, Chertova E, Hunte T, Barrett S, Ojong F, Sanders-Buell E, Zolla-Pazner S, McCutchan FE, Roser JD, Gabuzda D, Lifson JD, Mascola JR. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology. 2005;339:226–238. doi: 10.1016/j.virol.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Lovelace E, Xu H, Blish CA, Strong R, Overbaugh J. The role of amino acid changes in the human immunodeficiency virus type 1 transmembrane domain in antibody binding and neutralization. Virology. 2011;421:235–244. doi: 10.1016/j.virol.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann AM, Rusert P, Berlinger L, Kuster H, Günthard HF, Trkola A. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS. 2009;23:1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Louder MK, Winter C, Prabhakara R, De Rosa SC, Douek DC, Hill BJ, Gabuzda D, Roederer M. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. Journal of Virology. 2002;76:4810–4821. doi: 10.1128/JVI.76.10.4810-4821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annual Review of Immunology. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Rüker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. Journal of Virology. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JR, John GC, Carr JK, Lewis P, Kreiss JK, Jackson S, Nduati RW, Mbori-Ngacha D, Panteleeff DD, Bodrug S, Giachetti C, Bott MA, Richardson BA, Bwayo J, Ndinya-Achola J, Overbaugh J. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. Journal of Virology. 1999;73:4393–4403. doi: 10.1128/jvi.73.5.4393-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. Journal of Virology. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss M, Overbaugh J. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. Journal of Virology. 1999;73:5255–5264. doi: 10.1128/jvi.73.7.5255-5264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provine NM, Puryear WB, Wu X, Overbaugh J, Haigwood NL. The Infectious Molecular Clone and Pseudotyped Virus Models of Human Immunodeficiency Virus Type 1 Exhibit Significant Differences in Virion Composition with Only Moderate Differences in Infectivity and Inhibition Sensitivity. Journal of Virology. 2009;83:9002–9007. doi: 10.1128/JVI.00423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainwater SMJ, Wu X, Nduati R, Nedellec R, Mosier D, John-Stewart G, Mbori-Ngacha D, Overbaugh J. Cloning and characterization of functional subtype A HIV-1 envelope variants transmitted through breastfeeding. Current HIV research. 2007;5:189–197. doi: 10.2174/157016207780076986. [DOI] [PubMed] [Google Scholar]
- Raska M, Takahashi K, Czernekova L, Zachova K, Hall S, Moldoveanu Z, Elliott MC, Wilson L, Brown R, Jancova D, Barnes S, Vrbkova J, Tomana M, Smith PD, Mestecky J, Renfrow MB, Novak J. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. The Journal of biological chemistry. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer LS, Wrin MT, Crawford-Miksza L, Potts B, Wu Y, Weber PA, Alfonso RD, Hanson CV. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. Journal of Virology. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. Journal of Virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. Journal of Virology. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utachee P, Nakamura S, Isarangkura-Na-Ayuthaya P, Tokunaga K, Sawanpanyalert P, Ikuta K, Auwanit W, Kameoka M. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. Journal of Virology. 2010;84:4311–4320. doi: 10.1128/JVI.02619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Investigators PGP, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Willey R, Shibata R, Freed E, Cho M, Martin M. Differential glycosylation, virion incorporation, and sensitivity to neutralizing antibodies of human immunodeficiency virus type 1 envelope produced from infected primary T-lymphocyte and macrophage cultures. Journal of Virology. 1996;70:6431–6436. doi: 10.1128/jvi.70.9.6431-6436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SMJ, Overbaugh J. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. Journal of Virology. 2006;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Fredriksson R, McKeating JA, Fenyö EM. Passage of HIV-1 molecular clones into different cell lines confers differential sensitivity to neutralization. Virology. 1997;238:254–264. doi: 10.1006/viro.1997.8812. [DOI] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Tewari K, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. Journal of Virology. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The y-axis denotes IC50 for each viral isolate as calculated in Figure 2 for the HIV plasma pool or Figure 4 for the MAbs PG9 and VRC01. The x-axis is either the total Env / p24 or gp120 / p24 as calculated in Figure 5. The cell type used to produce each virus is listed vertically on the left of the figure, and the antibody/plasma used for the neutralization data is shown at the top of the figure. The R2 and p values were calculated by Pearson’s linear regresion analysis, and the line of best fit is shown on each graph.
Supplemental Table 1. 50% inhibitory concentration of HIV-1 positive plasma pool against 293T-derived pseudoviruses and chimeras and PBMC-derived chimeras
Supplemental Table 2. 50% inhibitory concentration of neutralizing antibodies b12, 4E10, and 2F5 against 293T-derived pseudoviruses and chimeras and PBMC-derived chimeras
Supplemental Table 3. 50% inhibitory concentration of broadly neutralizing antibodies VRC01 and PG9 against 293T-derived chimeras and PBMC-derived chimeras


